Abstract
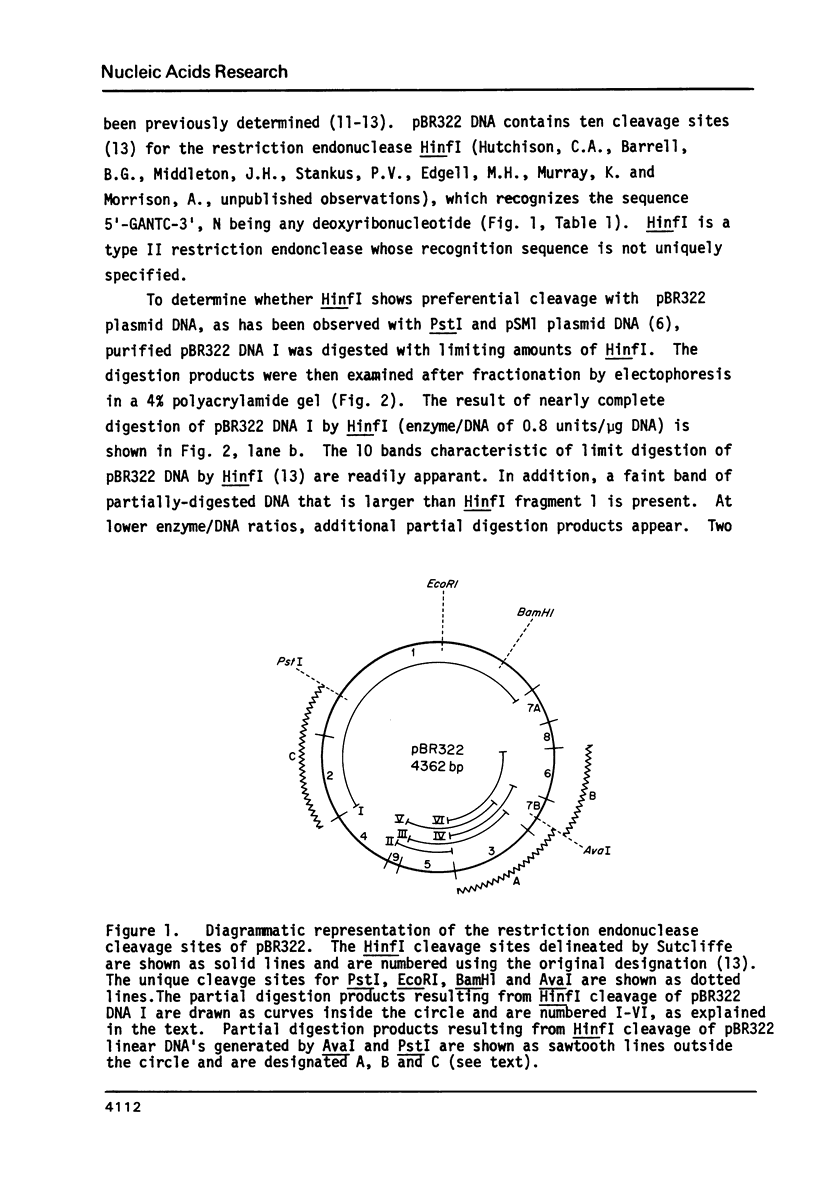
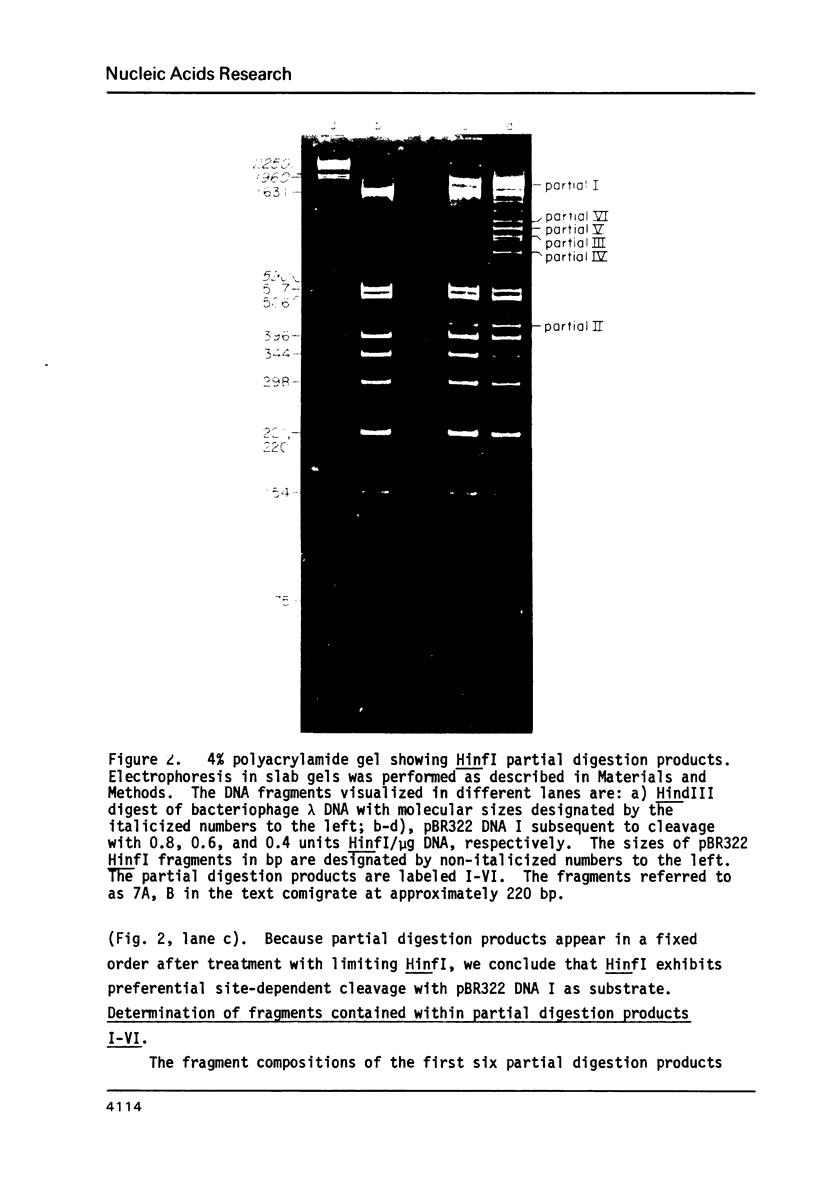
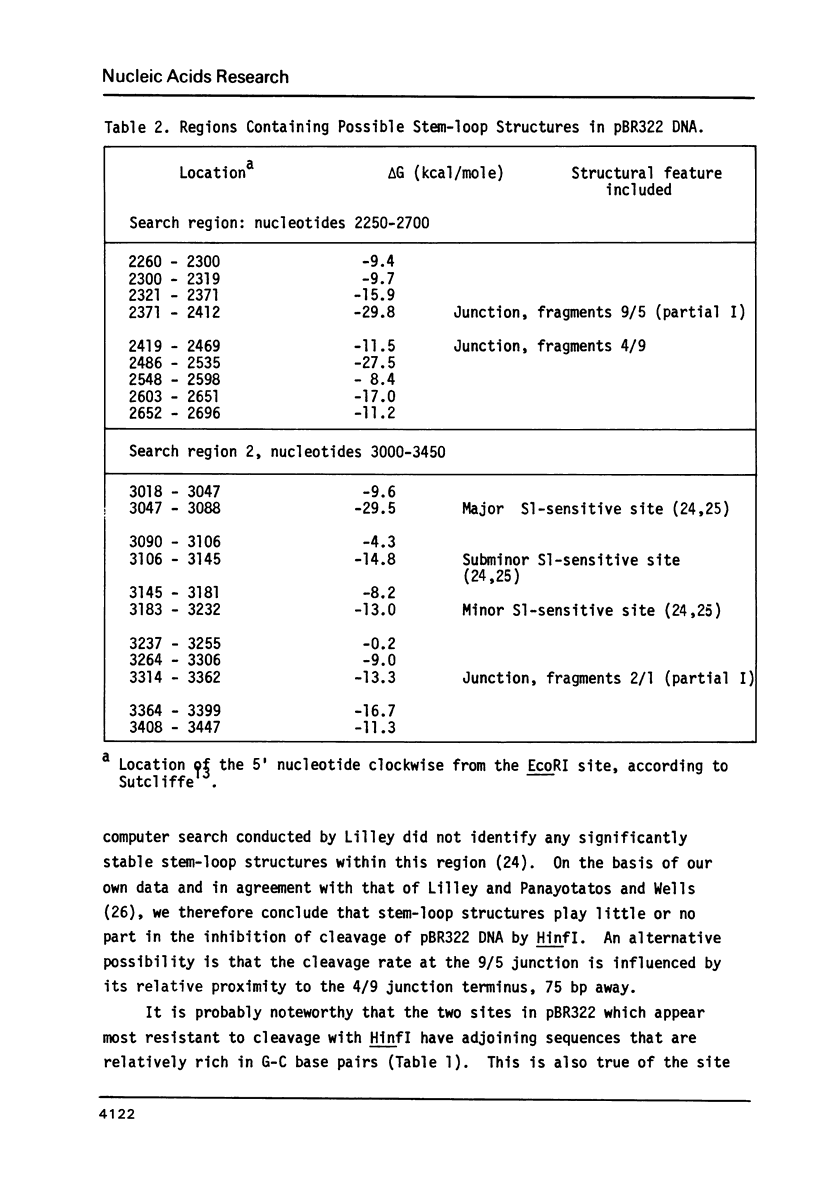
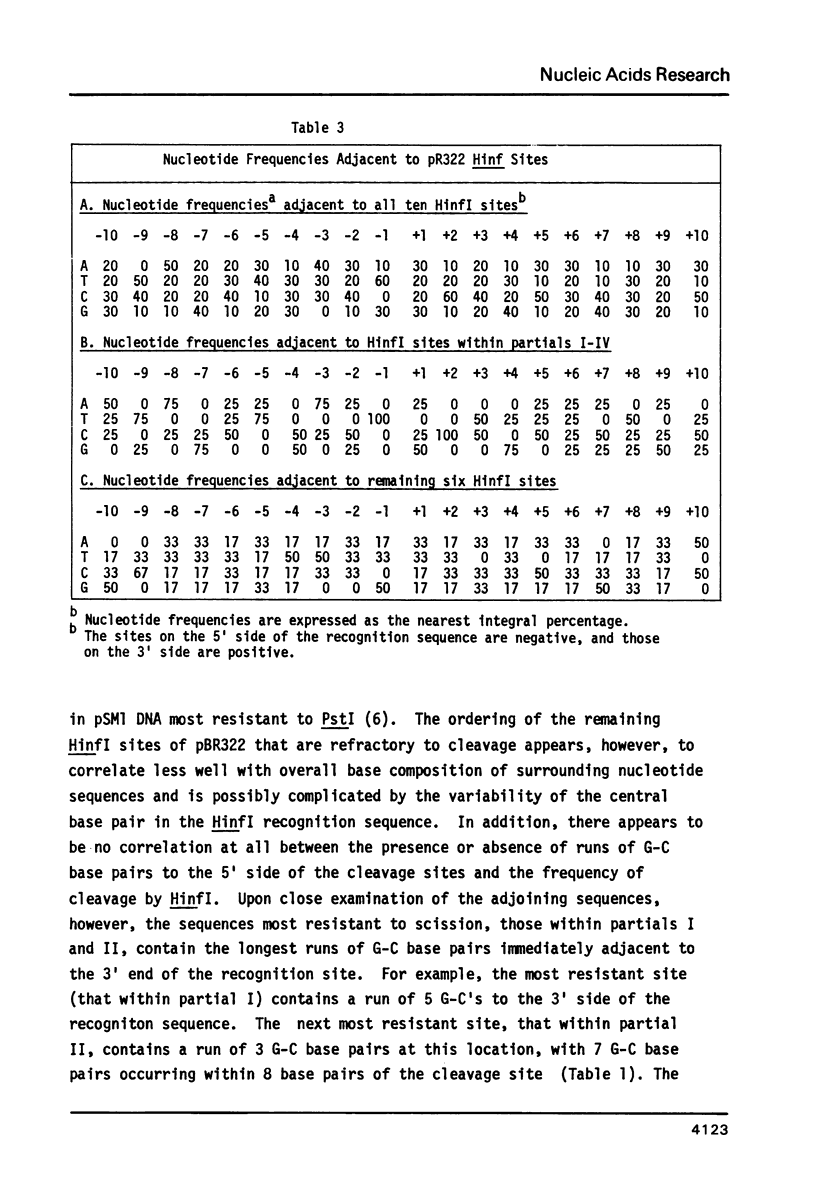
Cleavage of pBR322 DNA I by the restriction endonuclease HinfI is preferentially inhibited at specific HinfI cleavage sites. These sites in pBR322 DNA I have been identified and ordered with respect to the frequency with which they are cleaved. The HinfI site most resistant to cleavage in pBR322 DNA I is unique in that runs of G-C base pairs are immediately adjacent on both sites. Two differently permuted linear (DNA III) species were produced by cleavage with two different restriction endonucleases, PstI and AvaI. Only one of these linear molecules, that produced by PstI, exhibits the same preferential