STRUCTURED ABSTRACT
Purpose of review
It has only recently become apparent that mutations in epigenetic mechanisms and perturbation of epigenomic patterning are frequent events in B-cell lymphomas. The purpose of this review is to highlight these new findings and provide a conceptual framework for understanding how epigenetic modifications might contribute to lymphomagenesis.
Recent findings
Somatic mutations affecting histone methyltransferases such as EZH2 and MLL2, histone demethylases including UTX and JMJD2C and histone acetyltransferases including CBP and p300 are recurrent and common in lymphomas. These mutations result in disruption of chromatin structure and functions of other proteins, ultimately causing aberrant transcriptional programming affecting multiple gene networks. Widespread perturbation of cytosine methylation patterning now appears to be a hallmark of B-cell lymphomas and occurs in specific patterns that can distinguish disease subtypes. Therapeutic targeting strategies can overcome abnormal epigenetic mechanisms and potently kill lymphoma cells.
Summary
Newly discovered epigenetic lesions may provide critical insights into the genesis of B-cell lymphomas but further studies are required to understand how they affect biological mechanism. Epigenetic lesions offer tremendous opportunities for the development of improved biomarkers and treatments.
Keywords: DNA methylation, Chromatin modifications, Epigenetic programming, EZH2, Histone acetyltransferase
INTRODUCTION
Deregulated gene expression is a hallmark of cancer and is well documented in B-cell lymphomas. Expression patterning is reflective of the status of the various gene pathways that define cellular phenotype. For example, in activated B-cell (ABC) type diffuse large B-cell lymphoma (DLBCL) the presence of NFkB signatures indicated constitutive activation and biological dependence on this pathway. Likewise, DLBCLs with BCL6 target gene signatures are dependent on BCL6 for their survival. Gene expression is controlled not only by transcription factors but also by the regulatory state of chromatin. Chromatin modifications including cytosine methylation and histone modifications encode crucial DNA sequence independent (i.e. “epigenetic) information. Because chromatin architecture explains in large part how gene expression and cellular phenotypes are controlled, the elucidation of epigenetic mechanism is of great interest. This review highlights and explores how newly discovered mutations in chromatin modifying genes and perturbations in cytosine methylation patterning might contribute to lymphomagenesis.
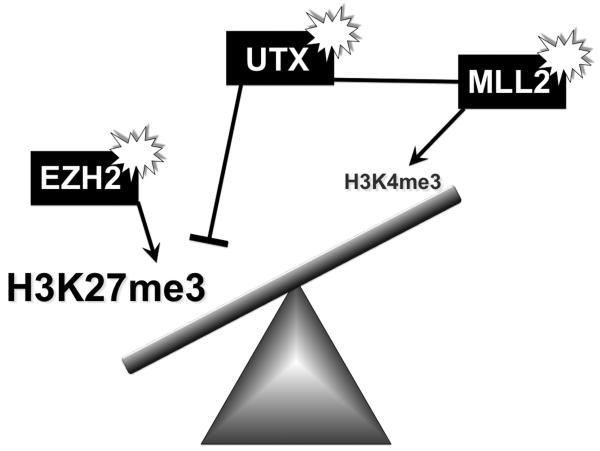
Disequilibrium of histone methylation marks: a key event in lymphomagenesis (Figure 1)?
Figure 1. Possible disequilibrium of H3K27 and H3K4 trimethylation in B-cell lymphomas.
EZH2 is a histone methyltransferase component of Polycomb Repressive Complex 2. Somatic heterozygous point mutations altering the catalytic activity of the catalytic SET domain of EZH2 occur in ~7-12% of follicular lymphomas, ~21% of GCB-DLBCLs and ~4% of PMBLs. Mutant EZH2 induces accumulation of the repressive H3K27 trimethylation chromatin mark in DLBCL cells. The MLL-trithorax histone methyltransferases oppose the actions of PRC2 and EZH2, and induce H3K4 trimethylation, an activating chromatin mark. Somatic heterozygous inactivating mutations of MLL2 occur in up to 89% of FL and 32% of DLBCL. MLL2 forms a complex with the H3K27 demethylase UTX, which is inactivated through mutations in multiple myeloma. These mutations would be expected to result in an increase in H3K27 trimethylation and reduction of H3K4 trimethylation. Presumably the consequence of this disequilibrium is oncogenic disruption of epigenetic programming facilitating lymphomagenesis. However these are all multifunctional proteins and may have numerous other downstream effects.
EZH2 is a histone methyltransferase component of Polycomb Repression Complex 2 (PRC2) that methylates histone 3 lysine 27(H3K27). EZH2 plays an essential role in programming pluripotency and self-renewal of embryonic stem cells [1]. During early B-cell development EZH2 is required for VDJ recombination [2], is subsequently downregulated in mature B-cells, but is highly expressed again after T-cell dependent activation in germinal center (GC) B-cells [3]. In GC B-cells EZH2 binds to genes involved in differentiation and suppressing cell growth and proliferation such as CDKN1A, CDKN1B and CDKN2A [3]. EZH2 targets displayed H3K27 trimethylation and were repressed. Many B-cell EZH2 targets (i.e.TGFβ pathway genes) are different from those in embryonic cells [3]. By epigenetically silencing these growth suppressive pathways EZH2 might facilitate the emergence of genetic mutations at these stages, which could lead to development of B-cell neoplasms. Diffuse large B-cell lymphomas (DLBCLs) and follicular lymphomas (FL) are derived from cells that have transited the GC reaction. EZH2 is often expressed in DLBCL where it represses its B-cell target genes [3].
