Abstract
Iron (Fe) deficiency anemia is a global health concern and Fe fortification and supplementation are common corrective strategies. Fe is essential not only for the human host but also for nearly all gut bacteria. We studied the impact of Fe deficiency and Fe repletion on the gut microbiota in rats. Weanling rats were fed an Fe-deficient diet for 24 d and then repleted for 13 d with FeSO4 (n = 15) or electrolytic Fe (n = 14) at 10 and 20 mg Fe · kg diet−1. In addition, one group of rats (n = 8) received the Fe-deficient diet and one group (n = 3) received a Fe-sufficient control diet for all 37 d. Fecal samples were collected at baseline and after the depletion and repletion periods, and colonic tissues were examined histologically. Microbial metabolite composition in cecal water was measured and fecal samples were analyzed for microbial composition with temporal temperature gradient gel electrophoresis and qPCR. Compared to Fe-sufficient rats, Fe-deficient rats had significantly lower concentrations of cecal butyrate (−87%) and propionate (−72%) and the abundance of dominant species was strongly modified, including greater numbers of lactobacilli and Enterobacteriaceae and a large significant decrease of the Roseburia spp./E. rectale group, a major butyrate producer. Repletion with 20 mg FeSO4 · kg diet−1 significantly increased cecal butyrate concentrations and partially restored bacterial populations compared to Fe-deficient rats at endpoint. The effects on the gut microbiota were stronger in rats repleted with FeSO4 than in rats repleted with electrolytic Fe, suggesting ferrous Fe may be more available for utilization by the gut microbiota than elemental Fe. Repletion with FeSO4 significantly increased neutrophilic infiltration of the colonic mucosa compared to Fe-deficient rats. In conclusion, Fe depletion and repletion strongly affect the composition and metabolic activity of rat gut microbiota.
Introduction
Fe is involved in many biological processes and is thus essential for nearly all prokaryotic and eukaryotic cells (1, 2). Fe deficiency is a leading global risk factor for disease, with >2 billion individuals affected worldwide in both industrialized and developing countries. The WHO estimates that 39% of <5-y-old children, 48% of 5- to 14-y-old children, 42% of all women, and 52% of pregnant women in developing countries are anemic (3). The prevalence of Fe deficiency anemia can be reduced by Fe fortification of foods, and electrolytic Fe and FeSO4 are widely used fortificants. Depending on dietary bioavailability, only ~5–15% of fortification Fe is absorbed and the remainder passes into the colon, where it is available for the gut microbiota (4).
The gut microbiota is a complex microbial ecosystem with many different species competing for nutrients. These organisms have a major impact on the nutrition and health of the human host by modifying nutrient supply, conversion of metabolites, and interactions with host cells (4). High bacterial density and the occupation of ecological niches produce a barrier effect that helps to protect the host from colonization by environmental bacteria (5). Indigestible dietary compounds can be metabolized by the gut microbiota into the SCFA acetate, propionate, and butyrate; these can have beneficial effects on gut health. Butyrate is a major energy source for the colonic mucosa and also may have antiinflammatory and antineoplastic properties (6–8). Molecular approaches based on 16S rDNA analysis have shown that the gut microbiota is mainly composed of the phyla Bacteroidetes (e.g., Bacteroides spp.), Firmicutes (e.g., Clostridium, Roseburia, Ruminococcus, or Lactobacillus spp.), Actinobacteria (e.g., bifidobacteria), and Proteobacteria (enterobacteria) (9–11).
Fe is an essential trace element for most gut bacteria and many have active Fe transport systems and other mechanisms to scavenge Fe (12); e.g., Bacteroides spp. are highly dependent on heme and Fe (13, 14). Many members of the Enterobacteriaceae have developed mechanisms, including siderophores, to acquire Fe in competition with other bacteria and the host (12). Only a few bacteria, including lactobacilli, do not require Fe. Lactobacilli are a large group in the gut microbiota that have beneficial effects on gut health (15).
Despite the crucial role of Fe for microorganisms, there are few data on the effect of Fe deficiency and repletion on the gut microbiota. In animal models, using culture methods to assess the gut microbiota, dietary Fe restriction in mice increased total colonic anaerobes, lactobacilli, and enterococci (16) and in weanling pigs, an Fe-fortified diet increased Enterobacteriaceae numbers (17). In human studies, infants receiving Fe fortified cow milk had higher counts of Enterobacteriaceae compared to bifidobacteria (18, 19). Using molecular methods in a controlled study, Zimmermann et al. (21) recently investigated the effect of 6 mo of Fe fortification with electrolytic Fe on the gut microbiota of African children. Fortification modified the fecal microbiota composition, increased the number of Enterobacteriaceae, and decreased the number of lactobacilli (20). Using similar molecular methods, low counts of fecal lactobacilli were found in women with Fe deficiency anemia in South India (21). The objective of this study was to investigate the impact of Fe deficiency and subsequent dietary Fe fortification on the gut microbiota composition and metabolic activity.
Materials and Methods
Rats and diets.
Male Sprague-Dawley rats (21 d old, n = 40; Charles River) were housed individually in stainless steel cages at 22 ± 1°C and a RH6 of 40 ± 3%, with a 12-h-light/-dark cycle. Body weight was measured twice weekly. Production of the diets was done by Dyets. The diets were equivalent and conformed to AIN-93G purified diets (22) and varied only in Fe compound and concentration (24). Food intake was assessed daily. Rats consumed Millipore water (Milli-Q UF Plus) ad libitum throughout the study.
