Abstract
Background
IL8RA and IL8RB, encoded by CXCR1 and CXCR2, are receptors for interleukin (IL)-8 and other CXC chemokines involved in chemotaxis and activation of polymorphonuclear neutrophils (PMN). Variants at CXCR1 and CXCR2 have been associated with susceptibility to cutaneous and mucocutaneous leishmaniasis in Brazil. Here we investigate the role of CXCR1/CXCR2 in visceral leishmaniasis (VL) in India.
Methods
Three single nucleotide polymorphisms (SNPs) (rs4674259, rs2234671, rs3138060) that tag linkage disequilibrium blocks across CXCR1/CXCR2 were genotyped in primary family-based (313 cases; 176 nuclear families; 836 individuals) and replication (941 cases; 992 controls) samples. Family- and population-based analyses were performed to look for association between CXCR1/CXCR2 variants and VL. Quantitative RT/PCR was used to compare CXCR1/CXCR2 expression in mRNA from paired splenic aspirates taken before and after treatment from 19 VL patients.
Results
Family-based analysis using FBAT showed association between VL and SNPs CXCR1_rs2234671 (Z-score = 2.935, P = 0.003) and CXCR1_rs3138060 (Z-score = 2.22, P = 0.026), but not with CXCR2_rs4674259. Logistic regression analysis of the case-control data under an additive model of inheritance showed association between VL and SNPs CXCR2_rs4674259 (OR = 1.15, 95%CI = 1.01-1.31, P = 0.027) and CXCR1_rs3138060 (OR = 1.25, 95%CI = 1.02-1.53, P = 0.028), but not with CXCR1_rs2234671. The 3-locus haplotype T_G_C across these SNPs was shown to be the risk haplotype in both family- (TRANSMIT; P = 0.014) and population- (OR = 1.16, P = 0.028) samples (combined P = 0.002). CXCR2, but not CXCR1, expression was down regulated in pre-treatment compared to post-treatment splenic aspirates (P = 0.021).
Conclusions
This well-powered primary and replication genetic study, together with functional analysis of gene expression, implicate CXCR2 in determining outcome of VL in India.
Background
Visceral leishmaniasis (VL), also known as Kala-azar, is a life-threatening disease caused by protozoans belonging to Leishmania donovani complex. Population based epidemiological surveys suggest that 80-90% of individuals infected with L. donovani show no clinical symptoms [1-3]. Familial clustering, and the range of clinical outcomes from asymptomatic to fatal disease within and between ethnic groups sharing similar risk factors, support a contribution of host genotype to susceptibility [4-8]. Resistance to VL, as determined by a positive antigen-specific DTH response without clinical symptoms, is ~80% heritable in family-based studies [9]. Genetic variability in ability to mount an innate immune response, such as in the influx of polymorphonuclear neutrophils (PMN) during initial hours of infection, could be important in innate killing of the parasite [10], and in providing the cytokine environment in which parasite-specific T cells are primed [11]. Pro-inflammatory responses elicited by PMN as part of the response to the bite of the sand fly vector are important in initiation of infection [12]. The arrival and maintenance of infiltrating cells at bite sites is thought to be mediated by sand fly-derived factors that mimic a tissue damage signal and/or activate chemokine/chemokine receptor pathways [13-15]. Recently [16] we examined the role of polymorphisms at CXCR1 and CXCR2, which act as receptors for CXC chemokines that attract PMN to inflammatory sites, in determining susceptibility to cutaneous forms of leishmaniasis caused by infection with L. braziliensis. We found that cutaneous (CL) and mucocutaneous (ML) forms of disease were associated with opposing alleles for SNP rs2854386 at CXCR1. Studies in mice [17] show that selective depletion of PMN has a dramatic effect on the course of infection with L. donovani. Here we use genetic and functional approaches to evaluate the role of PMN in VL caused by L. donovani in humans through analysis of the receptors CXCR1, which is a specific receptor for IL-8 (= CXCL8), and CXCR2, which is promiscuous in binding a variety of CXC chemokines (CXCL-1, 2, 3, 5, 6, 7) in addition to CXCL8.