Next generation sequencing of an FL patient identified a heterozygous somatic point mutation in EZH2 exon 15, replacing tyrosine 641 with a histidine residue [4]. Resequencing of 221 additional FL patients yielded a 7.2% incidence of heterozygous EZH2Y641 mutations with several amino acids substituting for tyrosine [4]. The incidence of EZH2Y641 point mutations was 12% in another cohort of 221 patients [5]. 9.7% of DLBCLs (n=320) had EZH2 mutations. However, these occurred only in GCB-DLBCLs (21.7%) and primary mediastinal B-cell lymphoma (PMBL, 4.2%) [4]. Presence of EZH2 mutations did not appear to affect clinical outcome [4]. The Y641 residue is located within the EZH2 SET (catalytic) domain, and hence would be expected to affect enzymatic activity. Although initially thought to confer loss of function [4], more detailed studies indicated that EZH2Y641 mutants display a shift in their histone methylation activity. While wild-type EZH2 readily catalyzes H3K27 mono and dimethylation, EZH2Y641 is markedly more efficient at trimethylating H3K27 [6][7]. EZH2Y641 DLBCL cells displayed greater levels of H3K27me3 and less H3K27me1 than wild-type EZH2 cells [6][7]. EZH2Y641 likely cooperates with and requires the wild-type allele to bring H3K27 to the fully trimethylated state. Although the biological impact of increased H3K27me3 is not known, it is likely significant since EZH2 siRNA induced profound growth arrest in an EZH2Y641 DLBCL cell line [3]. EZH2 also has cytoplasmatic functions in cell signaling and actin polymerization [8], which raise the possibility that EZH2Y641 might have effects beyond histone modification.
Methylation of H3K4 is associated with gene activation and acts as an opposing force to H3K27 methylation [9]. Although usually mutually exclusive, H3K4me3 and H3K27me3 can coexist in embryonic stem cells as “bivalent” marks primed to be either activated or repressed during subsequent lineage specification [9][10]. H3K4 trimethylation is mediated in part by the MLL/trithorax family of histone methyltransferases [9]. MLL1 is frequently translocated in acute leukemias, and the resulting fusion proteins aberrantly activate oncogenic target genes such as HOXA9 and EVI1 [9][11]. Strikingly, it was recently reported that 89% of FLs (n=35) and 32% of DLBCLs (n=37) display truncation and frameshift mutations of MLL2 [12]. All of these mutations disrupt the SET domain and so would be expected to result in deficient H3K4 methylation. Mutations in MLL, MLL2 and MLL3 were also recently identified in multiple myeloma [13]. Similar MLL2 mutations occur in Kabuki syndrome, a congenital disorder associated with multiple organ and skeletal malformations and intellectual disability [14]. One study suggested a link between Kabuki syndrome and neuroblastoma [15]. MLL is also involved with the S-phase DNA replication checkpoint controlled by ATR, and loss of MLL resulted in DNA damage independent DNA synthesis [16]. It is interesting to speculate whether MLL2 might have a similar function in B-cells and thus whether MLL2 mutations would lead lymphoma cells to tolerate genomic instability.
The frequent occurrence of MLL2 loss of function and EZH2 gain of function mutations underline the significance of H3K4me3 and H3K27me3 in aberrant epigenetic programming. Along these lines MLL2 forms a complex with UTX, an H3K27 demethylase that can oppose the actions of EZH2 [17]. In addition to potential reduction in H3K4me3, loss of MLL2 or other MLL proteins might also fail demethylate H3K27. In fact, homozygous or hemizygous inactivating mutations of UTX (which is located on the X chromosome) have been identified in multiple myeloma, and may represent an alternative way to alter H3K27 methylation in lymphoid malignancies [18]. UTX mutations were mutually exclusive with t(4;14) translocations, which overexpress another histone methyltransferase called MMSET. In contrast to EZH2 and UTX mutation, MMSET overexpression is associated with reduction in cellular H3K27 methylation [19], as well as with increased H3K36 and H4K20 methylation [19][20]. These considerations underline the fact that histone modifying enzymes truly function in an integrated manner, with disruption of one enzyme affecting the actions of many others [17]. Notably, ~12% of patients with myelodysplastic syndrome or myeloproliferative neoplasms feature mono or bi-allelic EZH2 loss of function mutations accompanied by a reduction in cellular H3K27me3 [21]. Therefore EZH2 and MLL family proteins may have opposing roles as tumor suppressors and oncogenes in myeloid development and mature B-cells.