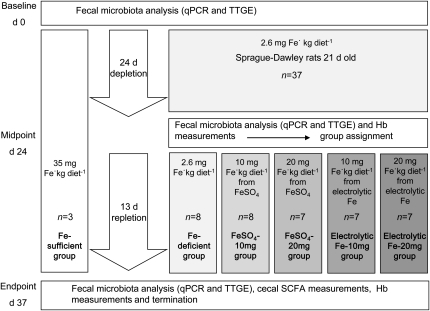
The study design was the standard Hb depletion-repletion assay (23) (Fig. 1). The rats (n = 37) were depleted of Fe for 24 d (2.6 mg Fe · kg diet−1). After depletion (mean Hb, 46.5 ± 3.9 g · L−1), one group continued to receive the Fe-deficient diet (n = 8, 2.6 mg Fe · kg diet−1) for 13 d and 4 other groups were fed a Fe-fortified diet with either 10 mg Fe · kg diet−1 from FeSO4 (FeSO4-10 mg, n = 8), 20 mg Fe · kg diet−1 from FeSO4-20mg, n = 7), or 10 mg Fe · kg diet−1 from electrolytic Fe (electrolytic Fe-10mg, n = 7) 20 mg Fe · kg diet−1 from electrolytic Fe-20mg, n = 7) for 13 d. Dr. Paul Lohmann GmbH provided dried FeSO4 (no. 501022005480) and electrolytic Fe powder (99% Fe, 325 Mesh) was obtained from Industrial Metal Powders. In parallel to this, a control group of 3 rats was fed with a Fe-sufficient diet (34.7 mg Fe · kg diet−1) for the entire study period for the assessment of gut microbiota stability. The Fe content of the diets was measured by atomic absorption spectroscopy (SpectrAA-240K with GTA-120 Graphite Tube Atomizer Varion Techtron). The Veterinary Office of the Canton Zurich, Switzerland, approved all procedures (authorization no. 101/208) (24).
FIGURE 1.
Study design according to the standard Hb depletion-repletion assay (23). Hb, hemoglobin; TTGE, temporal temperature gradient gel electrophoresis.
Sampling procedure.
Blood was collected at the end of the depletion (d 24) and repletion periods (d 37) by tail vein incision (25). The Hb concentration was measured in whole blood using a Scil vet abc counter (Scil Animal Care Company) (24). Fecal samples were collected from all rats at baseline (d 0), after depletion (d 24), and after repletion (d 37) and stored at −20°C.
Immediately after the rats were killed, tissue samples of the colon and cecum contents were collected and stored at −80°C. Cecal water was obtained by centrifuging the samples for 30 min at 14,000 × g and stored at −80°C. For light microscopy, colon tissue samples of Fe-sufficient rats (n = 3), Fe-deficient rats (n = 6), and rats of the FeSO4-20mg group (n = 6) were fixed by immersion in 4% buffered formaldehyde, dehydrated with xylene and a descending alcohol row (Tissue Tek VIP), paraffin embedded, and subsequently stained with hematoxylin-eosin. An Olympus Vanox-S AH-2 (Olympus Schweiz) microscope equipped with an Axio cam and the Axio Vision Program (Carl Zeiss MicroImaging) was used to count neutrophils (40× magnification). On each slide, four regions were selected and in each region five subregions were analyzed for neutrophils, resulting in 60, 120, and 120 subregions analyzed for Fe-sufficient, Fe-deficient, and FeSO4-20mg rats, respectively. The veterinary pathologist performing the histological examinations was unaware of the group assignment.
DNA extraction.
DNA was extracted from fecal samples using the Fast DNA Spin Kit for Soil (MP Biomedicals) and quantified with a Nanodrop ND-1000 Spectrophotometer (Witec) at a wavelength of 260 nm. DNA extracts were stored at −20°C until further analysis.
Analysis of fecal microbiota by qPCR.
For the in-depth analysis of the gut microbiota composition, qPCR was performed using specific primers for bacterial groups most prevalent in the gut. The enumeration of these bacterial groups was performed with a 7500 Fast Real-Time qPCR system (Applied Biosystems Europe). The specific primers to quantify total 16S rDNA, Firmicutes, Bacteroides spp., Lactobacillus/Leuconostoc/Pediococcus spp., and Enterobacteriaceae as well as the PCR conditions were applied as previously described (20, 26). Roseburia spp./E. rectale were enumerated using primers previously published by Ramirez-Farias et al. (27) (Rrec1) at a concentration of 0.2 μmol · L−1 and using Roseburia intestinalis DSM14610 as a standard strain. Duplicate sample analysis and standard curves were routinely performed in each run. Data were analyzed using the 7500 Fast System Sequence Detection Software (version 1.4, Applied Biosystems).
Analysis of fecal microbiota diversity by PCR/TTGE.
For the analysis of the top diversity and changes in the gut microbiota balance, TTGE of the microbial DNA isolated from feces was performed. TTGE gives a “fingerprint” of the V2-V3 regions of the 16S rDNA present in the complex bacterial community of the gut. To amplify the variable V2-V3 region of the 16S rDNA, 1 μL fecal DNA extract was amplified with a PCR using universal primers HDA-1GC (5′-CGC CCG GGG CGC GCC CCG GGC GGG GCG GGG GCA CGG GGGG AC TCC TAC GGG AGG CAG CAG T-3′) and HDA-2 (5′-GTA TTA CCG CGG CTG CTG GCA C-3′) (28). PCR reactions consisted of 0.4 μmol · L−1 of each primer and 2× Fermentas PCR Mastermix diluted 1:1 with sterile MilliQ-grade water (Millipore). Samples were amplified on a Biometra TPersonal Cycler: 94°C for 5 min, 35 cycles of 94°C for 3 min, 58°C for 30 s, 68°C for 1 min, and finally 68°C for 7 min. TTGE separation of PCR amplicons (V2-V3 region of 16S rDNA) was performed using a Dcode Universal Mutation system (Bio-Rad Laboratories) as previously described (21), but voltages of 45 and 70V were applied for 16 h. Gels were stained for 30 min in ethidium bromide and destained for 1 h in dH2O prior to imaging.