Methods
The study was conducted in the district of Muzaffarpur in Bihar State, India, where VL is highly endemic. Diagnosis of VL was made on the basis of clinical, parasitological and serological criteria as described [18,19]. Further epidemiological and demographic details relating to the study samples and study site are described elsewhere [20,21]. Informed written consent in Hindi was obtained from all participating individuals and from parents of children under 18 years old. Approval for the study was provided by the Ethical Committee of the Institute of Medical Sciences, Banaras Hindu University, Varanasi, India. Collection of families was undertaken between 2004 and 2006 while for the case-control study collection was undertaken during 2009-2010. Table 1 provides details of primary and replication samples, indicating the number of extended families that are decomposed into nuclear families for genetic analysis. For the family-based primary study, DNA was prepared from buccal swabs by whole genome amplification as described [19]. For the replication case-control study, genomic DNA was extracted from saliva using the Oragene technology (DNA Genotek, Ontario, Canada).
Table 1.
Baseline characteristics of (A) families for the primary sample of Indian multicase VL families, and (B) the Indian case-control cohorts
| (A) Family Structure | Number* |
|---|---|
| No families | 137 |
| No nuclear families | 176 |
| Nuclear families with 1 affected sib | 63 |
| Nuclear families with 2 affected sibs | 95 |
| Nuclear families with 3 affected sibs | 14 |
| Nuclear families with 4 affected sibs | 2 |
| Nuclear families with 5 affected sibs | 2 |
| No affected offspring | 313 |
| No affected parents | 63 |
| Total No affected individuals | 394 |
| Total No individuals | 836 |
| (B) Case-Control Sample | Number |
| Cases (no.) | 958 |
| Male | 571 |
| Female | 387 |
| Mean age at study encounter ± SD (yr) | 31.2 ± 16.7 |
| Range | 3-73 |
| Mean age at onset of VL ± SD (yr) | 26.8 ± 15.3 |
| Religious Group (no.) | |
| Hindu | 850 |
| Muslim | 108 |
| Controls (no.) | 1015 |
| Male | 570 |
| Female | 445 |
| Mean age at study encounter ± SD (yr) | 31.8 ± 15.9 |
| Religious Group (no.) | |
| Hindu | 885 |
| Muslim | 130 |
* Numbers are given for the individuals with DNA available for genotyping
CXCR1 and CXCR2 lie on a 26 kb region adjacent to each other on human Chromosome 2q35. CXCR2 is encoded on the positive strand and lies proximal to CXCR1 encoded on the negative strand. DNA samples were genotyped for SNPs (Table 2) CXCR2 (rs4674259) and CXCR1 (rs3138060) that tag major linkage disequilibrium (LD) blocks (r2 > 0.8; as determined for available data from HapMap populations CEU/JPT/CHB; Additional File 1; Figure S1) across the two genes, as well as the non-synonymous exon 1 SNP rs2234671 in CXCR1. For the primary family-based study, SNPs were genotyped in 836 individuals using ABI predesigned Taqman assays (ABI, Mulgrave, Victoria, Australia). For the replication study, SNPs were genotyped using Sequenom iPLEX platform (Sequenom, San Diego, CA, USA) for 2022 individuals comprising 990 cases, 1029 controls (1168 males, 850 females). All 3 SNPs met minimum quality control checks for call rate (> 99.5%) across all individuals, and all were in Hardy Weinberg equilibrium (HWE) in unrelated founders of families and in the control replication sample. Twelve individuals were removed from further analysis where data was missing for two or more SNPs.
Table 2.
Details of polymorphisms genotyped and the minor allele frequency (MAF) of variants in the Indian study population
| SNP Identity | Location | Amino Acid Change | Physical Position1 (bp) | Alleles2 | Strand | MAF |
|---|---|---|---|---|---|---|
| CXCR2_rs4674259 | 5'UTR | - | 218991005 | T/C | - | 0.43 |
| CXCR1_rs2234671 | Exon1 | S276T | 219029108 | G/C | - | 0.16 |
| CXCR1_rs3138060 | Intron1 | - | 219031500 | C/G | - | 0.12 |
1 Physical positions of markers are given according to Build 37.1 of the human genome; 2Major > minor alleles for this Indian population; All 3 SNPs were in HWE in both unaffected founders of families and in controls for the replication study.
