Abstract
Background
APE1 (apurinic/apyrimidinic endonuclease 1) is an important DNA repair protein in the base excision repair pathway. Polymorphisms in APE1 have been implicated in susceptibility to cancer; however, results from the published studies remained inconclusive. The objective of this study was to conduct a meta-analysis investigating the association between polymorphisms in APE1 and the risk for cancer.
Methods
The PubMed and Embase databases were searched for case-control studies published up to June, 2011 that investigated APE1 polymorphisms and cancer risk. Odds ratios (ORs) and 95% confidence intervals (CIs) were used to assess the strength of the associations.
Results
Two polymorphisms (−656 T > G, rs1760944 and 1349 T > G, rs1130409) in 37 case-control studies including 15, 544 cancer cases and 21, 109 controls were analyzed. Overall, variant genotypes (GG and TG/GG) of −656 T > G polymorphism were associated with significantly decreased cancer risk in homozygote comparison (OR = 0.81, 95%CI: 0.67-0.97), dominant model comparison (OR = 0.89, 95%CI: 0.81-0.97) and recessive model comparison (OR = 0.90, 95%CI: 0.82-0.98), whereas the 1349 T > G polymorphism had no effects on overall cancer risk. In the stratified analyses for −656 T > G polymorphism, there was a significantly decreased risk of lung cancer and among Asian populations.
Conclusions
Although some modest bias could not be eliminated, the meta-analysis suggests that APE1 −656 T > G polymorphism has a possible protective effect on cancer risk particularly among Asian populations whereas 1349 T > G polymorphism does not contribute to the development of cancer.
Keywords: cAPE1, Single nucleotide polymorphism, Cancer risk, Meta-analysis
Background
Cancer is a multifactor disease caused by complex interactions between environmental and genetic factors [1]. The incidence of different cancer varies widely in different populations which may be largely attributed to lifestyle and genetic background [2]. Environmental factors such as smoking and exposure to carcinogens lead to direct damage to DNA [3]. To a certain extent, these damages can be repaired by endogenous DNA repair systems [4]. However, genetic variations in these systems may result in reduced repair capability and the defect allow DNA damage to accumulate which leads to permanent mutations in the genome and contributes to carcinogenesis.
Among DNA repair systems, base excision repair (BER) pathway is responsible for repairing small lesions such as oxidative damage, alkylation, or methylation [5]. AP Endonuclease 1 (APE1, also known as APE, APEX, HAP1, and REF-1) is a multifunctional protein and plays a central role in the BER pathway through hydrolyzing the phosphodiester backbone immediately 5' to the AP site [5,6]. APE1 can act as a 3'-phosphodiesterase to initiate repair of DNA single strand breaks, which are produced either directly by reactive oxygen species or indirectly through the enzymatic removal of damaged bases [7,8]. APE1 is also known as a transcriptional coactivator for numerous transcription factors involved in cancer development [9] and is considered as a promising tool for anticancer therapy [10].
The human APE1 is located on chromosome 14q11.2-q12 and consists of five Exons spanning 2.21 kb [11]. Till now, many epidemiological studies suggested genetic variations in APE1 may confer individuals' susceptibility to cancer. A total of 18 single nucleotide polymorphisms (SNPs) in APE1 have been identified [11], of which, two functional SNPs (−656 T > G in the promoter region, rs1760944 and 1349 T > G in the fifth Exon, rs1130409) have been wildly investigated. In lung cancer, Lu et al. and Lo et al. reported that −656 T > G polymorphism influenced the transcriptional activity of APE1 and contributed to lung cancer susceptibility [12,13]. Although one study in bladder cancer supported the association [14], it has not been replicated in some others [15-17]. As to 1349 T > G polymorphism, functional studies suggested that the G allele may have altered endonuclease and DNA-binding activity, reduced ability to communicate with other BER proteins and decreased capacity to repair DNA oxidative damage [18,19]. Recently, numerous studies investigated the influence of 1349 T > G polymorphism on cancer risk; however, the results of these studies remain inconclusive.
To reveal a small effect of the polymorphisms on cancer risk, a single study might be underpowered, particularly for studies with relatively small sample size. Meta-analysis is a statistical technique for combining results from different studies to produce a single estimate of the major effect with enhanced precision. It is considered a powerful tool to summarize inconclusive results from different studies. To clarify the effect of the APE1 -656 T > G and 1349 T > G polymorphisms on cancer risk; we conducted a meta-analysis of all eligible case-control studies that have been published. Before analysis, we tested whether these two SNPs were in linkage disequilibrium with each other in Europeans and Asians, and the data from HapMap database suggested the two SNPs were not correlated (D' = 0.335, r2 = 0.071 for Europeans and D' = 0.409, r2 = 0.071 for Asians).
Methods
Identification of eligible studies
A literature search of the PubMed and EMBASE databases (updated to June, 2011) was conducted using combinations of the following terms: "APE1", "APEX", "HAP1", "REF-1", "polymorphism", and "cancer". The search was limited to English language papers. All studies that evaluated the association between polymorphisms of APE1 and cancer risk were retrieved, and their bibliographies were checked for other relevant publications. Studies that were included in the meta-analysis had to meet all of the following criteria: 1) evaluate the APE1 −656T > G and 1349T > G polymorphisms and cancer risk, 2) use a case-control design, 3) contain available genotypes frequency for estimating an odds ratio (OR) with a 95% confidence intervals (CIs), 4) genotype distributions in control consistent with Hardy-Weinberg equilibrium (HWE). Accordingly, abstracts and reviews studies that that did not report genotype frequency were excluded. When more than one of the same patient population was included in several publications, only the most recent or complete study was used in this meta-analysis.
Data extraction
Information was carefully extracted from all eligible publications independently by two of the authors according to the selection criteria. In case of disagreement, a third reviewer assessed the articles. The following items were collected: author name, year of publication, country of origin, ethnicity, source of the controls, genotyping method, cancer type, total number of cases and controls, and genotype distributions in cases and controls.