TTGE band cloning and sequencing.
TTGE bands were excised and stored overnight at 4°C in 10 mmol · L−1 Tris-EDTA buffer. DNA was precipitated overnight at −20°C by addition of 0.1 volume 3 mol · L−1 sodium acetate and 3 volumes 100% ethanol. The PCR amplification conditions of precipitated DNA were identical to those described above except the forward primer HDA-1 lacked the GC-clamp. Amplicons were then ligated into a vector using the pGEM-T easy Vector System (Promega) according to the manufacturers instructions. Then 1 μL ligation product was mixed with 50 μL XL-1 blue electrocompetent cells (Stratagene) on ice. After electroporation (45 s, 2500 V), cells were immediately transferred to 960 μL SOC media and incubated at 37°C for 1 h. Then 100 μL of serial dilutions was plated on Lysogeny Broth agar plates containing 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (final concentration of 80 mg · L−1, prepared in dimethylformamide) and isopropyl-1-thio-β-D-galactopyranoside (final concentration of 20 mmol · L−1, prepared in sterile dH2O). Plates were aerobically incubated overnight at 37°C. Multiple clones per TTGE band were picked and grown overnight in 3 mL Lysogeny Broth supplemented with 6% ampicillin. Plasmids were isolated using the GeneJET Plasmid Miniprep kit (Fermentas). Plasmid inserts were sequenced (Microsynth) using the T7 sequencing primer. Sequences were compared to the Ribosomal Database Project (29). Sequences with a percentage identity ≥ 97% were considered to represent the same species.
Analysis of cecal SCFA.
SCFA (mainly acetate, propionate, and butyrate) were determined in cecal water by using HPLC as previously described (30). Mean metabolite concentrations of fecal samples were calculated from duplicate sample analysis.
Statistical analysis.
Statistical analyses were done using JMP 8.0 and SPSS Statistics (version 19.0) (SAS Institute). Data were expressed as means ± SD. All variables were tested by the Shapiro-Wilk test for normal distribution. HPLC data were transformed to square roots for statistical analysis for a better fit. Comparisons were done using 1-way ANOVA and the Tukey-Kramer honestly significant difference test. For HPLC data, qPCR log change data, Hb change data, body weight gain data, and neutrophil data, comparisons were conducted between groups of rats at endpoint (d 37). For qPCR data, comparisons were done within groups of rats to analyze changes of each bacterial population from d 0 to 24 to 37 using 1-way repeated-measures ANOVA and post hoc Tukey’s tests. P values < 0.05 were considered significant. For the microbiota analyses, results were expressed as number of 16S rDNA copies · g−1 of feces and the data were not transformed to number of bacteria.
Results
Food intake, body weight, Hb, and colonic inflammation.
After repletion, the increase in Hb was significantly greater in the FeSO4-20mg group compared to the other Fe-fortified groups of rats (Table 1). The Fe-deficient group had significantly lower weight gain and food intake compared to all other groups. At the end of the repletion period, there was a greater number of neutrophils infiltrating the colonic mucosa in the FeSO4-20mg group (5.4 ± 2.3) compared to the Fe-deficient group (2.0 ± 0.7) (P < 0.01).
TABLE 1.
Fe fortification level, Fe intake, body weight gain, Hb change, and food intake of the rats during the 13-d Fe repletion period after an initial Fe-depletion period of 24 d1
| Groups of rats | n | Fortification level | Fe intake | Body weight gain | Hb after depletion | Hb change from depletion to killing | Food intake |
| mg Fe · kg diet−1 | μg · d−1 | g · 15 d−1 | g · L−1 | g · L−1 | g ·13 d−1 | ||
| Fe-deficient | 8 | 2.6 ± 0.3 | 32.1 ± 4d | 40 ± 8d | 46.0 ± 3.6b | −4.1 ± 4.6d | 163 ± 18d |
| FeSO4-10mg | 8 | 10.6 ± 2.4 | 195 ± 19c | 87 ± 10bc | 46.6 ± 3.6b | 20.6 ± 6.6bc | 216 ± 21bc |
| FeSO4-20mg | 7 | 21.4 ± 1.7 | 415 ± 32b | 104 ± 10ab | 46.8 ± 3.7b | 52.3 ± 6.2a | 252 ± 19ab |
| Electrolytic Fe-10mg | 7 | 10.9 ± 0.4 | 170 ± 29c | 79 ± 15c | 47.4 ± 4.6b | 9.2 ± 4.3c | 203 ± 34c |
| Electrolytic Fe-20mg | 7 | 20.8 ± 2.5 | 402 ± 46b | 92 ± 17bc | 45.7 ± 4.8b | 27.4 ± 12.5b | 235 ± 27bc |
| Fe-sufficient | 3 | 34.7 ± 1.8 | 797 ± 51a | 118 ± 11a | 140 ± 2.1a | 12.5 ± 6.5bc | 299 ± 19a |
Values are means ± SD. Means in a column with superscripts without a common letter differ, < 0.05. FeSO4-10mg, rats repleted with 10 mg Fe · kg diet−1 from FeSO4; FeSO4-20mg, rats repleted with 20 mg Fe · kg diet−1 from FeSO4; electrolytic Fe-10mg, rats repleted with 10 mg Fe · kg diet−1 from electrolytic Fe; electrolytic Fe-20mg, rats repleted with 20 mg Fe · kg diet−1 from electrolytic Fe; Hb, hemoglobin.