Family-based allelic association tests were performed within FBAT which is based on the transmission disequilibrium test (TDT) but allows for different genetic model analyses with incomplete parental data [22,23]. Analyses were performed using an additive model and under the null hypothesis of "no linkage and no association". Family-based haplotype TDT was performed using TRANSMIT [24]. Robust tests were performed to take account of multiple trios within some pedigrees (Table 1). Family based TDT power approximations [25] show that the 313 VL trios had 49% power to detect an odds ratio ≥ 1.5 at P = 0.01 for markers with MAF ≥ 0.1. Single marker and haplotype association tests for the case-control sample were carried out using logistic regression analysis performed in PLINK [26] under an additive model. Inclusion of caste as a covariate, which we have shown to provide a good surrogate for genetic substructure in genome-wide analyses in this population (unpublished data), was used to take account of population substructure. Religion was also analysed as a covariate. The 1933 individuals (941 cases and 992 controls) which passed quality control had 93.5% power to detect associations with an odds ratio of 1.5 for markers with MAF ≥ 0.1 at P = 0.01. Nominal two-tailed P-values are presented throughout, i.e. without correction for multiple testing. Application of a strict Bonferroni correction for 3 independent (r2 < 0.8) SNPs provides a significance cut-off of P ≤ 0.017 (i.e. P = 0.05/3). Combined P-values using Fisher's Trend Test were calculated using MetaP [27]. LD plots for D' and r2 were generated in Haploview [28].
Splenic biopsies were taken as part of routine diagnostic procedure at the Kala Azar Medical Research Centre, Muzaffarpur, Bihar State, India. Paired pre- (Day-0) and post- (Day-30) treatment splenic samples were collected from 19 VL patients in 5xRNA Later (AMBION Inc., Austin, Texas, USA) during 2009-2010, transported to Varanasi at 4°C and stored at -80°C until RNA was isolated. Details regarding age and sex, splenic parasites and drug administered were recorded for each patient. Total RNA was isolated using RNeasy tissue kit (Qiagen, GmbH, Hilden, Germany) according to the manufacturer's instructions. Sample quality and integrity was assessed by ND-2000 spectrophotometer (Thermo Fischer Scientific, Wilmington, DE, USA) and agarose (Sigma Aldrich Chemicals, St Louis, MO, USA) gel electrophoresis. 1 μg of RNA was reverse transcribed using the High Capacity cDNA synthesis kit (Applied Biosystems, Foster City, CA, USA). Taqman predesigned gene expression assays (CXCR1_Hs00174146_m1 and CXCR2_Hs01011557_m1) were used to perform expression studies (7500 HT Real Time PCR system, Applied Biosystems, Foster City CA, USA) with 18S rRNA (P/N 4319413E) being used as an endogenous control to normalize the expression data. No RT and no template controls were included in each plate. All samples were run in duplicate. Results were analysed by 7500 software v.2.0.1 and GraphPad Prism (version 5.00 for Windows, Graph Pad Software, San Diego California USA, http://www.graphpad.com). The significance of differences between pre- and post-treatment groups was determined using paired Student's T test. Power calculations showed that N = 19 paired samples had 92.2% power to detect a difference in mean values pre- and post-treatment of 0.9 with standard deviation of 0.9 at alpha level 0.01; 98.2% power at alpha level 0.05.
Results and Discussion
To test the hypothesis that polymorphisms at CXCR1/CXCR2 might influence susceptibility to VL in India we initially genotyped 3 tagging SNPs (Table 2) in 176 nuclear families (Table 1) used in our previous studies [19,21] that contain 313 offspring with VL collected in the area of Muzaffarpur, Bihar State, India, where L. donovani is endemic. Using the family-based association test (FBAT) [29,30] in this primary family dataset (Table 3) we found evidence (nominal P-values ≤ 0.03) for associations between VL and SNPs CXCR1_rs2234671 (Z-score = 2.935, P = 0.003) and CXCR1_rs3138060 (Z-score = 2.22, P = 0.026), but not with CXCR2_rs4674259. Since these two positive markers are in quite strong LD with each other (Additional File 2; Figure S2: D' = 0.84; r2 = 0.46), these associations are likely to be measuring a single effect. The association at CXCR1_rs2234671 is robust to application of a strict Bonferroni correction for 3 independent (i.e. r2 < 0.8) SNPs genotyped, which requires a significance cut-off of P ≤ 0.017 (i.e. P = 0.05/3).
Table 3.