Another example of epigenetic cooperativity involves the H3K9 demethylase JMJD2C, which is localized near the JAK2 locus on a region of chromosome 9p24 amplified in PMBL and Hodgkin lymphoma (HL). JMJD2C demethylation of H3K9 and JAK2 phosphorylation of H3Y41 both inhibit binding of heterochromatin silencing protein HP1α [22]. Inhibition of JAK2 plus JMJD2C cooperatively increased H3K9 methylation and HP1α heterochromatin foci in PMBL cells [22]. One of the loci silenced in this manner was MYC, which is an important downstream mediator of JAK2 and JMJD2C [22]. Blockade or silencing of JAK2 and JMJD2C also resulted in enhanced death of PMBL and HL cell lines [22]. Collectively these instances offer some of the first proof that disruption of the proposed combinatorial “histone code” [17] is deleterious for the homeostasis of transcriptional regulation in B-cells and facilitates lymphomagenesis.
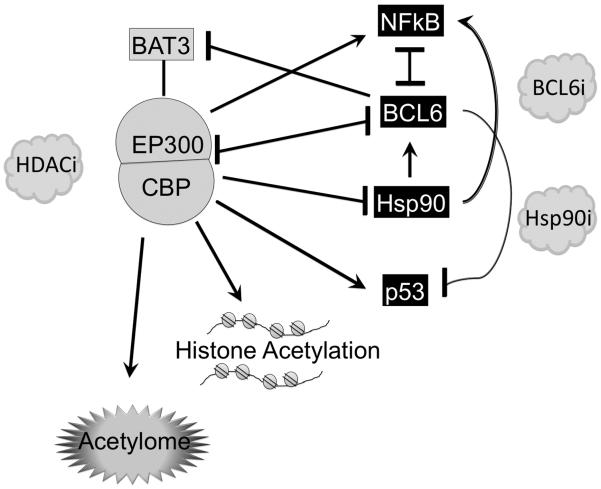
Histone acetyltransferases as tumor suppressors in B-cells (Figure 2)
Figure 2. Disruption of CBP and p300 acetyltransferases in B-cell lymphomas.
CBP and p300 have both overlapping and unique functions. p300 acts as a co-activator for NFkB, and activates p53 and on the other hand can attenuate Hsp90 chaperone functions and BCL6 transcriptional repressor functions. P300 and CBP also acetylate histones and potentially hundreds of other proteins in the acetylome. Deacetylated Hsp90 maintains BCL6 expression, which in turn suppresses p300 and its essential cofactor BAT3 and also represses p53. This vicious circle can be disrupted by treating lymphoma cells with BCL6 inhibitors, Hsp90 inhibitors or HDAC inhibitors, and these agents are synergistic when administered in combination. Blockade of p300 rescues DLBCL cells from being killed by BCL6 inhibitors and HDAC inhibitors. CBP also acetylates p53 and BCL6. Somatic heterozygous mutations or deletions of the CREBBP locus occur in 41.5% of GCB-DLBCLs, 12% of ABC-DLBCLs, and 32.6% of FL cases. EP300 mutations occur in 10% of DLBCLs. All cases seem to have in common disruption of the histone acetyltransferase catalytic domain, and the resulting truncated or mutant proteins may have dominant negative or gain of function properties, or may simply result in a reduced dosage of histone acetyltransferases. These effects would presumably lock DLBCL cells in a state with inactivated p53, and activated BCL6 and Hsp90, in addition to many other possible downstream effects.
Histone lysine residues are acetylated by histone acetyltransferases (HATs). Histone acetylation induces a more “open” chromatin structure and recruits bromodomain protein “readers” to facilitate transcriptional activation. At least 1750 proteins in addition to histones are modified by lysine acetylation in leukemia cells [17], indicating that HATs have broad cellular activities. Several recent studies implicate the HAT proteins CBP and p300 as tumor suppressors in B-cell neoplasms. CBP and p300 function as co-activators of transcription factors and acetylate proteins relevant to lymphomagenesis such as p53, NFkB, BCL6 and Hsp90 [24][25][26][27]In lymphoma, heterozygous p300 truncation mutants deleting the lysine acetyltransferase (KAT) domain were first revealed in the lymphoma cell line RCK8[28]. Truncated p300 disrupted the function of c-Rel, which normally uses p300 as a co-activator[29]. P300 is a direct target gene of the transcriptional repressor BCL6, a key lymphoma oncoprotein and BCL6 and p300 levels were inversely correlated in primary DLBCLs [26]. BCL6 inhibitors induce p300 protein expression and acetyltransferase activity in DLBCL cells, with subsequent acetylation of p53 (which induces p53 transcriptional functions) and Hsp90 (which suppresses Hsp90 chaperone activity) [26]. Blockade of p300 activity in DLBCL cells rescued them from being killed by BCL6 inhibitors [26] and lymphoma cells with mutant p300 were resistant to BCL6 inhibitors. Induction of BAT3, a critical cofactor of p300 required to acetylate p53, was also required for BCL6 inhibitors to kill DLBCL cells [26]. Combining BCL6 and HDAC inhibitors led to even higher p300 activity and synergistic killing of lymphoma cells in vitro and in vivo [26]. Suppression of p300 either through BCL6 or inactivating mutations thus plays a key role in DLBCL.