TTGE analysis.
The profiles of the Fe-sufficient rats were consistent over the entire trial period and did not show major changes in appearance or disappearance of bands (Supplemental Fig. 1A,B). In the Fe-deficient rats, a loss of bands with high GC concentrations (Supplemental Fig. 1A,B, Fe-deficient rats; bottom of gel) was observed and cloning sequencing of one of the lost bands (band 3) revealed a decrease in Barnesiella spp. (29). In Fe-deficient rats, the major bands corresponding to dominant species present at baseline (d 0) disappeared on d 24 and 37. At the same time, several bands increased in intensity (e.g., Supplemental Fig. 1, band 1). We further analyzed this band by cloning sequencing and found that its V2-V3 region belonged to the species Citrobacter freundii of the family Enterobacteriaceae. In the rats that were fed the Fe-deficient diet, the same band appeared on d 24 [Supplemental Fig. 1A, repleted rat (a) band 1]; when they were repleted with the 20-mg FeSO4 diet, the band disappeared on d 37. Also, repletion with electrolytic Fe promoted the growth of some bacterial groups such as Allobaculum spp. (Supplemental Fig. 1B, band 2) belonging to the phylum Firmicutes.
qPCR analysis.
There were no significant changes in the total 16S rDNA copy numbers over the entire trial period in any of the groups (Table 2). However, dominant populations such as Bacteroides spp., Roseburia spp./E. rectale, and Lactobacillus/Leuconostoc/Pediococcus spp. as well as subdominant bacterial groups such as Enterobacteriaceae were affected by Fe deficiency and subsequent Fe repletion.
TABLE 2.
Bacterial populations over time in the gut microbiota of 24-d Fe-depleted rats repleted with 10 or 20 mg Fe · kg diet−1 from FeSO4 for 13 d and Fe-deficient rats1
| d |
||||
| Bacterial population | 0 | 24 | 37 | P value |
| log number of 16S rDNA copies · g rat feces−1 | ||||
| Fe-deficient group | ||||
| Total 16S rDNA | 11.3 ± 0.5 | 10.8 ± 0.6 | 10.7 ± 0.6 | 0.07 |
| Bacteroides spp. | 10.0 ± 0.9a | 9.1 ± 0.9b | 8.6 ± 1.0b | <0.001 |
| Firmicutes | 10.3 ± 0.5a | 9.9 ± 0.6ab | 9.7 ± 0.8b | 0.009 |
| Enterobacteriaceae | 6.4 ± 0.4b | 6.8 ± 0.2ab | 6.9 ± 0.3a | 0.021 |
| Lactobacillus/Leuconostoc/Pediococcus spp. | 9.4 ± 0.7 | 9.8 ± 0.8 | 9.5 ± 1.0 | 0.24 |
| Roseburia spp./E. rectale | 9.0 ± 0.8a | 4.5 ± 0.7b | 4.3 ± 0.9b | <0.001 |
| FeSO4-20mg group | ||||
| Total 16S rDNA | 11.1 ± 0.3 | 11.3 ± 0.2 | 11.3 ± 0.3 | 0.14 |
| Bacteroides spp. | 10.2 ± 0.6 | 9.6 ± 0.6 | 9.9 ± 0.6 | 0.15 |
| Firmicutes | 9.8 ± 0.5b | 10.1 ± 0.5ab | 10.4 ± 0.4a | 0.012 |
| Enterobacteriaceae | 6.4 ± 0.4b | 7.3 ± 0.4a | 5.6 ± 0.4c | 0.001 |
| Lactobacillus/Leuconostoc/Pediococcus spp. | 9.8 ± 0.5b | 10.6 ± 0.3a | 9.7 ± 0.5b | <0.001 |
| Roseburia spp./E. rectale | 6.1 ± 1.6a | 3.6 ± 0.2b | 3.0 ± 0.2b | 0.012 |
| FeSO4-10mg group | ||||
| Total 16S rDNA | 10.9 ± 0.2 | 11.2 ± 0.2 | 10.9 ± 1.0 | 0.18 |
| Bacteroides spp. | 10.4 ± 0.6a | 9.6 ± 0.7b | 9.6 ± 0.6b | 0.01 |
| Firmicutes | 9.8 ± 0.2b | 10.1 ± 0.4a | 10.3 ± 0.3a | 0.006 |
| Enterobacteriaceae | 6.0 ± 0.6b | 6.9 ± 0.3a | 6.1 ± 0.3b | <0.001 |
| Lactobacillus/Leuconostoc/Pediococcus spp. | 9.3 ± 0.4c | 10.6 ± 0.3a | 10.2 ± 0.1b | <0.001 |
| Roseburia spp./E. rectale | 7.8 ± 1.2a | 4.3 ± 0.8b | 4.6 ± 1.5b | 0.007 |
Values are means ± SD, = 7 or 8 (Fe-deficient). Means in a row with superscripts without a common letter differ, P < 0.05. FeSO4-10mg, rats repleted with 10 mg Fe · kg diet−1 from FeSO4; FeSO4-20mg, rats repleted with 20 mg Fe · kg diet−1 from FeSO4.
Fe deficiency.