Family-based association analysis between CXCR1/CXCR2 and VL
| Common Designation | Allele | Allele frequency |
# Fam | S | E(S) | Var(S) | Z | P |
|---|---|---|---|---|---|---|---|---|
| CXCR2_rs4674259 | T | 0.53 | 105 | 196 | 194 | 57.313 | 0.216 | 0.829 |
| CXCR2_rs4674259 | C | 0.47 | 105 | 184 | 186 | 57.313 | -0.216 | 0.829 |
| CXCR1_rs2234671 | G | 0.85 | 69 | 181 | 165 | 31.519 | 2.935 | 0.003 |
| CXCR1_rs2234671 | C | 0.15 | 69 | 61 | 77 | 31.519 | -2.935 | 0.003 |
| CXCR1_rs3138060 | G | 0.12 | 49 | 49 | 59 | 22.187 | -2.222 | 0.026 |
| CXCR1_rs3138060 | C | 0.88 | 49 | 133 | 122 | 22.187 | 2.222 | 0.026 |
Single point FBAT analysis under an additive model of inheritance for associations between CXCR1/CXCR2 polymorphisms and VL in the primary family-based sample set. # Fam = number of families informative for the FBAT analysis; S and E(S) represent the observed and expected transmissions for that allele, Var(S) is the variance. A positive Z score indicates association with disease; a negative Z score indicates the non-associated or protective allele or genotype. Bold indicates significant associations at nominal P ≤ 0.05. The corrected P-value required to achieve significance taking account of 3 independent SNPs is P ≤ 0.017.
Evidence for an association between Indian VL and SNPs at CXCR1, which was consistent with data for CL/ML from Brazil [16], prompted us to pursue two further avenues of investigation. In so doing, we did not discount the possibility that CXCR2 might play a role, since the D' measure (Additional File 2; Figure S2) indicates strong LD across the CXCR1/2 SNPs for this sample (D' = 0.98 and 0.91). First, we looked at expression levels of CXCR1/CXCR2 in mRNA from splenic aspirates from 19 patients as paired samples taken pre- and post-treatment for VL (Figure 1). This demonstrated that CXCR2, but not CXCR1, is significantly (P = 0.021) reduced in expression in pre-treatment samples compared to post-treatment recovery samples, indicating that a deficiency in expression of CXCR2 might contribute to VL disease. Down regulation of CXCR2, but not CXCR1, was similarly observed in PMN from patients with pulmonary tuberculosis [31]. Secondly, we carried out a comprehensive replication of the association study in a much larger population-based case-control sample (Table 1) from the same region of Bihar State in India. Logistic regression analysis (Table 4) under an additive model of inheritance showed association between VL and SNPs CXCR2_rs4674259 (OR = 1.15, 95%CI = 1.01-1.31, P = 0.027) and CXCR1_rs3138060 (OR = 1.25, 95%CI = 1.02-1.53, P = 0.028), but not with CXCR1_rs2234671. Effect sizes (OR) are small, as is common in complex communicable or non-communicable diseases [32]. Significance was retained when either religion (CXCR2_rs4674259: OR = 1.15, 95%CI = 1.02-1.31, P = 0.028; CXCR1_rs3138060: OR = 1.26, 95%CI = 1.03-1.53, P = 0.026) or caste (CXCR2_rs4674259: OR = 1.15, 95%CI = 1.01-1.32, P = 0.039; CXCR1_rs3138060: OR = 1.25, 95%CI = 1.01-1.54, P = 0.037) was used as a covariate to take account of population substructure. Interestingly, although CXCR1_rs3138060 was the only SNP directly replicated across primary and replication samples, the same 3-SNP haplotype T_G_C (called on the negative strand; frequency 0.42) was shown to be the risk haplotype in both family- (Table 5; TRANSMIT; P = 0.014) and population- (Table 5; PLINK; OR = 1.16, P = 0.028) samples (combined P = 0.002). The 2- and 3-SNP over-transmitted haplotypes were more apparent in the family-based sample, which likely reflects over-relatedness for this set of families from this region of India where we showed previously that consanguineal marriages were common [19]. These haplotype results indicate that associations seen at CXCR1 might be due to LD with regulatory polymorphisms that influence CXCR2 expression. Together, the genetic and functional analyses favour a role for CXCR2 in contributing to susceptibility to VL in this region of India, although a role for CXCR1 cannot be discounted.
Figure 1.