Resequencing studies in patients with B-cell neoplasms provided definitive evidence that CBP and p300 are bona fide tumor suppressors [26][30][31]. Sequencing and copy number analysis of the CREBBP locus that encodes CBP in 134 DLBCLs identified truncation or HAT domain mutations, and monoallelic deletions in 29% of patients [30]. Like EZH2 mutations, these more frequently occurred in GCB DLBCLs (41.5%) vs. ABC DLBCLs (12%) [30]. HAT domain inactivating mutations were also present in 15/46 FL cases (32.6 %) [30]. 18.3% of relapsed pediatric B-acute lymphoblastic leukemia cases harbored CREBBP mutations, and only half of these were present at diagnosis [31]. Only 1% of non-relapsing patients displayed CREBBP mutations at diagnosis [31], suggesting that these mutations contribute to chemotherapy resistance. Somatic mutations truncating or disrupting the HAT domain of p300 were identified in 9.7% of a cohort of 97 DLBCL patients[26**], in 10% of the above cohort of 134 DLBCL cases [30], and in 8.7% of the 46 FL cases [30]. CREBBP mutants affecting the HAT domain were deficient in binding acetylCoA, in acetylating p53, BCL6 and H3K18, and in reconstituting cyclic AMP signaling in Crebbp/Ep300 knockout MEFs [30][31].
Similar to the case of EZH2 and MLL2, mutations of CREBBP and EP300 are usually heterozygous [26][30][31], suggesting that mutant protein could function as dominant negative or that the reduction in active protein level is sufficient to disrupt biological functions. Along these lines transduction of two different HAT domain deleted p300 mutants in DLBCL cell lines rescued cells from BCL6 inhibitors, consistent with a dominant negative effect [26]. Conversely transfecting wild type p300 into RCK8 cells, which express truncated p300, partially sensitized them to the actions of BCL6 inhibitor. Therefore, the stoichiometry between wild type and mutant alleles may be important in determining the biological impact of mutant alleles. Importantly, CBP and p300 are not identical in their biological functions and so it is not clear whether their loss of function in DLBCL is completely bio-equivalent. While Cebbp+/− mice display hematopoietic defects and eventually develop hematologic tumors, Ep300+/− do not reproduce either phenotype [32]. CBP and p300 also mediate different effects on the biological output of WNT signaling and other pathways (reviewed in [33]). Careful study will thus be required to parse out the various mechanisms through which these mutant alleles contribute to lymphomagenesis.
Aberrant DNA methylation patterning in B-cell lymphomas
DNA methylation patterning contains much of the epigenetic information that determines the phenotype of normal and malignant cells [34]. Along these lines the methylome of multipotent and lineage-committed progenitors revealed numerous differentially methylated regions and a greater burden of methylation associated with lymphoid commitment [35]. Hypermethylation of gene regulatory regions is generally associated with silencing and hypomethylation with gene expression [34], although In reality DNA methylation patterning is more complex. For example, hypermethylation of a CpG-rich region within the first intron of BCL6 was reported to maintain high levels of BCL6 expression, at least in part by blocking binding of a negative regulator of this locus (CTCF) [36]. BCL6 levels decreased in lymphoma cell lines exposed to the DNA methyltransferase inhibitor decitabine [36]. Aberrant hypomethylation of intergenic regions can lead to genomic instability and contribute to malignant transformation [34].
DNA methylation profiling studies indicate that cytosine methylation distribution is perturbed in lymphomas vs. normal B-cells, and that promoter methylation is generally inversely correlated with gene expression [3][37][38][39][40][41][42][43]. Several groups observed that certain aberrantly methylated genes in lymphomas are known targets of the PRC2 polycomb complex (which includes EZH2) in embryonic stem cells [3][39][40][41][42]. However, cytosine methylation and EZH2/H3K27me3 binding are almost entirely mutually exclusive in normal GC B-cells [3]. Another report found little overlap of SUZ12 (a PRC2 component) and H3K27me3 with differentially methylated regions [41]. In contrast, examination of methylation profiles of DLBCL patients revealed that EZH2 B-cell target genes frequently become hypermethylated [3] implying breakdown in lymphomas of the epigenetic barrier that separates PRC2 and cytosine methylation in normal B-cells. It is not known whether these events are linked to mutations in EZH2 and MLL2, but is intriguing to consider that perturbations in chromatin structure could lead to mislocalization of cytosine methylation and consequent aberrant epigenetic silencing. It has also been speculated that methylation of PRC2 genes in lymphoma could represent oncogenic events occurring during early lymphoid development [39].
DNA methylation profiling can also used to classify lymphoma subtypes. Integrated DNA methylation and gene expression profiling of 69 DLBCL cases identified 239 genes differentially methylated in ABC vs. GCB DLBCLs indicating that these are epigenetically distinct entities [37]. Differentially methylated genes in ABC and GCB DLBCLs were centered on a TNFa/cytokine network implicating differential methylation of these genes as distinguishing ABC and GCB lymphoma biology [37]. The sixteen most differentially methylated and expressed genes between ABC and GCB predicted DLBCL subtype in an independent cohort of 203 DLBCL cases with 92% accuracy [37]. In a different study, comparison of DNA methylation profiles distinguished mediastinal gray zone lymphoma from the related PMBL and Hodgkin lymphomas and revealed small sets of differentially methylated genes associated with each subtype [43]. Although requiring further validation, diagnosis of morphologically difficult to distinguish lymphomas may thus be enhanced using DNA methylation biomarkers.