Fe deficiency induced significant changes from baseline (d 0) to endpoint (d 37) in bacterial population levels in the gut microbiota of rats of the Fe-deficient group (Table 2). Although the total 16S rDNA number of copies per gram of feces after 24 and 37 d did not significantly differ from baseline (d 0), the enumeration of different bacterial groups revealed a significant reorganization of the gut microbiota composition. In the Fe-deficient group, Bacteroides spp. were significantly decreased (~1.5 log) from baseline after 37 d of depletion. There was an even larger significant decrease of ~4.7 log of Roseburia spp./E. rectale 16S rDNA copy numbers (a member of Cluster XIVa and a butyrate producer) from d 0 to 37 of Fe depletion (P < 0.05). Fe deficiency also promoted the growth of some bacteria: in the Fe-deficient group there was a significant increase in Enterobacteriaceae (~0.5 log) (d 37) and after depletion (d 24), in the FeSO4-20 mg and the FeSO4-10 mg groups, Lactobacillus / Pediococcus/Leuconostoc spp. copy numbers were significantly higher compared to baseline.
Fe repletion.
In the FeSO4-20 mg and FeSO4-10 mg groups, the numbers of Roseburia spp./E. rectale did not recover from d 24 to 37 (Table 2).
Fe repletion from d 24 to 37 in the FeSO4-20 mg group restored a portion of the original gut microbiota composition as seen in TTGE profiles. qPCR results confirmed these findings (Table 2) and in this group of rats, Lactobacillus/Leuconostoc/Pediococcus spp. and Enterobacteriaceae significantly decreased to their baseline levels, whereas Bacteroides spp. were increased. Also, Fe repletion with 10 mg FeSO4 · kg diet−1 (FeSO4-10mg group) significantly decreased the Enterobacteriaceae population compared to the end of the depletion period (d 24).
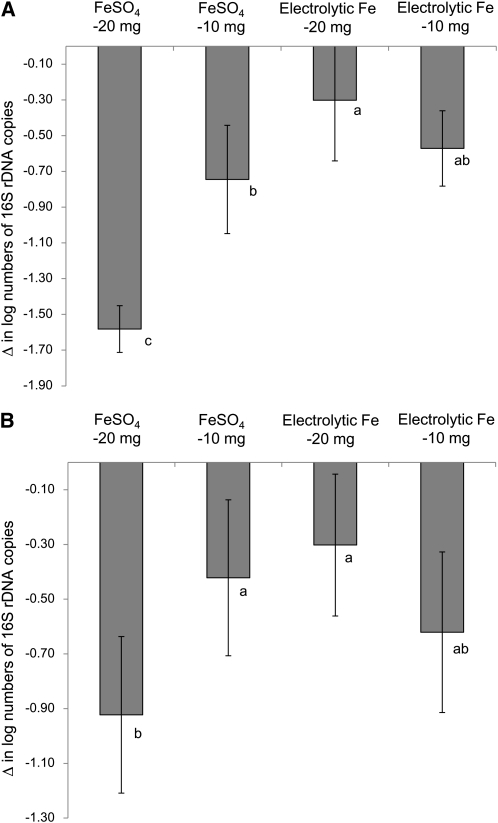
During repletion with both levels of FeSO4 and electrolytic Fe, Enterobacteriaceae (Fig. 2A) and Lactobacillus/Leuconostoc/Pediococcus spp. (Fig. 2B) populations decreased. However, the effects of repletion with 20 mg FeSO4 · kg diet−1 were significantly stronger than with 20 mg electrolytic Fe · kg diet−1.
FIGURE 2.
Changes in numbers of Enterobacteriaceae (A) and Lactobacillus/Pediococcus/Leuconostoc spp. (B) per gram feces of rats repleted for 13 d with 10 or 20 mg Fe·kg diet−1 from FeSO4 or electrolytic Fe after 24-d Fe depletion. Bars are means ± SD, n = 7, 6 (electrolytic Fe-20mg), or 4 (electrolytic Fe-10mg). Means without a common letter differ, P < 0.05.
HPLC analysis of cecum contents.
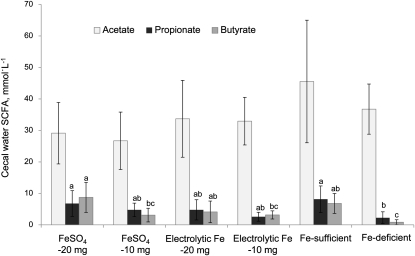
Acetate concentrations of all rats did not significantly differ from each other or the Fe-sufficient group, but the concentration in the Fe-repleted rats (30.5 ± 9.7 mmol · L−1; mean ± SD) tended to be lower than in the Fe-sufficient group (P = 0.06) (Fig. 3). In contrast, butyrate and propionate concentrations were strongly affected by Fe deficiency and subsequent Fe fortification. The cecal concentration of butyrate, the main metabolite of Roseburia spp./E. rectale, was 87% lower and that of propionate was 72% lower compared to the Fe-sufficient group (P < 0.05). The ratio of SCFA in the Fe-deficient group was significantly modified, with acetate comprising 92% of the total. Fe repletion with both Fe compounds partially restored the butyrate and propionate concentrations. Fortification with 20 mg FeSO4 · kg diet−1 significantly increased the propionate and butyrate concentrations to levels comparable to the Fe-sufficient group and restored the ratio of acetate:propionate:butyrate to 65:15:20 (control group 75:14:11), with a particularly strong effect on butyrate production. Fe fortification partially restored the SCFA concentrations in the FeSO4-10mg group as well as the electrolytic Fe-20mg and electrolytic Fe-10mg groups.
FIGURE 3.