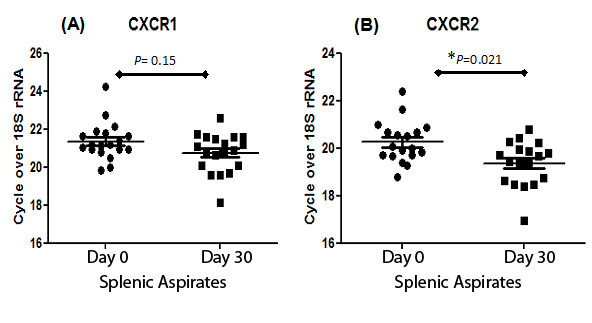
Relative expression of CXCR1 (A) and CXCR2 (B) mRNA in paired splenic aspirates from VL patients before (Day 0) and after (Day 30) antileishmanial treatment. Individual as well as mean (± SEM) relative expression for each group is indicated. Paired Student's t tests show significant differences in expression of CXCR2 but not CXCR1 when Day 0 values were compared to Day 30 values.
Table 4.
Population-based association analysis between CXCR1/CXCR2 and VL
| Common Designation | Allele | Affected | Unaffected | OR | L95 | U95 | P-values |
|---|---|---|---|---|---|---|---|
| CXCR2_rs4674259 | T | 840/1036 | 816/1160 | 1.15 | 1.01 | 1.31 | 0.027 |
| CXCR1_rs2234671 | G | 286/1592 | 321/1657 | 0.92 | 0.77 | 1.10 | 0.384 |
| CXCR1_rs3138060 | C | 197/1685 | 252/1732 | 1.25 | 1.02 | 1.53 | 0.028 |
Logistic regression analyses under an additive model for the replication sample of VL cases and controls. OR = odds ratio; L95 and U95 are lower and upper 95% confidence intervals. Allele counts are shown for minor/major allele for affected and unaffected individuals. Bold indicates associations significant at nominal P ≤ 0.05. The corrected P-value required to achieve significance taking account of 3 independent SNPs is P ≤ 0.017.
Table 5.
Haplotype association analyses between CXCR1/CXCR2 and VL
| Haplo | Family Analysis | Freq | CXCR2_rs4674259 | CXCR1_rs2234671 | CXCR1_rs3138060 |
|---|---|---|---|---|---|
| T.G | TRANSMIT | 0.41 | χ2 = 4.61; 1df; P = 0.032 | ||
| _ G.C | TRANSMIT | 0.83 | χ2 = 10.27; 1df; P = 0.001 | ||
| T.G.C | TRANSMIT | 0.40 | χ2 = 5.98; 1df; P = 0.014 | ||
| Haplo | Case-Control | Freq | CXCR2_rs4674259 | CXCR1_rs2234671 | CXCR1_rs3138060 |
| T.G | PLINK | 0.43 | χ2 = 4.34; 1df; P = 0.037 | ||
| _ G.C | PLINK | 0.83 | χ2 = 1.29; 1df; P = 0.256 | ||
| T.G.C | PLINK | 0.42 | χ2 = 4.99; 1df; P = 0.025 | ||
Chi-squared (χ2), degrees of freedom (df) and P-values for risk associated (over-transmitted) haplotypes for the 3 SNPs as determined in TRANSMIT for the family-based primary sample, and in PLINK for the case-control replication sample. Bold indicates significant associations at nominal P ≤ 0.05. Haplotypes are called on the negative strand across the 3 SNPs.
The involvement of either CXCR1 or CXCR2 in VL is of interest given recent observations on the different roles they may play in chemotaxis and activation of PMN in sites of infection [33], and the increasing recognition of the importance of PMN in the VL disease process [12,17]. Both are high affinity receptors for the CXC chemokine IL-8, but CXCR2 is more promiscuous in also binding a range of other CXC chemokines (CXCL-1, 2, 3, 5, 6, and 7) which may play important roles in directing a broader array of immune cells to sites of infection in the visceral organs. Recent studies [34] also show that, while neutralizing antibodies against CXCR2 abolish neutrophil extracellular trap (NET) formation, antibodies against CXCR1 have no effect. NETs are formed by DNA fibers decorated with antimicrobial proteins released from PMN upon activation. The release of DNA NETs, decorated with elastases and histones, by human PMN upon interaction with Leishmania parasites has been shown to ensnare the parasite and is leishmanicidal [10], adding to the potential importance of PMN in innate immunity to leishmanial infection.