Cytosine methylation profiles of 22 primary mantle cell lymphomas (MCL) were compared to 10 sets of purified tonsilar naïve B-cells. Remarkably, MCLs displayed twice as many aberrantly hypomethylated as hypermethylated loci, suggesting that aberrant loss of methylation is a dominant feature of this disease [38]. The gene networks most heavily affecting by aberrant DNA methylation revolved around NFkB and HDAC1 [38]. Gene expression was inversely correlated with methylation at over 1400 loci [38]. CD37, an aberrantly hypomethylated and overexpressed gene in MCL, was expressed on the surface of MCL cells. CD37-SMIP therapeutic antibodies killed MCL cells suggesting targeting of a hypomethylated gene as a therapeutic approach for MCL [38]. Conversely, several tumor suppressors including CDKN2B, HOXD8, MLF1 and PCDH8 were hypermethylated and silenced in MCL. Treatment with decitabine and the HDAC inhibitor SAHA reactivated these genes and potently killed MCL cells [38].
CpG dinucleotides are methylated by DNMT1, DNMT3A and DNMT3B. DNMT1 is predominantly involved in maintaining, while DNMT3A and 3B mediate de novo cytosine methylation[44]. DNMT1 also plays a critical role in replication, repair of double strand break and stem cell self renewal [45][46]. Analysis of DNMTs by immunohistochemistry in 81 DLBCL cases identified expression of DNMT1, 3A and 3B in 48%, 13% and 45% of DLBCLs respectively [47]. Of these DNMT3B expression was independently associated with worse overall and progression free survival [47]. Interestingly, tumor cells often produce an aberrantly spliced isoform (DNMT3B7) lacking the C-terminal catalytic domain [48]. Transgenic mice expressing Dnmt3b7 in an Eμ-Myc background develop mediastinal B-cell lymphomas with markedly increased proliferative rates, increased genomic instability and altered methylation patterning [48]. The data suggest possible dominant negative and gain of function features of DNMT3B7 warranting exploration of this isoform in lymphoma patients.
CONCLUSION
Mutations affecting epigenetic and transcriptional modifiers now appear to be an almost universal feature of B-cell lymphomas. Large-scale disruptions of DNA methylation and histone modification patterning are emerging as a hallmark of these diseases. The challenge for the next years will be to understand the nature of the lymphoma epigenome and the biochemical and biological effects of mutant epigenetic factors. It will also be necessary to ascertain the utility of DNA methylation classifiers as biomarkers for diagnostic accuracy and therapeutic stratification. Because epigenetic marks are potentially reversible, the development of genuine epigenetic-targeted therapy drugs holds great promise. Already agents are emerging that target transcription factors, chromatin modifying enzymes and epigenetic reader proteins (e.g. [49][50]). In the long-term such agents might significantly advance the treatment of B-cell lymphomas with less toxicity to the normal immune system and other tissues.
KEY POINTS.
Gain of function mutations of EZH2 and loss of function mutations of MLL family members suggest that abundance and localization of histone methylation plays a fundamental role in lymphomagenesis.
Frequent genetic lesions disrupting the histone acetyltransferases CBP and p300 implicate protein acetylation as a critical barrier preventing malignant transformation of B-cells.
Aberrant DNA methylation patterning is emerging as a hallmark of B-cell lymphomas, and may cooperate with altered polycomb complexes to deregulate gene expression.
Epigenetic lesions are promising biomarkers to improve diagnosis and predict outcomes in B-cell lymphomas.
ACKNOWLEDGEMENTS
None
Funding: RS is supported by K08 CA127353 and the Leukemia and Lymphoma Society Translational Research Program 6304-11. AM is supported by NCI R01 CA 104348, the Leukemia and Lymphoma Society, the Chemotherapy Foundation and the Burroughs Wellcome Foundation.
ABBREVIATIONS
- DLBCL
Diffuse Large B-cell Lymphoma
- PMBL
Primary Mediastinal B-cell Lymphoma
- FL
Follicular Lymphoma
- MCL
Mantle Cell Lymphoma
- BCL6
B Cell Lymphoma 8
- EZH2
Enhancer of Zeste 2
- MLL
Mixed Lineage Leukemia
- CREBBP
CREB Binding Protein
- BAT3
HLA-B Associated Transcript 3 / BCL2-associated athanogene 6
- MMSET
Multiple Myeloma SET domain protein / Wolf-Hirschhorn syndrome candidate 1
- UTX
Ubiquitously-transcribed X chromosome tetratricopeptide repeat
- WNT
Wingless-type MMTV integration site
- TGFβ
Transforming Growth Factor Beta
- SET domain
Suppressor of variegation 3-9, Enhancer of zeste, Trithorax- chromatin regulator
- KAT domain
Lysine Acetyl Transferase
- HAT
Histone Acetyl Transferase
- JMJD2C
Jumonji Domain Containing 2C
- JAK2
Janus Kinase 2
- CTCF
CCCTC-binding factor
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Lee TI, Jenner RG, Boyer LA, et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell. 2006;125(2):301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Su IH, Basavaraj A, Krutchinsky AN, et al. Ezh2 controls b cell development through histone h3 methylation and igh rearrangement. Nat Immunol. 2003;4(2):124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- [3] *.Velichutina I, Shaknovich R, Geng H, et al. Ezh2-mediated epigenetic silencing in germinal center b cells contributes to proliferation and lymphomagenesis. Blood. 2010;116(24):5247–5255. doi: 10.1182/blood-2010-04-280149. The first paper identifying EZH2 target genes and biological function in normal and malignant B-cells, and the association of EZH2, H3K27me3 and DNA methylation in lymphomas.