Acetate, propionate, and butyrate concentrations in cecal water on d 37 of rats repleted for 13 d with 10 or 20 mg Fe · kg diet−1 from FeSO4 or electrolytic Fe after 24-d Fe depletion and of Fe-deficient and Fe-sufficient rats. Bars are means ± SD, n = 7, 8 (FeSO4-10mg, Fe deficient), or 3 (Fe sufficient). Means without a common letter differ, P < 0.05.
Discussion
In this study, we used the standardized rat Hb depletion-repletion assay (23) with a parallel Fe-sufficient control group to study changes in the gut microbiota and its metabolites. We chose this rat model and design to assess changes under well-controlled conditions where potential confounders (e.g. dietary, environmental, and host factors) are minimized. Potential confounding by these variables would have been much more likely in a human study; moreover, the use of a rat model allowed us access to different portions of the digestive tract during surgical dissection and allowed us to more completely investigate the metabolic activity of the gut microbiota.
We repleted the rats with two commonly used Fe fortificants, FeSO4 and electrolytic Fe. We chose these two compounds, because FeSO4 is water soluble and provides highly bioavailable ferrous Fe, whereas, in contrast, electrolytic Fe contains elemental Fe that is very poorly water soluble; its bioavailability in humans is only ~60% of FeSO4 (24). Overall, as discussed below, our data suggest that at equivalent levels of dietary Fe, Fe repletion with FeSO4 not only is more bioavailable to the host but also has a greater impact on the gut microbiota than electrolytic Fe.
This study demonstrates for the first time, to our knowledge, the profound impact of Fe depletion and subsequent dietary Fe repletion on key features of the gut microbiota. Compared to the Fe-sufficient group where the microbiota profile over the trial period was stable, Fe depletion and repletion modified gut microbiota composition, diversity, and metabolic activity. In particular, Fe deficiency induced major shifts in the gut microbiota balance and diversity. Especially notable was the decrease of Roseburia spp./E. rectale species belonging to the Clostridial Cluster XIVa during Fe deficiency. Many members of this bacterial group are butyrate producers (6) and a marked decrease of butyrate was observed in Fe-deficient rats. The butyrate production pathway in bacteria belonging to the Clostridial Cluster XIVa group includes pyruvate:ferredoxin oxidoreductases and hydrogenases, which are strongly dependent on Fe as a cofactor (6, 31, 32). Not much is known about the Fe-scavenging abilities of Roseburia spp. in a low-Fe environment and the lack of Fe may disrupt the butyrate production pathway needed for the survival of this species. However, during Fe repletion with 20 mg FeSO4 · kg diet−1, butyrate production was restored but Roseburia spp./E. rectale copy numbers did not increase (Table 2). This suggests other butyrate producers were promoted by Fe repletion, such as Allobaculum spp.; these species were prominent in rats repleted with electrolytic Fe and were identified by TTGE analysis followed by cloning sequencing. The end products of the glucose metabolism of Allobaculum spp. are mainly lactate and butyrate (33). Wei et al. (34) found Allobaculum as dominant species in the colon of rats that were exposed to a carcinogenic compound, but little is known about the abundance of this species in humans.
The potential benefits of butyrate on gut health include antiinflammatory and antineoplastic activities (8). In patients with ulcerative colitis, administration of butyrate reduced inflammation of the mucosa (35). Moreover, butyrate strongly affects colonocyte cell growth and function due to its ability to influence gene expression, especially in apoptosis and the cell cycle (7).
Along with the butyrate producers, other dominant bacterial groups were also profoundly affected by Fe depletion and repletion. In particular, Fe depletion affected the gut microbiota; Bacteroides spp. and Roseburia spp./E. rectale were decreased, whereas Lactobacillus/Leuconostoc/Pediococcus spp. and Enterobacteriaceae increased. Based on the TTGE results, Fe depletion caused a loss of the microbial diversity, because fewer bands were present in Fe-deficient rats compared to the Fe-sufficient rats. However, the total 16S rDNA copy numbers were stable over the entire trial period. Bacterial density in the gut is high and the decrease of one bacterial group will allow growth of other bacterial species better adapted to changes in environmental conditions. Bacteroides spp. are strongly dependent on heme (13, 14) and in Fe depletion, heme availability in the gut lumen is likely to be limited. The decrease of this bacterial group in the rat gut may have opened a niche for the growth of lactobacilli, which do not require Fe for growth (15, 36).
A previous study in mice reported higher lactobacilli counts during Fe depletion, a finding supported by our data found in FeSO4-fortified rats after Fe depletion (16). A recent controlled intervention in children in Côte d’Ivoire reported a significant decrease in lactobacilli after Fe fortification (20); this pattern is evident in our study in the rats of the FeSO4-20mg group (Table 2; Fig. 2B). In contrast, a recent cross sectional study in Indian women reported lower numbers of the Lactobacillus acidophilus group in fecal samples of women with anemia (21). It seems that lactobacilli are one of the bacterial groups affected by Fe deficiency and fortification, but the mechanisms behind this pattern still need to be investigated.
The numbers of Enterobacteriaceae also increased during Fe depletion. Many members of this group have strong Fe scavenging abilities due to the excretion of siderophores and the expression of special Fe transport systems (12). These allow this bacterial group to effectively compete for colonic Fe in a low-Fe environment. In our study, Enterobacteriaceae decreased after repletion with FeSO4. This may have been an indirect effect due to the growth promotion of other bacterial groups such as Allobaculum spp. However, in contrast to our results, 6 mo of dietary fortification with electrolytic Fe increased the number of Enterobacteriaceae in African children (20). Also, in a study in weanling pigs, dietary Fe fortification increased the number of coliform bacteria (17). Differences in these studies suggest other factors interact with colonic Fe supply to affect the growth of Enterobacteriaceae, such as varying diets and varying population dynamics in host bacterial ecosystems.