Statistical analysis
The strength of associations between APE1 polymorphisms and cancer risk were measured by ORs with 95%CIs. We examined the association between the APE1 −656 T > G and 1349 T > G polymorphisms and cancer risk in homozygote comparison (GG vs. TT), heterozygote comparison (TG vs. TT), the dominant genetic model (TG/GG vs. TT), and the recessive genetic model (GG vs. TT/TG). Stratified analyses were also carried out by ethnicity, cancer type and source of controls.
Heterogeneity assumption was evaluated with a chi-square-based Q-test. If the P value is greater than 0.05 of the Q-test, which indicates a lack of heterogeneity among studies, the summary OR estimate of each study was calculated by a fixed effects model (the Mantel-Haenszel method) [20]; otherwise, the random-effects model (the DerSimoniane and Laird method) [21] was performed. Sensitivity analyses were also performed to assess the stability of the results. Funnel plots and Egger's linear regression test were used to provide diagnosis of the potential publication bias. All statistical analysis were done with the Stata software (version 11.0; StataCorp LP, College Station, TX), using two-sided P values.
Results
Study selection and characteristics in the meta-analysis
A total of 37 eligible studies from 32 articles met the inclusion criteria [12-17,22-47]. In total, 5,139 cases and 5,201 controls from 8 studies for −656 T > G polymorphism [12-17], and 14,222 cases and 19,746 controls from 35 studies for 1349 T > G polymorphism were included in the pooled analyses [12-14,16,17,22-47]. The characteristics of selected studies are presented in Table 1. The distribution of genotypes in the controls of all studies was consistent with HWE. Of the 8 studies for −656 T > G polymorphism, 3 investigated lung cancer and 2 investigated bladder cancer, and the other 3 articles investigated individually brain cancer, renal cell cancer and colorectal adenoma. Six of these studies were conducted in Asian populations and 2 in European populations. Among the 35 studies for 1349 T > G polymorphism, there were 7 bladder cancer studies, 4 breast cancer studies, 4 colorectal cancer studies, 2 head and neck cancer studies, 9 lung cancer studies and 9 others studies including oesophageal cancer, pancreatic cancer, leukaemia, prostate cancer, biliary tract cancer, thyroid cancer, renal cell cancer and gastric cancer. Ten of these studies were from Asians, 23 were from Europeans and 2 were studies of mixed populations.
Table 1.
Characteristics of populations and cancer types of the studies included in the meta-analysis
| Study | Country | Ethnicity | Cancer type | Sample size case/control | Source of controls | Genotyping method | Polymorphisms |
|---|---|---|---|---|---|---|---|
| Misra 2003 20 | Finland | European | Lung cancer | 310/302 | Population | TaqMan | 1349 T > G |
| Popanda 2004 21 | Germany | European | Lung cancer | 459/457 | Hospital | Rapid capillary PCR | 1349 T > G |
| Hao 2004 22 | China | Asian | Oesophageal cancer | 409/478 | Population | PCR-RFLP | 1349 T > G |
| Ito 2004 23 | Japan | Asian | Lung cancer | 178/449 | Hospital | PCR-RFLP | 1349 T > G |
| Shen 2005 24 | China | Asian | Lung cancer | 117/113 | Population | TaqMan | 1349 T > G |
| Moreno 2006 25 | Spain | European | Colorectal cancer | 359/312 | Hospital | Arrayed primer extension | 1349 T > G |
| Jiao 2006 26 | USA | European | Pancreatic cancer | 367/330 | Hospital | Allele-specific PCR | 1349 T > G |
| Terry 2006 27 | USA | European | Bladder cancer | 229/207 | Hospital | MALDI-TOF | 1349 T > G |
| Wu 2006 28 | USA | European | Bladder cancer | 596/590 | Hospital | TaqMan | 1349 T > G |
| Zhang 2006 29 | USA | European | Breast cancer | 1529/1207 | Population | TaqMan | 1349 T > G |
| Matullo 2006 30 | Multi-country | European | Breast cancer | 124/1094 | Population | TaqMan | 1349 T > G |
| Lung cancer | 116/1094 | Population | TaqMan | 1349 T > G | |||
| Head and neck cancer | 82/1094 | Population | TaqMan | 1349 T > G | |||
| Leukemia | 169/1094 | Population | TaqMan | 1349 T > G | |||
| Chen 2006 31 | USA | Mixed | Prostate cancer | 351/329 | Hospital | PCR-RFLP | 1349 T > G |
| Li 2007 32 | USA | European | Head and neck cancer | 830/854 | Hospital | PCR-RFLP | 1349 T > G |
| Berndt 2007 14 | USA | European | Colorectal cancer | 767/773 | Population | TaqMan | 1349 T > G, −656 T > G |
| Figueroa 2007 15 | Spain | European | Bladder cancer | 1150/1149 | Hospital | TaqMan | 1349 T > G, −656 T > G |
| Andrew 2008 33 | USA/Italy | European | Bladder cancer | 911/1165 | Population | SNP mass-tagging system | 1349 T > G |
| Pardini 2008 34 | Czech | European | Colorectal cancer | 531/530 | Hospital | TaqMan | 1349 T > G |
| Mitra 2008 35 | India | European | Bladder cancer | 150/225 | Population | PCR-RFLP | 1349 T > G |
| Tse 2008 36 | USA | European | Colorectal cancer | 311/454 | Hospital | TaqMan | 1349 T > G |
| Huang 2008 37 | China | Asian | Biliary tract cancer | 409/783 | Population | TaqMan | 1349 T > G |
| Chiang 2008 38 | China | Asian | Thyroid cancer | 283/469 | Hospital | TaqMan | 1349 T > G |
| Smith 2008 39 | USA | Asian | Breast cancer | 372/480 | Hospital | Arrayed primer extension | 1349 T > G |
| Gangwar 2009 40 | India | European | Bladder cancer | 206/250 | Hospital | TaqMan | 1349 T > G |
| Agachan 2009 41 | Turkey | European | Lung cancer | 98/67 | Hospital | PCR-RFLP | 1349 T > G |
| Lo 2009 11 | China | Asian | Lung cancer | 729/722 | Hospital | Arrayed primer extension | 1349 T > G, −656 T > G |
| Lu 2009 10 | China | Asian | Lung cancer | 500/517 | Population | Illumina | 1349 T > G, −656 T > G |
| Lung cancer | 572/547 | Hospital | SNPscan | −656 T > G | |||
| Wang 2010 12 | China | Asian | Bladder cancer | 234/253 | Hospital | PCR-RFLP | 1349 T > G, -656 T > G |
| Deng 2010 42 | China | Asian | Lung cancer | 315/314 | Population | RT-PCR | 1349 T > G |
| Palli 2010 43 | Italy | European | Gastric cancer | 298/546 | Population | TaqMan | 1349 T > G |
| Jelonek 2010 44 | Poland | European | Colorectal cancer | 113/153 | Hospital | PCR-RFLP | 1349 T > G |
| Breast cancer | 91/412 | Hospital | PCR-RFLP | 1349 T > G | |||
| Zhou 2011 13 | China | Asian | Brain cancer | 750/816 | Hospital | MassARRAY | −656 T > G |
| Cao 2011 45 | China | Asian | Renal cell carcinoma | 612/632 | Hospital | TaqMan | 1349 T > G, -656 T > G |
RFLP, restriction fragment length polymorphism; PCR, polymerase chain reaction; MALDI-TOF, matrix assisted laser desorption/ionization time-of-flight; SNP, single nucleotide polymorphism; C, cytosine; T, thymine; PCR-CTPP, polymerase chain reaction with confronting two-pair primers.