- [4] **.Morin RD, Johnson NA, Severson TM, et al. Somatic mutations altering ezh2 (tyr641) in follicular and diffuse large b-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. doi: 10.1038/ng.518. Genomic resequencing identified the first point mutation in the EZH2 SET domain. EZH2 was then sequenced in large cohorts of FL and DLBCL patients showing for the first time recurrent mutation of this histone methyltransferase in a human disease. Functional assays showing that the EZHY641 mutation inactivated EZH2 were subsequently shown to be incorrect.
- [5] *.Bodor C, O’Riain C, Wrench D, et al. Ezh2 y641 mutations in follicular lymphoma. Leukemia. 2011 doi: 10.1038/leu.2010.311. Confirmation of the frequent occurrence of EZH2Y641 mutations in follicular lymphoma in an independent cohort of patients.
- [6] *.Yap DB, Chu J, Berg T, et al. Somatic mutations at ezh2 y641 act dominantly through a mechanism of selectively altered prc2 catalytic activity, to increase h3k27 trimethylation. Blood. 2011;117(8):2451–2459. doi: 10.1182/blood-2010-11-321208. The demonstration that EZH2Y641 mutations result in a gain of function favoring H3K27 trimethylation over mono and dimethylation.
- [7] *.Sneeringer CJ, Scott MP, Kuntz KW, et al. Coordinated activities of wild-type plus mutant ezh2 drive tumor-associated hypertrimethylation of lysine 27 on histone h3 (h3k27) in human b-cell lymphomas. Proc Natl Acad Sci U S A. 2010;107(49):20980–20985. doi: 10.1073/pnas.1012525107. The demonstration that EZH2Y641 mutations result in a gain of function favoring H3K27 trimethylation over mono and dimethylation. This paper and the one above were published very close together.
- [8].Su IH, Dobenecker MW, Dickinson E, et al. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121(3):425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- [9] *.Mills AA. Throwing the cancer switch: Reciprocal roles of polycomb and trithorax proteins. Nat Rev Cancer. 2010;10(10):669–682. doi: 10.1038/nrc2931. An detailed review comparing and contrasting the function of Polycomb proteins like EZH2 and Trithorax proteins like MLL and their functional interactions.
- [10] *.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. A very useful review charting the functional significance of chromatin modifications throughout the genome.
- [11].Arai S, Yoshimi A, Shimabe M, et al. Evi-1 is a transcriptional target of mll oncoproteins in hematopoietic stem cells. Blood. 2010 doi: 10.1182/blood-2009-07-234310. [DOI] [PubMed] [Google Scholar]
- [12] *.Mendez-Lago M, Morin RD, Mungall AJ, et al. Mutations in mll2 and mef2b genes in follicular lymphoma and diffuse large b-cell lymphoma. Blood. 2010;116(21):473. This abstract presented at the American Society of Hematology described for the first time the large scale genomic resequencing of follicular lymphoma and diffuse large B-cell lymphoma and high incidence of MLL2 mutations.
- [13] *.Chapman MA, Lawrence MS, Keats JJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471(7339):467–472. doi: 10.1038/nature09837. This manuscript details the first genomic resequencing in multiple myeloma including the identification of mutations in MLL family members.
- [14] *.Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies mll2 mutations as a cause of kabuki syndrome. Nat Genet. 2010;42(9):790–793. doi: 10.1038/ng.646. The discovery that MLL2 mutations cause a congenital malformation disorder.
- [15].Merks JH, Caron HN, Hennekam RC. High incidence of malformation syndromes in a series of 1,073 children with cancer. Am J Med Genet A. 2005;134A(2):132–143. doi: 10.1002/ajmg.a.30603. [DOI] [PubMed] [Google Scholar]
- [16] *.Liu H, Takeda S, Kumar R, et al. Phosphorylation of mll by atr is required for execution of mammalian s-phase checkpoint. Nature. 2010;467(7313):343–346. doi: 10.1038/nature09350. The discovery of a novel function for MLL in ATR dependent replication checkpoint, with potential implications for genomic stability in leukemias and lymphomas with mutant forms of MLL.
- [17] *.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10(7):457–469. doi: 10.1038/nrc2876. An outstanding review discussing how the various protein components of the epigenomic machinery can be disrupted in cancer.
- [18].van Haaften G, Dalgliesh GL, Davies H, et al. Somatic mutations of the histone h3k27 demethylase gene utx in human cancer. Nat Genet. 2009;41(5):521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19] *.Martinez-Garcia E, Popovic R, Min DJ, et al. The mmset histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood. 2011;117(1):211–220. doi: 10.1182/blood-2010-07-298349. The first demonstration that MMSET overexpression induces reduction in H3K27 levels in myeloma cells.
- [20] *.Pei H, Zhang L, Luo K, et al. Mmset regulates histone h4k20 methylation and 53bp1 accumulation at DNA damage sites. Nature. 2011;470(7332):124–128. doi: 10.1038/nature09658. This manuscript shows that MMSET is recruited to sites of DNA damage where it induces H4K20 methylation and contributes to repair.