Here we examined CXCR1 and CXCR2 as candidate genes for susceptibility to VL in India. Whilst SNPs at both loci were associated with VL, functional analysis of expression in splenic aspirates together with a common risk haplotype favour CXCR2 as the etiological gene regulating susceptibility to disease. Our data contribute to increasing evidence for an important role for PMN in directing the outcome of leishmanial infections in humans.
Conclusions
From the results of this well-powered primary and replication genetic study, together with functional analysis of gene expression, we conclude that CXCR2 plays a role in determining outcome of VL in India.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
AM, MF and SM carried out the field collection and/or preparation of the samples. SM and JO performed the genotyping, and participated in the statistical analysis and interpretation of the data. MF cross-checked statistical analyses and carried out additional statistical tests, including the haplotype analyses. SEJ trained SM in the laboratory for genotyping techniques, in database entry and use of the genetic database GenIE in Perth, and in genetic statistical analysis methods. MR oversaw laboratory-based work in Varanasi. DSR and KT oversaw the Sequenom genotyping undertaken by SM in Hyderabad. MS and PT assisted with RNA preparation. SM designed and carried out the QRT/PCR. SS helped conceive the study, was responsible for clinical care of cases at the Kala Azar Medical Research Centre, Muzaffarpur, Bihar State, India, and provided the logistical support to make the study possible. SM prepared the first draft of the manuscript. JMB designed the study, conceived the specific hypothesis to be tested, made the final interpretation of the data, and prepared the final manuscript. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Supplementary Material
Figure S1. Graphical representation of pairwise D' and r2 LD measures across CXCR1 and CXCR2 in the HapMap CHB/JPT populations demonstrating large LD blocks tagged by SNPs CXCR1_rs3138060 and CXCR2_rs4674259 genotyped as tag-SNPs in the study.
Figure S2. Graphical representation of pairwise D' and r2 LD measures across 3 SNPs genotyped in the study.
Contributor Information
Sanjana Mehrotra, Email: anniebhu19@gmail.com.
Michaela Fakiola, Email: mf300@cam.ac.uk.
Joyce Oommen, Email: joommen@ichr.uwa.edu.au.
Sarra E Jamieson, Email: sjamieson@ichr.uwa.edu.au.
Anshuman Mishra, Email: anshuindia@gmail.com.
Medhavi Sudarshan, Email: medhavisudarshan@gmail.com.
Puja Tiwary, Email: tiwarypuja@gmail.com.
Deepa Selvi Rani, Email: deepa@ccmb.res.in.
Kumarasamy Thangaraj, Email: thangs@ccmb.res.in.
Madhukar Rai, Email: upicon2007@gmail.com.
Shyam Sundar, Email: drshyamsundar@hotmail.com.
Jenefer M Blackwell, Email: jmb37@cam.ac.uk.
Acknowledgements
We would like to thank the families from the state of Bihar, northeast India for their participation in this study. The authors declare no conflict of interest. This research was funded by grants from The Wellcome Trust in the UK (grant numbers: 074196/Z/04/Z and 085475/Z/08/Z) and The National Institutes of Health in the USA (grant numbers: R01 AI076233-01 and 1P50AI074321-01). Authors SM, MS and PT are thankful to Council of Scientific and Industrial Research (CSIR), New Delhi for providing financial assistance.