The −656 T > G polymorphism
The frequency of the G allele varied widely across the 8 studies, ranging from 0.44 to 0.64. The average frequency of the G allele in Asian populations was 0.45, which was lower than that in European populations (0.62). The difference was statistical significant (P < 0.001).
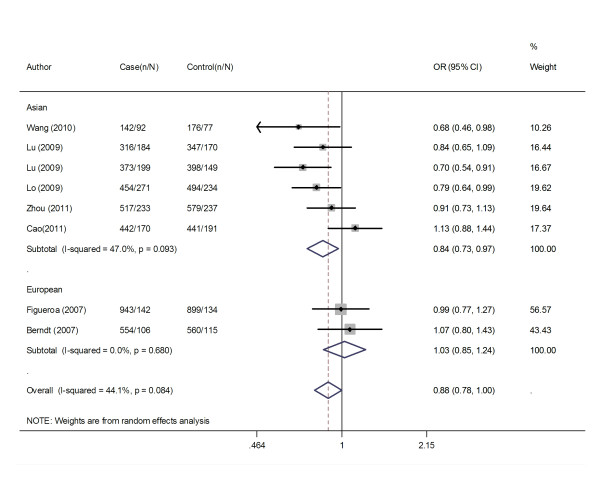
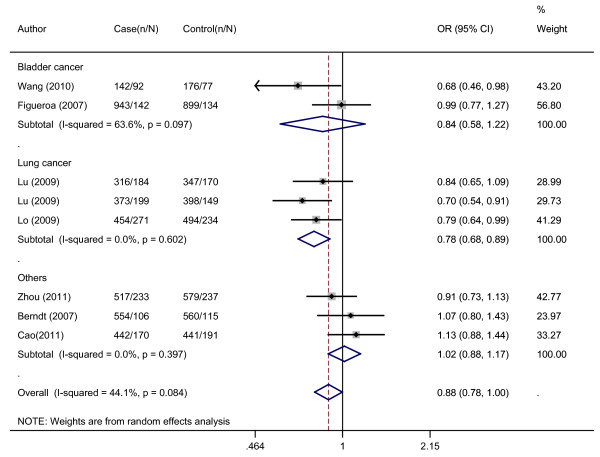
Overall, there was evidence of an association between decreased cancer risk and the variant genotypes in different genetic models when all the eligible studies were pooled into the meta-analysis. As show in Table 2, compared with the wild-type homozygote genotype, the homozygote variant genotype GG was associated with a statistically significant decreased risk of all types of cancer (OR = 0.81, 95%CI = 0.67-0.97). Besides, significant main effects were also observed both in dominant and recessive models (OR = 0.89, 95%CI = 0.81-0.97 and OR = 0.90, 95%CI = 0.82-0.98). In the stratified analysis by populations, as shown in Table 2 and Figure 1, the effect was remain in studies of Asian populations (homozygote comparison: OR = 0.75, 95%CI = 0.65-0.86; heterozygote comparison: OR = 0.89, 95%CI = 0.80-0.99; dominant model: OR = 0.85, 95%CI = 0.77-0.94 and recessive model: OR = 0.81, 95%CI = 0.71-0.91). In the stratified analysis by cancer type, as shown in Table 2 and Figure 2, the effect was remain in lung cancer studies (homozygote comparison: OR = 0.65, 95%CI = 0.54-0.79; heterozygote comparison: OR = 0.83, 95%CI = 0.72-0.97; dominant model: OR = 0.78, 95%CI = 0.68-0.89 and recessive model: OR = 0.72, 95%CI = 0.61-0.86). Further subgroup analyses were not performed because of limited data for this polymorphism.
Table 2.