Greater neutrophil infiltration of the colonic mucosa was found in the rats of the FeSO4-20mg group compared to the Fe-deficient rats. Neutrophil infiltration of the gut mucosa is a marker of inflammation that may be associated with pathogen colonization and inflammatory bowel disease (35, 37). An inflammatory response to Fe fortification was recently reported in a human Fe fortification study in Côte d’Ivoire, where fecal calprotectin (a neutrophil protein that is a marker of gut inflammation) was elevated in children receiving electrolytic Fe (20).
The effects of Fe depletion on the gut microbiota in this study could have adverse effects on host health. For example, risk for gut inflammation and colonic neoplasia may be increased due to the markedly lower amount of colonic butyrate in Fe depletion. Shifting equilibrium within the gut microbiota could alter human host immune responses and open niches for the establishment of environmental bacteria in the gut. For example, the significant decrease in numbers of lactobacilli (beneficial bacteria) during Fe repletion with FeSO4 could encourage growth of potential gut pathogens (20). However, with the exception of the Roseburia/E. rectale group, our TTGE profile data suggest that Fe repletion in depleted rats results in a partial recovery of the diversity of the dominant populations in the gut microbiota. Future in vivo studies in humans, including the use of pyrosequencing methods and metabolomics, will be valuable to further define the complex effects of Fe on the human gut microbiota.
Supplementary Material
Acknowledgments
M.B.Z., F.M.H., C.L., and C.C. designed the research; A.D., F.M.H., T.J., and S.R. conducted the research; A.D., C.L., C.C., M.B.Z., F.M.H., T.J., and S.R. analyzed the data; and A.D., C.L., C.C., and M.B.Z. wrote the paper. All authors read and approved the final manuscript.
Footnotes
Supported by the Swiss National Science Foundation (project no. 310030_127272, Bern, Switzerland) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (award no. U01HD064921). The content is solely the responsibility of the authors and does not necessarily represent the official views of the Swiss National Science Foundation or the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Supplemental Figure 1 is available from the “Online Supporting Material” link in the online posting of the article and from the same link in the online table of contents at jn.nutrition.org.
Abbreviations used: FeSO4-10mg, rats repleted with 10 mg Fe · kg diet−1 from FeSO4; FeSO4-20mg, rats repleted with 20 mg Fe · kg diet−1 from FeSO4; electrolytic Fe-10mg, rats repleted with 10 mg Fe · kg diet−1 from electrolytic Fe; electrolytic Fe-20mg, rats repleted with 20 mg Fe · kg diet−1 from electrolytic Fe; Hb, hemoglobin; RH, relative humidity; TTGE, temporal temperature gradient gel electrophoresis.
Literature Cited
- 1.MacKenzie EL, Iwasaki K, Tsuji Y. Intracellular iron transport and storage: from molecular mechanisms to health implications. Antioxid Redox Signal. 2008;10:997–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hentze MW, Muckenthaler MU, Andrews NC. Balancing acts: molecular control of mammalian iron metabolism. Cell. 2004;117:285–97 [DOI] [PubMed] [Google Scholar]
- 3. WHO. Iron deficiency anemia: assessment, prevention, and control; a guide for programme managers. Geneva: WHO; 2002.
- 4.Louis P, McCrae SI, Charrier C, Flint HJ. Organization of butyrate synthetic genes in human colonic bacteria: phylogenetic conservation and horizontal gene transfer. FEMS Microbiol Lett. 2007;269:240–7 [DOI] [PubMed] [Google Scholar]
- 5.Guarner F. Enteric flora in health and disease. Digestion. 2006;73 Suppl 1:5–12 [DOI] [PubMed] [Google Scholar]
- 6.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8 [DOI] [PubMed] [Google Scholar]
- 7.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–19 [DOI] [PubMed] [Google Scholar]
- 8.Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217:133–9 [DOI] [PubMed] [Google Scholar]
- 9.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007;9:1101–11 [DOI] [PubMed] [Google Scholar]
- 11.Wu GD, Lewis JD, Hoffmann C, Chen YY, Knight R, Bittinger K, Hwang J, Chen J, Berkowsky R, Nessel L, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–37 [DOI] [PubMed] [Google Scholar]
- 13.Otto BR, Sparrius M, Verweij-van Vught AM, MacLaren DM. Iron-regulated outer membrane protein of Bacteroides fragilis involved in heme uptake. Infect Immun. 1990;58:3954–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sperry JF, Appleman MD, Wilkins TD. Requirement of heme for growth of Bacteroides fragilis. Appl Environ Microbiol. 1977;34:386–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imbert M, Blondeau R. On the iron requirement of lactobacilli grown in chemically defined medium. Curr Microbiol. 1998;37:64–6 [DOI] [PubMed] [Google Scholar]
- 16.Tompkins GR, O'Dell NL, Bryson IT, Pennington CB. The effects of dietary ferric iron and iron deprivation on the bacterial composition of the mouse intestine. Curr Microbiol. 2001;43:38–42 [DOI] [PubMed] [Google Scholar]