- [21] *.Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyltransferase gene ezh2 in myeloid disorders. Nat Genet. 2010;42(8):722–726. doi: 10.1038/ng.621. The first demonstration of EZH2 loss of function mutations, occuring in myelodysplasia and myeloproliferative disorders in contrast to the gain of function mutations occuring in B-cell lymphomas
- [22] **.Rui L, Emre NC, Kruhlak MJ, et al. Cooperative epigenetic modulation by cancer amplicon genes. Cancer Cell. 2010;18(6):590–605. doi: 10.1016/j.ccr.2010.11.013. The demonstration that two genes overexpressed from the same amplified chromosomal region have cooperating effects in the epigenetic modification of genes, leading to loss of repressive chromatin and the activation of cytokine loops and MYC expression.
- [23].Choudhary C, Kumar C, Gnad F, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- [24].Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 c-terminal domain. Cell. 1997;90(4):595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- [25].Rothgiesser KM, Fey M, Hottiger MO. Acetylation of p65 at lysine 314 is important for late nf-kappab-dependent gene expression. BMC Genomics. 2010;11(22) doi: 10.1186/1471-2164-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26] **.Cerchietti LC, Hatzi K, Caldas-Lopes E, et al. Bcl6 repression of ep300 in human diffuse large b cell lymphoma cells provides a basis for rational combinatorial therapy. J Clin Invest. 2010 doi: 10.1172/JCI42869. The demonstration that p300 suppression plays a critical role in lymphomagenesis and that therapeutic reactivation of p300 is lethal to lymphoma cells. The manuscript is also the first report of p300 loss of function mutations in B-cell lymphoma patients.
- [27].Yang Y, Rao R, Shen J, et al. Role of acetylation and extracellular location of heat shock protein 90alpha in tumor cell invasion. Cancer Res. 2008;68(12):4833–4842. doi: 10.1158/0008-5472.CAN-08-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28] *.Garbati MR, Alco G, Gilmore TD. Histone acetyltransferase p300 is a coactivator for transcription factor rel and is c-terminally truncated in the human diffuse large b-cell lymphoma cell line rc-k8. Cancer Lett. 2010;291(2):237–245. doi: 10.1016/j.canlet.2009.10.018. The first demonstration of a p300 in a DLBCL cell line, and the potential implications for NFkB signaling.
- [29] *.Garbati MR, Thompson RC, Haery L, et al. A rearranged ep300 gene in the human b-cell lymphoma cell line rc-k8 encodes a disabled transcriptional co-activator that contributes to cell growth and oncogenicity. Cancer Lett. 2011;302(1):76–83. doi: 10.1016/j.canlet.2010.12.018. This manuscript shows that mutant p300 can disrupt the actions of c-Rel in DLBCL cells.
- [30] **.Pasqualucci L, Dominguez-Sola D, Chiarenza A, et al. Inactivating mutations of acetyltransferase genes in b-cell lymphoma. Nature. 2011;471(7337):189–195. doi: 10.1038/nature09730. This manuscript details for the first time inactivating mutations of CREBBP in DLBCL and FL as well as confirming the incidence of p300 mutations in DLBCL. The manuscript also shows that mutant CREBBP is functionally deficient in mediating protein acetylation.
- [31] *.Mullighan CG, Zhang J, Kasper LH, et al. Crebbp mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471(7337):235–239. doi: 10.1038/nature09727. The first demonstration that CREBBP is mutated in B-ALL. Most significantly, CREBBP mutations were associated with relapsed disease suggesting a role in chemoresistance.
- [32].Kung AL, Rebel VI, Bronson RT, et al. Gene dose-dependent control of hematopoiesis and hematologic tumor suppression by cbp. Genes Dev. 2000;14(3):272–277. [PMC free article] [PubMed] [Google Scholar]
- [33].Teo JL, Kahn M. The wnt signaling pathway in cellular proliferation and differentiation: A tale of two coactivators. Adv Drug Deliv Rev. 2010;62(12):1149–1155. doi: 10.1016/j.addr.2010.09.012. [DOI] [PubMed] [Google Scholar]
- [34] *.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: How cellular identity goes awry. Dev Cell. 2010;19(5):698–711. doi: 10.1016/j.devcel.2010.10.005. A detailed review of the perturnations and the impact of epigenetic programming in cancer.
- [35] *.Ji H, Ehrlich LI, Seita J, et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature. 2010;467(7313):338–342. doi: 10.1038/nature09367. The demonstration that DNA methylation patterning shifts during hematopoiesis and that lymphoid lineage commitment is associated with greater DNA methylation levels.
- [36] *.Lai AY, Fatemi M, Dhasarathy A, et al. DNA methylation prevents ctcf-mediated silencing of the oncogene bcl6 in b cell lymphomas. J Exp Med. 2010;207(9):1939–1950. doi: 10.1084/jem.20100204. A novel function for DNA methylation in sustaining high levels of BCL6 in lymphoma cells, by preventing the binding of CTCF to sites within the first intron of the BCL6 locus.