Meta-analysis of the APE1 −656 T > G polymorphism and cancer risk
| Variables | n * | GG vs. TT | TG vs. TT | GG/TG vs. TT | GG vs. TT/TG | ||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) † | P ‡ | OR (95% CI) † | P ‡ | OR (95% CI) † | P ‡ | OR (95% CI) † | P ‡ | ||
| Total | 8 | 0.81 (0.67-0.97) | 0.015 | 0.92 (0.83-1.01) | 0.340 | 0.89 (0.81-0.97) | 0.084 | 0.90 (0.82-0.98) | 0.059 |
| Ethnicities | |||||||||
| Asian | 6 | 0.75 (0.65-0.86) | 0.057 | 0.89 (0.80-0.99) | 0.255 | 0.85 (0.77-0.94) | 0.093 | 0.81(0.71-0.91) | 0.252 |
| European | 2 | 1.04 (0.85-1.28) | 0.703 | 1.01 (0.83-1.24) | 0.692 | 1.03 (0.85-1.24) | 0.680 | 1.03 (0.90-1.18) | 0.891 |
| Cancer types | |||||||||
| Lung cancer | 3 | 0.65 (0.54-0.79) | 0.857 | 0.83 (0.72-0.97) | 0.469 | 0.78 (0.68-0.89) | 0.602 | 0.72 (0.61-0.86) | 0.799 |
| Bladder cancer | 2 | 0.88 (0.70-1.12) | 0.045 | 0.89 (0.72-1.11) | 0.583 | 0.88 (0.71-1.08) | 0.097 | 0.97 (0.82-1.14) | 0.086 |
| Others | 3 | 1.00 (0.83-1.19) | 0.276 | 1.03 (0.88-1.19) | 0.340 | 1.02 (0.88-1.15) | 0.397 | 0.98 (0.85-1.13) | 0.437 |
* Number of comparisons
† Random-effects model was used when P value for heterogeneity test < 0.05; otherwise, fix-effects model was used
‡ P value of Q-test for heterogeneity test
Figure 1.
Meta-analysis with a random-effects model for the association between cancer risk and the APE1 −656 T > G polymorphism stratified by ethnicity (TG/GG vs. TT). OR, odds ratio; CI, confidence interval; I-squared, measure to quantify the degree of heterogeneity in meta-analyses.
Figure 2.
Meta-analysis with a random-effects model for the association between cancer risk and the APE1 -656 T > G polymorphism stratified by cancer types (TG/GG vs. TT). OR, odds ratio; CI, confidence interval; I-squared, measure to quantify the degree of heterogeneity in meta-analyses.
Heterogeneity was observed in homozygote comparison (P heterogeneity = 0.015) for the main effects. Therefore, we conducted sensitivity analyses to locate the source of heterogeneity, and the results suggested that two studies [16,47] were the main origin of the heterogeneity. The heterogeneity was effectively removed after exclusion of these studies (P heterogeneity = 0.123, 0.355 and 0.163, respectively for homozygote comparison, dominant and recessive model). The results were not materially altered after exclusion of these studies, suggesting the results of this meta-analysis were stable.
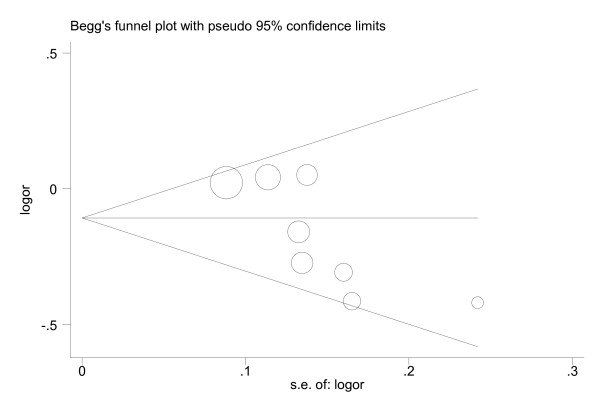
As shown in Figure 3, the shape of the funnel plots seemed asymmetrical in the recessive model, suggesting the presence of publication bias. The Egger's test showed obvious evidence of publication bias (t = −3.01, P = 0.024, recessive model). To adjust for this bias, a trim-and-fill method developed by Duval and Tweedie [48] was conducted, the adjusted estimates obtained by using random-effects model was OR of 0.87 (0.76-0.99). Meta-analysis with or without the trim-and-fill method did not draw different conclusions, indicating that our results were statistically robust.
Figure 3.
Begg's funnel plot of publication bias test (−656 T > G; GG vs. TT/TG). Each point represents a separate study for the indicated association. Log (OR), natural logarithm of OR; Horizontal line, mean effect size.
The 1349 T > G polymorphism
The frequency of the G allele varied widely across the 39 studies, ranging from 0.20 to 0.52. However, no significant difference in the frequency distribution of the G allele between Asian and European populations were observed (frequencies of G allele were 0.42 and 0.44 in Asian and European populations, respectively; P = 0.514).
As shown in Table 3, the results did not suggest any statistical evidence of an association between the 1349 T > G polymorphism and overall cancer risk (Table 3). In the subgroup analysis by ethnicities, source of controls and cancer types, still no significant association was observed (Table 3). No publication bias was detected by either the funnel plot or the Egger test (GG vs. TT: t = 0.33, P = 0.745; TG vs.TT: t = 1.22, P = 0.233; TG/GG vs. TT: t = 1.29, P = 0.244; GG vs. TT/TG: t = 0.02, P = 0.985).
Table 3.