- 17.Lee SH, Shinde P, Choi J, Park M, Ohh S, Kwon IK, Pak SI, Chae BJ. Effects of dietary iron levels on growth performance, hematological status, liver mineral concentration, fecal microflora, and diarrhea incidence in weanling pigs. Biol Trace Elem Res. 2008;126 Suppl 1:S57–68 [DOI] [PubMed] [Google Scholar]
- 18.Mevissen-Verhage EA, Marcelis JH, Harmsen-van Amerongen WC, de Vos NM, Berkel J, Verhoef J. Effect of iron on neonatal gut flora during the first week of life. Eur J Clin Microbiol. 1985;4:14–8 [DOI] [PubMed] [Google Scholar]
- 19.Mevissen-Verhage EA, Marcelis JH, Harmsen-Van Amerongen WC, de Vos NM, Verhoef J. Effect of iron on neonatal gut flora during the first three months of life. Eur J Clin Microbiol. 1985;4:273–8 [DOI] [PubMed] [Google Scholar]
- 20.Zimmermann MB, Chassard C, Rohner F, N'Goran EK, Nindjin C, Dostal A, Utzinger J, Ghattas H, Lacroix C, Hurrell RF. The effects of iron fortification on the gut microbiota in African children: a randomized controlled trial in Cote d'Ivoire. Am J Clin Nutr. 2010;92:1406–15 [DOI] [PubMed] [Google Scholar]
- 21.Balamurugan R, Mary RR, Chittaranjan S, Jancy H, Shobana Devi R, Ramakrishna BS. Low levels of faecal lactobacilli in women with iron-deficiency anaemia in south India. Br J Nutr. 2010;104:931–4 [DOI] [PubMed] [Google Scholar]
- 22.Reeves PG, Nielsen FH, Fahey GC., Jr AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr. 1993;123:1939–51 [DOI] [PubMed] [Google Scholar]
- 23.Forbes AL, Arnaud MJ, Chichester CO, Cook JD, Harrison BN, Hurrell RF, Kahn SG, Morris ER, Tanner JT, Whittaker P, et al. Comparison of in vitro, animal, and clinical determinations of iron bioavailability: International Nutritional Anemia Consultative Group Task Force report on iron bioavailability. Am J Clin Nutr. 1989;49:225–38 [DOI] [PubMed] [Google Scholar]
- 24.Hilty FM, Arnold M, Hilbe M, Teleki A, Knijnenburg JT, Ehrensperger F, Hurrell RF, Pratsinis SE, Langhans W, Zimmermann MB. Iron from nanocompounds containing iron and zinc is highly bioavailable in rats without tissue accumulation. Nat Nanotechnol. 2010;5:374–80 [DOI] [PubMed] [Google Scholar]
- 25.Fluttert M, Dalm S, Oitzl MS. A refined method for sequential blood sampling by tail incision in rats. Lab Anim. 2000;34:372–8 [DOI] [PubMed] [Google Scholar]
- 26.Zihler A, Gagnon M, Chassard C, Hegland A, Stevens MJ, Braegger CP, Lacroix C. Unexpected consequences of administering bacteriocinogenic probiotic strains for Salmonella populations, revealed by an in vitro colonic model of the child gut. Microbiology. 2010;156:3342–53 [DOI] [PubMed] [Google Scholar]
- 27.Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr. 2009;101:541–50 [DOI] [PubMed] [Google Scholar]
- 28.Ogier JC, Son O, Gruss A, Tailliez P, Delacroix-Buchet A. Identification of the bacterial microflora in dairy products by temporal temperature gradient gel electrophoresis. Appl Environ Microbiol. 2002;68:3691–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole JR, Chai B, Farris RJ, Wang Q, Kulam SA, McGarrell DM, Garrity GM, Tiedje JM. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 2005;33:D294–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cleusix V, Lacroix C, Vollenweider S, Le Blay G. Glycerol induces reuterin production and decreases Escherichia coli population in an in vitro model of colonic fermentation with immobilized human feces. FEMS Microbiol Ecol. 2008;63:56–64 [DOI] [PubMed] [Google Scholar]
- 31.Falony G, Verschaeren A, De Bruycker F, De Preter V, Verbeke K, Leroy F, De Vuyst L. In vitro kinetics of prebiotic inulin-type fructan fermentation by butyrate-producing colon bacteria: implementation of online gas chromatography for quantitative analysis of carbon dioxide and hydrogen gas production. Appl Environ Microbiol. 2009;75:5884–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrmann G, Jayamani E, Mai G, Buckel W. Energy conservation via electron-transferring flavoprotein in anaerobic bacteria. J Bacteriol. 2008;190:784–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greetham HL, Gibson GR, Giffard C, Hippe H, Merkhoffer B, Steiner U, Falsen E, Collins MD. Allobaculum stercoricanis gen. nov., sp. nov., isolated from canine feces. Anaerobe. 2004;10:301–7 [DOI] [PubMed] [Google Scholar]
- 34.Wei H, Dong L, Wang T, Zhang M, Hua W, Zhang C, Pang X, Chen M, Su M, Qiu Y, et al. Structural shifts of gut microbiota as surrogate endpoints for monitoring host health changes induced by carcinogen exposure. FEMS Microbiol Ecol. 2010;73:577–86 [DOI] [PubMed] [Google Scholar]
- 35.Lührs H, Gerke T, Muller JG, Melcher R, Schauber J, Boxberge F, Scheppach W, Menzel T. Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand J Gastroenterol. 2002;37:458–66 [DOI] [PubMed] [Google Scholar]
- 36.Elli M, Zink R, Rytz A, Reniero R, Morelli L. Iron requirement of Lactobacillus spp. in completely chemically defined growth media. J Appl Microbiol. 2000;88:695–703 [DOI] [PubMed] [Google Scholar]
- 37.Yu Y, Sitaraman S, Gewirtz AT. Intestinal epithelial cell regulation of mucosal inflammation. Immunol Res. 2004;29:55–68 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.