References
- Desjeux P. The increase in risk factors for leishmaniasis worldwide. Trans R Soc Trop Med Hyg. 2001;95(3):239–243. doi: 10.1016/S0035-9203(01)90223-8. [DOI] [PubMed] [Google Scholar]
- Badaro R, Jones TC, Lorenco R, Cerf BJ, Sampaio D, Carvalho EM, Rocha H, Teixeira R, Johnson WD Jr. A prospective study of visceral leishmaniasis in an endemic area of Brazil. J Infect Dis. 1986;154(4):639–649. doi: 10.1093/infdis/154.4.639. [DOI] [PubMed] [Google Scholar]
- Topno RK, Das VN, Ranjan A, Pandey K, Singh D, Kumar N, Siddiqui NA, Singh VP, Kesari S, Bimal S, Kumar AJ, Meena C, Kumar R, Das P. Asymptomatic infection with visceral leishmaniasis in a disease-endemic area in bihar, India. Am J Trop Med Hyg. pp. 502–506. [DOI] [PMC free article] [PubMed]
- Peacock CS, Collins A, Shaw MA, Silveira F, Costa J, Coste CH, Nascimento MD, Siddiqui R, Shaw JJ, Blackwell JM. Genetic epidemiology of visceral leishmaniasis in northeastern Brazil. Genet Epidemiol. 2001;20(3):383–396. doi: 10.1002/gepi.8. [DOI] [PubMed] [Google Scholar]
- Cabello PH, Lima AM, Azevedo ES, Krieger H. Familial aggregation of Leishmania chagasi infection in northeastern Brazil. Am J Trop Med Hyg. 1995;52(4):364–365. doi: 10.4269/ajtmh.1995.52.364. [DOI] [PubMed] [Google Scholar]
- Zijlstra EE, el-Hassan AM, Ismael A, Ghalib HW. Endemic kala-azar in eastern Sudan: a longitudinal study on the incidence of clinical and subclinical infection and post-kala-azar dermal leishmaniasis. Am J Trop Med Hyg. 1994;51(6):826–836. doi: 10.4269/ajtmh.1994.51.826. [DOI] [PubMed] [Google Scholar]
- Blackwell JM, Mohamed HS, Ibrahim ME. Genetics and visceral leishmaniasis in the Sudan: seeking a link. Trends Parasitol. 2004;20(6):268–274. doi: 10.1016/j.pt.2004.04.003. [DOI] [PubMed] [Google Scholar]
- El-Safi S, Kheir MM, Bucheton B, Argiro L, Abel L, Dereure J, Dedet JP, Dessein A. Genes and environment in susceptibility to visceral leishmaniasis. C R Biol. 2006;329(11):863–870. doi: 10.1016/j.crvi.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Jeronimo SM, Duggal P, Ettinger NA, Nascimento ET, Monteiro GR, Cabral AP, Pontes NN, Lacerda HG, Queiroz PV, Gomes CE, Pearson RD, Blackwell JM, Beaty TH, Wilson ME. Genetic predisposition to self-curing infection with the protozoan Leishmania chagasi: a genomewide scan. J Infect Dis. 2007;196(8):1261–1269. doi: 10.1086/521682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes-Costa AB, Nascimento MT, Froment GS, Soares RP, Morgado FN, Conceicao-Silva F, Saraiva EM. Leishmania amazonensis promastigotes induce and are killed by neutrophil extracellular traps. Proc Natl Acad Sci USA. 2009;106(16):6748–6753. doi: 10.1073/pnas.0900226106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Mackay CR, Lanzavecchia A. The role of chemokine receptors in primary, effector, and memory immune responses. Annu Rev Immunol. 2000;18:593–620. doi: 10.1146/annurev.immunol.18.1.593. [DOI] [PubMed] [Google Scholar]
- Peters NC, Egen JG, Secundino N, Debrabant A, Kimblin N, Kamhawi S, Lawyer P, Fay MP, Germain RN, Sacks D. In vivo imaging reveals an essential role for neutrophils in leishmaniasis transmitted by sand flies. Science. 2008;321(5891):970–974. doi: 10.1126/science.1159194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira CR, Teixeira MJ, Gomes RB, Santos CS, Andrade BB, Raffaele-Netto I, Silva JS, Guglielmotti A, Miranda JC, Barral A, Brodskyn C, Barral-Netto M. Saliva from Lutzomyia longipalpis induces CC chemokine ligand 2/monocyte chemoattractant protein-1 expression and macrophage recruitment. J Immunol. 2005;175(12):8346–8353. doi: 10.4049/jimmunol.175.12.8346. [DOI] [PubMed] [Google Scholar]
- Teixeira MJ, Teixeira CR, Andrade BB, Barral-Netto M, Barral A. Chemokines in host-parasite interactions in leishmaniasis. Trends Parasitol. 2006;22(1):32–40. doi: 10.1016/j.pt.2005.11.010. [DOI] [PubMed] [Google Scholar]