Meta-analysis of the APE1 1349 T > G polymorphism and cancer risk
| Variables | n * | GG vs. TT | TG vs. TT | GG/TG vs. TT | GG vs. TT/TG | ||||
|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) † | P ‡ | OR (95% CI) † | P ‡ | OR (95% CI) † | P‡ | OR (95% CI) † | P‡ | ||
| Total | 35 | 1.05 (0.95-1.16) | 0.001 | 1.04 (0.96-1.12) | 0.004 | 1.04 (0.97-1.13) | 0.000 | 1.03 (0.96-1.11) | 0.022 |
| Ethnicities | |||||||||
| Asian | 10 | 1.05 (0.93-1.20) | 0.051 | 0.97 (0.88-1.07) | 0.657 | 0.99 (0.90-1.08) | 0.427 | 1.08 (0.92-1.28) | 0.035 |
| European | 23 | 1.05 (0.92-1.19) | 0.001 | 1.09 (0.99-1.20) | 0.003 | 1.09 (0.98-1.21) | 0.000 | 1.00 (0.94-1.07) | 0.062 |
| Source of controls | |||||||||
| Population-based | 15 | 1.01 (0.92-1.11) | 0.130 | 1.07 (0.95-1.21) | 0.010 | 1.06 (094-1.20) | 0.004 | 0.98 (0.90-1.07) | 0.335 |
| Hospital-based | 20 | 1.07 (0.92-1.24) | 0.000 | 1.02 (0.93-1.11) | 0.048 | 1.03 (0.93-1.15) | 0.002 | 1.06 (0.95-1.20) | 0.014 |
| Cancer types | |||||||||
| Lung cancer | 9 | 1.08 (0.85-1.38) | 0.017 | 0.99 (0.84-1.17) | 0.059 | 1.03 (0.85-1.25) | 0.007 | 1.04 (0.92-1.19) | 0.086 |
| Bladder cancer | 7 | 0.96 (0.83-1.09) | 0.312 | 1.01 (0.91-1.12) | 0.376 | 0.99 (0.90-1.10) | 0.453 | 0.95 (0.85-1.07) | 0.360 |
| Colorectal cancer | 4 | 0.93 (0.55-1.57) | 0.001 | 1.05 (0.78-1.41) | 0.025 | 1.01 (0.72-1.43) | 0.003 | 0.93 (0.63-1.39) | 0.003 |
| Breast cancer | 4 | 1.20 (0.82-1.76) | 0.042 | 1.19 (0.76-1.86) | 0.000 | 1.23 (0.79-1.92) | 0.000 | 1.06 (0.92-1.23) | 0.345 |
| Head and neck cancer | 2 | 1.03 (0.80-1.32) | 0.306 | 1.00 (0.81-1.23) | 0.242 | 1.01 (0.83-1.23) | 0.202 | 1.03 (0.83-1.27) | 0.621 |
| Others | 9 | 1.07 (0.94-1.23) | 0.156 | 1.05 (0.94-1.16) | 0.754 | 1.05 (0.96-1.16) | 0.609 | 1.05 (0.93-1.18) | 0.111 |
* Number of comparisons
† Random-effects model was used when P value for heterogeneity test <0.05; otherwise, fix-effects model was used
‡ P value of Q-test for heterogeneity test
Discussion
To data, numerous studies have been carried out to investigate whether polymorphisms in APE1 are associated with the risk of cancer; however, the data have yielded conflicting results. In the present study, to derive a more precise estimation of the relationship, we performed a meta-analysis of 37 published studies including 5,139 cases and 5,201 controls for APE1 −656 T > G polymorphism, and 14,222 cases and 19,746 controls for APE1 1349 T > G polymorphism. Our meta-analysis indicated that variant genotypes (GG and TG/GG) of −656 T > G polymorphism was associated with a significant decrease in the overall risk of cancer, especially for lung cancer and Asians. However, the 1349 T > G polymorphism did not appear to have significant influence on the overall risk of cancer.
Apurinic/apyrimidinic (AP) sites are common mutagenic and cytotoxic DNA lesions which caused by the loss of normal bases [49]. APE1 had been extensively studied as the major AP endonuclease involved in the repair of AP sites through both its endonuclease and phosphodiesterase activities [7]. Alteration in the expression of APE1 may influence its capacity to repair DNA damage. Several lines of evidence support that the −656 T > G polymorphism plays a role in influencing the promoter activity of APE1. Using in vitro promoter assay, Lo et al. found that the −656 G allele had a significantly higher transcriptional activity than that of the −656 T allele and individuals with the −656 G allele were at a decreased risk for lung cancer [13]. In their view, the "higher production" genotype for APE1 might offer protection against the development of lung cancer [13]. Nearly at the same time, Lu et al. also reported a similar protective effect of the −656 G allele against lung cancer risk in two independent studies [12]. However, they showed that the G allele attenuated the binding of Oct-1 to the promoter region of APE1 and resulted in decreased transcriptional activity. Given the disparity between these functional studies, the mechanism underlying the regulation of APE1 by this polymorphism may be still required further investigation. Nevertheless, our meta-analysis showed that individuals with the variant genotypes (GG or GG/TG) of −656 T > G polymorphism were associated with a decreased cancer risk than those with the TT genotype. Although publication bias was observed in the recessive genetic model, our meta-analysis with or without trim-and-fill method did not draw different conclusions, suggesting that our results were statistically robust.
In the subgroup analysis by cancer type, we found that individuals with the variant genotypes of −656 T > G polymorphism were associated with decreased lung cancer risk. However, the result should be interpreted with caution. Because, only four types of cancer (lung cancer, bladder cancer, brain tumor and colorectal adenoma) were included and the studies of each type were very limited, which may have insufficient power to reveal a reliable association. In the subgroup analysis by ethnicity, the results suggested that the association was more apparent among Asian populations. Several reasons may lead to the ethnic difference. First, differences in genetic background may cause the difference. Then, environmental or life style context which strongly vary between populations may play a role. Besides, other factors such as selection bias and different matching criteria may also result in the difference. In considering of the limited numbers of studies, in the future, large numbers of studies will be required to validate these associations.
Although it has been suggested the 1349 T > G polymorphism could influence the sensitivity to ionizing radiation [50], it does not result in reduced endonuclease activity [18]. A previous meta-analysis which investigated the association of 1349 T > G polymorphism and cancer risk suggested that the variant genotypes were associated with a moderately increased risk of all cancer types (OR = 1.09, 95% CI = 1.01-1.18 for TG versus TT; OR = 1.08, 95% CI = 1.00-1.18 for GG/TG versus TT) [51]. Compared with the previous study, we excluded four studies [46,52-54] in which the 1349 T > G genotype distributions in controls did not conform to HWE, since deviations from HWE in control subjects may bias the estimates of genetic effects in genetic association studies and meta-analysis. Besides, four additional studies with 2, 307 cases and 3, 184 control subjects in total were included in the present meta-analysis which could provide more solid evidence on the specific lack of association between APE1 1349 T > G polymorphism and cancer risk.. Given the power of this meta-analysis which included 14,740 cases and 20,533 controls, a false-negative finding is unlikely. However, further studies may be still needed to investigate the interactions between APE1 polymorphism and environmental factors which play important role in carcinogenesis but were not assessed in the present meta-analysis due to lack of original data. Besides, more attention drew to the prospective −656 T > G polymorphism in the further studies may be helpful to reveal the role of APE1 in the etiology of cancer.