- [37] *.Shaknovich R, Geng H, Johnson NA, et al. DNA methylation signatures define molecular subtypes of diffuse large b-cell lymphoma. Blood. 2010;116(20):e81–89. doi: 10.1182/blood-2010-05-285320. This manuscript revealed the extent of epigenetic differences between ABC and GCB DLBCLs providing new information about the biological differences between these lymphoma subtypes.
- [38] **.Leshchenko VV, Kuo PY, Shaknovich R, et al. Genomewide DNA methylation analysis reveals novel targets for drug development in mantle cell lymphoma. Blood. 2010;116(7):1025–1034. doi: 10.1182/blood-2009-12-257485. The first comprehsive description of the MCL methylome, surprisingly showing that these tumors exhibit extensive aberrant hypomethylation compared to normal B-cells. This paper also shows how identification of aberrantly methylated and expressed genes can be used to design targeted therapies.
- [39].Martin-Subero JI, Kreuz M, Bibikova M, et al. New insights into the biology and origin of mature aggressive b-cell lymphomas by combined epigenomic, genomic, and transcriptional profiling. Blood. 2009;113(11):2488–2497. doi: 10.1182/blood-2008-04-152900. [DOI] [PubMed] [Google Scholar]
- [40].Bennett LB, Schnabel JL, Kelchen JM, et al. DNA hypermethylation accompanied by transcriptional repression in follicular lymphoma. Genes Chromosomes Cancer. 2009;48(9):828–841. doi: 10.1002/gcc.20687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41] *.Choi JH, Li Y, Guo J, et al. Genome-wide DNA methylation maps in follicular lymphoma cells determined by methylation-enriched bisulfite sequencing. PLoS One. 2010;5(9) doi: 10.1371/journal.pone.0013020. The first methylation sequencing of a lymphoma cell line, revealing extensive aberrant methylation and including an interesting comparison of DNA methylation with SUZ12 and H3K27 genomic localization.
- [42].O’Riain C, O’Shea DM, Yang Y, et al. Array-based DNA methylation profiling in follicular lymphoma. Leukemia. 2009;23(10):1858–1866. doi: 10.1038/leu.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43] *.Eberle FC, Rodriguez-Canales J, Wei L, et al. Methylation profiling of mediastinal gray zone lymphoma reveals a distinctive signature with elements shared by classical hodgkin’s lymphoma and primary mediastinal large b-cell lymphoma. Haematologica. 2011;96(4):558–566. doi: 10.3324/haematol.2010.033167. The demonstration that DNA methylation profiles can be used to distinguish lymphoma subtypes that might be dificult to identify using morphologic studies.
- [44].Ferguson LR, Tatham AL, Lin Z, et al. Epigenetic regulation of gene expression as an anticancer drug target. Curr Cancer Drug Targets. 2011;11(2):199–212. doi: 10.2174/156800911794328510. [DOI] [PubMed] [Google Scholar]
- [45] *.Ha K, Lee GE, Palii SS, et al. Rapid and transient recruitment of dnmt1 to DNA double-strand breaks is mediated by its interaction with multiple components of the DNA damage response machinery. Hum Mol Genet. 2011;20(1):126–140. doi: 10.1093/hmg/ddq451. DNMT1 is shown to be invovled in the repair of double strand breaks, further expanding its functional repertoire.
- [46] *.Sen GL, Reuter JA, Webster DE, et al. Dnmt1 maintains progenitor function in self-renewing somatic tissue. Nature. 2010;463(7280):563–567. doi: 10.1038/nature08683. DNMT1 is revealed to play a role in self renewal and proliferation, which has important implications in the 50% of lymphomas that express this factor.
- [47] *.Amara K, Ziadi S, Hachana M, et al. DNA methyltransferase DNMT3b protein overexpression as a prognostic factor in patients with diffuse large B-cell lymphomas. Cancer Sci. 2010;101(7):1722–1730. doi: 10.1111/j.1349-7006.2010.01569.x. The authors measured levels of DNMT1, DNMT3A and DNMT3B by immunohistochemistry in DLBCL patient specimens and characterize their frequency of expression. DNMT3B is shown to be a negative prognostic indicator in a multivariate analysis.
- [48] *.Shah MY, Vasanthakumar A, Barnes NY, et al. Dnmt3b7, a truncated dnmt3b isoform expressed in human tumors, disrupts embryonic development and accelerates lymphomagenesis. Cancer Res. 2010;70(14):5840–5850. doi: 10.1158/0008-5472.CAN-10-0847. The first demonstration that the cancer associated DNMT3B7 isoform, which seems to function as a dominant negative, can contribute to lymphomagenesis.
- [49] **.Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of bet bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. The first inhibitor of an epigenetic reader protein, in this case a bromodomain protein translocated in BRD4-NUT midline squamous carcinomas. The inhibitor suppressed human carcinoma cells in vitro and in vivo.
- [50] **.Cerchietti LC, Ghetu AF, Zhu X, et al. A small-molecule inhibitor of bcl6 kills dlbcl cells in vitro and in vivo. Cancer Cell. 2010;17(4):400–411. doi: 10.1016/j.ccr.2009.12.050. The first rationally designed small molecule therapeutic inhibitor of a transcriptional repressor, in this case BCL6. The inhibitor specifically blocked BCL6 and killed lymphoma cells without toxicity to other tissues.