- Teixeira MJ, Fernandes JD, Teixeira CR, Andrade BB, Pompeu ML, Santana da Silva J, Brodskyn CI, Barral-Netto M, Barral A. Distinct Leishmania braziliensis isolates induce different paces of chemokine expression patterns. Infect Immun. 2005;73(2):1191–1195. doi: 10.1128/IAI.73.2.1191-1195.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellucci L, Jamieson SE, Miller EN, Menezes E, Oliveira J, Magalhaes A, Guimaraes LH, Lessa M, de Jesus AR, Carvalho EM, Blackwell JM. CXCR1 and SLC11A1 polymorphisms affect susceptibility to cutaneous leishmaniasis in Brazil: a case-control and family-based study. BMC Med Genet. 2010;11:10. doi: 10.1186/1471-2350-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane E, Perez C, Charmoy M, Allenbach C, Carter KC, Alexander J, Tacchini-Cottier F. Neutrophils contribute to development of a protective immune response during onset of infection with Leishmania donovani. Infect Immun. 2008;76(2):532–541. doi: 10.1128/IAI.01388-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundar S, Benjamin B. Diagnosis and treatment of Indian visceral leishmaniasis. J Assoc Physicians India. 2003;51:195–201. [PubMed] [Google Scholar]
- Fakiola M, Mishra A, Rai M, Singh SP, O'Leary RA, Ball S, Francis RW, Firth MJ, Radford BT, Miller EN, Sundar S, Blackwell JM. Classification and regression tree and spatial analyses reveal geographic heterogeneity in genome wide linkage study of Indian visceral leishmaniasis. PLoS ONE. 2010;5(12):e15807. doi: 10.1371/journal.pone.0015807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Reddy DC, Mishra RN, Sundar S. Knowledge, attitude, and practices related to Kala-azar in a rural area of Bihar state, India. Am J Trop Med Hyg. 2006;75(3):505–508. [PubMed] [Google Scholar]
- Mehrotra S, Oommen J, Mishra A, Sudharshan M, Tiwary P, Jamieson SE, Fakiola M, Rani DS, Thangaraj K, Rai M, Sundar S, Blackwell JM. No evidence for association between SLC11A1 and visceral leishmaniasis in India. BMC Med Genet. 2011;12:71. doi: 10.1186/1471-2350-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19(Suppl 1):S36–42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype--phenotype associations. Eur J Hum Genet. 2001;9(4):301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- Clayton D, Jones H. Transmission disequilibrium tests for extended marker haplotypes. AmJHumGenet. 1999;65(4):1161–1169. doi: 10.1086/302566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp M. A note on power approximations for the transmission disequilibrium test. Am J Hum Genet. 1999;64(4):1177–1185. doi: 10.1086/302334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. Journal of evolutionary biology. 2005;18(5):1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol. 2000;19(Suppl 1):S36–42. doi: 10.1002/1098-2272(2000)19:1+<::AID-GEPI6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird NM. The family based association test method: strategies for studying general genotype--phenotype associations. Eur J Hum Genet. 2001;9(4):301–306. doi: 10.1038/sj.ejhg.5200625. [DOI] [PubMed] [Google Scholar]
- Juffermans NP, Dekkers PE, Peppelenbosch MP, Speelman P, van Deventer SJ, van Der Poll T. Expression of the chemokine receptors CXCR1 and CXCR2 on granulocytes in human endotoxemia and tuberculosis: involvement of the p38 mitogen-activated protein kinase pathway. J Infect Dis. 2000;182(3):888–894. doi: 10.1086/315750. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillie R, Farooq SM, Gordon JR, Stadnyk AW. The functional significance behind expressing two IL-8 receptor types on PMN. J Leukoc Biol. 2009;86(3):529–543. doi: 10.1189/jlb.0208125. [DOI] [PubMed] [Google Scholar]
- Marcos V, Zhou Z, Yildirim AO, Bohla A, Hector A, Vitkov L, Wiedenbauer EM, Krautgartner WD, Stoiber W, Belohradsky BH, Rieber N, Kormann M, Koller B, Roscher A, Roos D, Griese M, Eickelberg O, Doring G, Mall MA, Hartl D. CXCR2 mediates NADPH oxidase-independent neutrophil extracellular trap formation in cystic fibrosis airway inflammation. Nat Med. 2010;16(9):1018–1023. doi: 10.1038/nm.2209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Graphical representation of pairwise D' and r2 LD measures across CXCR1 and CXCR2 in the HapMap CHB/JPT populations demonstrating large LD blocks tagged by SNPs CXCR1_rs3138060 and CXCR2_rs4674259 genotyped as tag-SNPs in the study.
Figure S2. Graphical representation of pairwise D' and r2 LD measures across 3 SNPs genotyped in the study.