Some limitations of our meta-analysis should be addressed. Firstly, the numbers of published studies collected in our analysis were not large enough for the comprehensive analysis, especially for the APE1 −656 T > G polymorphism. Secondly, lacking the original data of the included studies limited our study to further evaluate the potential interactions, since gene-environment and gene-gene interactions and even different polymorphic loci of the same gene may also modulate cancer risk. Besides, our results were based on unadjusted estimates, while a more precise analysis needs to be conducted if individual data such as age and sex are available. Nevertheless, advantages in our meta-analysis should also be acknowledged. First, a systematic review of the association of APE1 polymorphisms with cancer risk is statistically more powerful than any single study. Second, the studies included in our meta-analysis strictly and satisfactory met our selection criteria.
Conclusions
In conclusion, the results from this meta-analysis suggest that the APE1 −656 T > G polymorphism, but not the APE1 1349 T > G polymorphism may contribute to genetic susceptibility to cancer. Further prospective studies with larger numbers of unbiased-matched homogeneous patients and well-matched controls are required to validate our results and to clarify the gene-gene and gene-environment interactions between APE1 polymorphisms and cancer risk.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
BZ carries out the meta-analysis study and drafted the manuscript. HS participates in the design of the study and performs the statistical analysis. YS and KX collect and extract the data. XS and WM have been involved in revising the manuscript critically for important intellectual content. QS conceives of the study, and participates in its design and coordination and helps to draft the manuscript. All authors read and approve the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Bin Zhou, Email: zhoubin_jy@126.com.
Hailin Shan, Email: jy_hailinshan@126.com.
Ying Su, Email: jy_suying@126.com.
Kai Xia, Email: jy_kaixia@126.com.
Xiaxia Shao, Email: jy_xiaxiashao@126.com.
Weidong Mao, Email: jy_weidongmao@126.com.
Qing Shao, Email: jy_zhoubin@126.com.
References
- Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004;4(11):850–860. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- Iscovich J, Howe GR. Cancer incidence patterns (1972-91) among migrants from the Soviet Union to Israel. Cancer Causes Control. 1998;9(1):29–36. doi: 10.1023/A:1008893102428. [DOI] [PubMed] [Google Scholar]
- Hecht SS. Cigarette smoking and lung cancer: chemical mechanisms and approaches to prevention. Lancet Oncol. 2002;3(8):461–469. doi: 10.1016/S1470-2045(02)00815-X. [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- Barzilay G, Hickson ID. Structure and function of apurinic/apyrimidinic endonucleases. Bioessays. 1995;17(8):713–719. doi: 10.1002/bies.950170808. [DOI] [PubMed] [Google Scholar]
- Tell G, Quadrifoglio F, Tiribelli C, Kelley MR. The many functions of APE1/Ref-1: not only a DNA repair enzyme. Antioxid Redox Signal. 2009;11(3):601–620. doi: 10.1089/ars.2008.2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Hazra TK, Boldogh I, Tomkinson AE, Park MS, Ikeda S, Mitra S. Requirement for human AP endonuclease 1 for repair of 3'-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis. 2000;21(7):1329–1334. doi: 10.1093/carcin/21.7.1329. [DOI] [PubMed] [Google Scholar]
- Fortini P, Pascucci B, Parlanti E, D'Errico M, Simonelli V, Dogliotti E. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003;85(11):1053–1071. doi: 10.1016/j.biochi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Bhakat KK, Mantha AK, Mitra S. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid Redox Signal. 2009;11(3):621–638. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tell G, Fantini D, Quadrifoglio F. Understanding different functions of mammalian AP endonuclease (APE1) as a promising tool for cancer treatment. Cell Mol Life Sci. 2010;67(21):3589–3608. doi: 10.1007/s00018-010-0486-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi T, Jones IM, Mohrenweiser HW. Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics. 2004;83(6):970–979. doi: 10.1016/j.ygeno.2003.12.016. [DOI] [PubMed] [Google Scholar]
- Lu J, Zhang S, Chen D, Wang H, Wu W, Wang X, Lei Y, Wang J, Qian J, Fan W. et al. Functional characterization of a promoter polymorphism in APE1/Ref-1 that contributes to reduced lung cancer susceptibility. FASEB J. 2009;23(10):3459–3469. doi: 10.1096/fj.09-136549. [DOI] [PubMed] [Google Scholar]
- Lo YL, Jou YS, Hsiao CF, Chang GC, Tsai YH, Su WC, Chen KY, Chen YM, Huang MS, Hu CY. et al. A polymorphism in the APE1 gene promoter is associated with lung cancer risk. Cancer Epidemiol Biomarkers Prev. 2009;18(1):223–229. doi: 10.1158/1055-9965.EPI-08-0749. [DOI] [PubMed] [Google Scholar]
- Wang M, Qin C, Zhu J, Yuan L, Fu G, Zhang Z, Yin C. Genetic variants of XRCC1, APE1, and ADPRT genes and risk of bladder cancer. DNA Cell Biol. 2010;29(6):303–311. doi: 10.1089/dna.2009.0969. [DOI] [PubMed] [Google Scholar]
- Zhou K, Hu D, Lu J, Fan W, Liu H, Chen H, Chen G, Wei Q, Du G, Mao Y. et al. A genetic variant in the APE1/Ref-1 gene promoter -141 T/G may modulate risk of glioblastoma in a Chinese Han population. BMC Cancer. 2011;11:104. doi: 10.1186/1471-2407-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt SI, Huang WY, Fallin MD, Helzlsouer KJ, Platz EA, Weissfeld JL, Church TR, Welch R, Chanock SJ, Hayes RB. Genetic variation in base excision repair genes and the prevalence of advanced colorectal adenoma. Cancer Res. 2007;67(3):1395–1404. doi: 10.1158/0008-5472.CAN-06-1390. [DOI] [PubMed] [Google Scholar]
- Figueroa JD, Malats N, Real FX, Silverman D, Kogevinas M, Chanock S, Welch R, Dosemeci M, Tardon A, Serra C. et al. Genetic variation in the base excision repair pathway and bladder cancer risk. Hum Genet. 2007;121(2):233–242. doi: 10.1007/s00439-006-0294-y. [DOI] [PubMed] [Google Scholar]
- Hadi MZ, Coleman MA, Fidelis K, Mohrenweiser HW, Wilson DM. Functional characterization of Ape1 variants identified in the human population. Nucleic Acids Res. 2000;28(20):3871–3879. doi: 10.1093/nar/28.20.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WW, Salama SA, Sierra-Torres CH. Functional characterization of polymorphisms in DNA repair genes using cytogenetic challenge assays. Environ Health Perspect. 2003;111(15):1843–1850. doi: 10.1289/ehp.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Mitra AK, Singh N, Singh A, Garg VK, Agarwal A, Sharma M, Chaturvedi R, Rath SK. Association of polymorphisms in base excision repair genes with the risk of breast cancer: a case-control study in North Indian women. Oncol Res. 2008;17(3):127–135. doi: 10.3727/096504008785055567. [DOI] [PubMed] [Google Scholar]
- Misra RR, Ratnasinghe D, Tangrea JA, Virtamo J, Andersen MR, Barrett M, Taylor PR, Albanes D. Polymorphisms in the DNA repair genes XPD, XRCC1, XRCC3, and APE/ref-1, and the risk of lung cancer among male smokers in Finland. Cancer Lett. 2003;191(2):171–178. doi: 10.1016/S0304-3835(02)00638-9. [DOI] [PubMed] [Google Scholar]
- Popanda O, Schattenberg T, Phong CT, Butkiewicz D, Risch A, Edler L, Kayser K, Dienemann H, Schulz V, Drings P. et al. Specific combinations of DNA repair gene variants and increased risk for non-small cell lung cancer. Carcinogenesis. 2004;25(12):2433–2441. doi: 10.1093/carcin/bgh264. [DOI] [PubMed] [Google Scholar]
- Hao B, Wang H, Zhou K, Li Y, Chen X, Zhou G, Zhu Y, Miao X, Tan W, Wei Q. et al. Identification of genetic variants in base excision repair pathway and their associations with risk of esophageal squamous cell carcinoma. Cancer Res. 2004;64(12):4378–4384. doi: 10.1158/0008-5472.CAN-04-0372. [DOI] [PubMed] [Google Scholar]
- Ito H, Matsuo K, Hamajima N, Mitsudomi T, Sugiura T, Saito T, Yasue T, Lee KM, Kang D, Yoo KY. et al. Gene-environment interactions between the smoking habit and polymorphisms in the DNA repair genes, APE1 Asp148Glu and XRCC1 Arg399Gln, in Japanese lung cancer risk. Carcinogenesis. 2004;25(8):1395–1401. doi: 10.1093/carcin/bgh153. [DOI] [PubMed] [Google Scholar]
- Shen M, Berndt SI, Rothman N, Mumford JL, He X, Yeager M, Welch R, Chanock S, Keohavong P, Donahue M. et al. Polymorphisms in the DNA base excision repair genes APEX1 and XRCC1 and lung cancer risk in Xuan Wei, China. Anticancer Res. 2005;25(1B):537–542. [PubMed] [Google Scholar]
- Moreno V, Gemignani F, Landi S, Gioia-Patricola L, Chabrier A, Blanco I, Gonzalez S, Guino E, Capella G, Canzian F. Polymorphisms in genes of nucleotide and base excision repair: risk and prognosis of colorectal cancer. Clin Cancer Res. 2006;12(7 Pt 1):2101–2108. doi: 10.1158/1078-0432.CCR-05-1363. [DOI] [PubMed] [Google Scholar]
- Jiao L, Bondy ML, Hassan MM, Wolff RA, Evans DB, Abbruzzese JL, Li D. Selected polymorphisms of DNA repair genes and risk of pancreatic cancer. Cancer Detect Prev. 2006;30(3):284–291. doi: 10.1016/j.cdp.2006.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry PD, Umbach DM, Taylor JA. APE1 genotype and risk of bladder cancer: evidence for effect modification by smoking. Int J Cancer. 2006;118(12):3170–3173. doi: 10.1002/ijc.21768. [DOI] [PubMed] [Google Scholar]
- Wu X, Gu J, Grossman HB, Amos CI, Etzel C, Huang M, Zhang Q, Millikan RE, Lerner S, Dinney CP. et al. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am J Hum Genet. 2006;78(3):464–479. doi: 10.1086/500848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Newcomb PA, Egan KM, Titus-Ernstoff L, Chanock S, Welch R, Brinton LA, Lissowska J, Bardin-Mikolajczak A, Peplonska B. et al. Genetic polymorphisms in base-excision repair pathway genes and risk of breast cancer. Cancer Epidemiol Biomarkers Prev. 2006;15(2):353–358. doi: 10.1158/1055-9965.EPI-05-0653. [DOI] [PubMed] [Google Scholar]
- Matullo G, Dunning AM, Guarrera S, Baynes C, Polidoro S, Garte S, Autrup H, Malaveille C, Peluso M, Airoldi L. et al. DNA repair polymorphisms and cancer risk in non-smokers in a cohort study. Carcinogenesis. 2006;27(5):997–1007. doi: 10.1093/carcin/bgi280. [DOI] [PubMed] [Google Scholar]
- Chen L, Ambrosone CB, Lee J, Sellers TA, Pow-Sang J, Park JY. Association between polymorphisms in the DNA repair genes XRCC1 and APE1, and the risk of prostate cancer in white and black Americans. J Urol. 2006;175(1):108–112. doi: 10.1016/S0022-5347(05)00042-X. discussion 112. [DOI] [PubMed] [Google Scholar]
- Li C, Hu Z, Lu J, Liu Z, Wang LE, El-Naggar AK, Sturgis EM, Spitz MR, Wei Q. Genetic polymorphisms in DNA base-excision repair genes ADPRT, XRCC1, and APE1 and the risk of squamous cell carcinoma of the head and neck. Cancer. 2007;110(4):867–875. doi: 10.1002/cncr.22861. [DOI] [PubMed] [Google Scholar]
- Andrew AS, Karagas MR, Nelson HH, Guarrera S, Polidoro S, Gamberini S, Sacerdote C, Moore JH, Kelsey KT, Demidenko E. et al. DNA repair polymorphisms modify bladder cancer risk: a multi-factor analytic strategy. Hum Hered. 2008;65(2):105–118. doi: 10.1159/000108942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardini B, Naccarati A, Novotny J, Smerhovsky Z, Vodickova L, Polakova V, Hanova M, Slyskova J, Tulupova E, Kumar R. et al. DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech Republic. Mutat Res. 2008;638(1-2):146–153. doi: 10.1016/j.mrfmmm.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Tse D, Zhai R, Zhou W, Heist RS, Asomaning K, Su L, Lynch TJ, Wain JC, Christiani DC, Liu G. Polymorphisms of the NER pathway genes, ERCC1 and XPD are associated with esophageal adenocarcinoma risk. Cancer Causes Control. 2008;19(10):1077–1083. doi: 10.1007/s10552-008-9171-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WY, Gao YT, Rashid A, Sakoda LC, Deng J, Shen MC, Wang BS, Han TQ, Zhang BH, Chen BE. et al. Selected base excision repair gene polymorphisms and susceptibility to biliary tract cancer and biliary stones: a population-based case-control study in China. Carcinogenesis. 2008;29(1):100–105. doi: 10.1093/carcin/bgm247. [DOI] [PubMed] [Google Scholar]
- Chiang FY, Wu CW, Hsiao PJ, Kuo WR, Lee KW, Lin JC, Liao YC, Juo SH. Association between polymorphisms in DNA base excision repair genes XRCC1, APE1, and ADPRT and differentiated thyroid carcinoma. Clin Cancer Res. 2008;14(18):5919–5924. doi: 10.1158/1078-0432.CCR-08-0906. [DOI] [PubMed] [Google Scholar]
- Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, Hoang KN, Liu-Mares W, Hu JJ. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis. 2008;29(11):2132–2138. doi: 10.1093/carcin/bgn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangwar R, Ahirwar D, Mandhani A, Mittal RD. Influence of XPD and APE1 DNA repair gene polymorphism on bladder cancer susceptibility in north India. Urology. 2009;73(3):675–680. doi: 10.1016/j.urology.2008.09.043. [DOI] [PubMed] [Google Scholar]
- Agachan B, Kucukhuseyin O, Aksoy P, Turna A, Yaylim I, Gormus U, Ergen A, Zeybek U, Dalan B, Isbir T. Apurinic/apyrimidinic endonuclease (APE1) gene polymorphisms and lung cancer risk in relation to tobacco smoking. Anticancer Res. 2009;29(6):2417–2420. [PubMed] [Google Scholar]
- Deng Q, Sheng L, Su D, Zhang L, Liu P, Lu K, Ma S. Genetic polymorphisms in ATM, ERCC1, APE1 and iASPP genes and lung cancer risk in a population of southeast China. Med Oncol. 2011;28(3):667–672. doi: 10.1007/s12032-010-9507-2. [DOI] [PubMed] [Google Scholar]
- Palli D, Polidoro S, D'Errico M, Saieva C, Guarrera S, Calcagnile AS, Sera F, Allione A, Gemma S, Zanna I. et al. Polymorphic DNA repair and metabolic genes: a multigenic study on gastric cancer. Mutagenesis. 2010;25(6):569–575. doi: 10.1093/mutage/geq042. [DOI] [PubMed] [Google Scholar]
- Jelonek K, Gdowicz-Klosok A, Pietrowska M, Borkowska M, Korfanty J, Rzeszowska-Wolny J, Widlak P. Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a Polish population. J Appl Genet. 2010;51(3):343–352. doi: 10.1007/BF03208865. [DOI] [PubMed] [Google Scholar]
- Cao Q, Qin C, Meng X, Ju X, Ding Q, Wang M, Zhu J, Wang W, Li P, Chen J. et al. Genetic polymorphisms in APE1 are associated with renal cell carcinoma risk in a chinese population. Mol Carcinog. 2011;50(11):863–870. doi: 10.1002/mc.20791. [DOI] [PubMed] [Google Scholar]
- Duval S, Tweedie R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics. 2000;56(2):455–463. doi: 10.1111/j.0006-341X.2000.00455.x. [DOI] [PubMed] [Google Scholar]
- Lindahl T. Recognition and processing of damaged DNA. J Cell Sci Suppl. 1995;19:73–77. doi: 10.1242/jcs.1995.supplement_19.10. [DOI] [PubMed] [Google Scholar]
- Hu JJ, Smith TR, Miller MS, Lohman K, Case LD. Genetic regulation of ionizing radiation sensitivity and breast cancer risk. Environ Mol Mutagen. 2002;39(2-3):208–215. doi: 10.1002/em.10058. [DOI] [PubMed] [Google Scholar]
- Gu D, Wang M, Zhang Z, Chen J. The DNA repair gene APE1 T1349G polymorphism and cancer risk: a meta-analysis of 27 case-control studies. Mutagenesis. 2009;24(6):507–512. doi: 10.1093/mutage/gep036. [DOI] [PubMed] [Google Scholar]
- Zienolddiny S, Campa D, Lind H, Ryberg D, Skaug V, Stangeland L, Phillips DH, Canzian F, Haugen A. Polymorphisms of DNA repair genes and risk of non-small cell lung cancer. Carcinogenesis. 2006;27(3):560–567. doi: 10.1093/carcin/bgi232. [DOI] [PubMed] [Google Scholar]
- De Ruyck K, Szaumkessel M, De Rudder I, Dehoorne A, Vral A, Claes K, Velghe A, Van Meerbeeck J, Thierens H. Polymorphisms in base-excision repair and nucleotide-excision repair genes in relation to lung cancer risk. Mutat Res. 2007;631(2):101–110. doi: 10.1016/j.mrgentox.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Canbay E, Agachan B, Gulluoglu M, Isbir T, Balik E, Yamaner S, Bulut T, Cacina C, Eraltan IY, Yilmaz A. et al. Possible associations of APE1 polymorphism with susceptibility and HOGG1 polymorphism with prognosis in gastric cancer. Anticancer Res. 2010;30(4):1359–1364. [PubMed] [Google Scholar]