cleavage pattern as DNA I. The second linear species, that arising from AvaI digestion, shows pronounced relative inhibition of cleavage at the HinfI sites nearest the ends of the molecule (100 to 120 base pairs away, respectively). This result suggest that proximity to the termini of a linear DNA molecule might also influence preferential cleavage. The possibility of formation of stem-loop structures does not appear to influence preferential cleavage by HinfI.
Full text
PDF












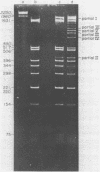
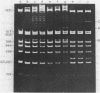
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong K., Bauer W. R. Preferential site-dependent cleavage by restriction endonuclease PstI. Nucleic Acids Res. 1982 Feb 11;10(3):993–1007. doi: 10.1093/nar/10.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Betlach M. C., Heyneker H. L., Shine J., Rodriguez R. L., Boyer H. W. Origin of replication of pBR345 plasmid DNA. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5265–5269. doi: 10.1073/pnas.74.12.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Brown N. L., Smith M. The mapping and sequence determination of the single site in phiX174am3 replicative form DNA cleaved by restriction endonuclease Pst I. FEBS Lett. 1976 Jun 15;65(3):284–287. doi: 10.1016/0014-5793(76)80130-5. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Edwards K. A., Halligan B. D., Davis J. L., Nivera N. L., Liu L. F. Recognition sites of eukaryotic DNA topoisomerase I: DNA nucleotide sequencing analysis of topo I cleavage sites on SV40 DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2565–2576. doi: 10.1093/nar/10.8.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiers W., Contreras R., Haegemann G., Rogiers R., Van de Voorde A., Van Heuverswyn H., Van Herreweghe J., Volckaert G., Ysebaert M. Complete nucleotide sequence of SV40 DNA. Nature. 1978 May 11;273(5658):113–120. doi: 10.1038/273113a0. [DOI] [PubMed] [Google Scholar]
- Forsblom S., Rigler R., Ehrenberg M., Philipson L. Kinetic studies on the cleavage of adenovirus DNA by restriction endonuclease Eco RI. Nucleic Acids Res. 1976 Dec;3(12):3255–3269. doi: 10.1093/nar/3.12.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garfin D. E., Boyer H. W., Goodman H. M. Sequences spanning the EcoRI substrate site. Nucleic Acids Res. 1975 Oct;2(10):1851–1865. doi: 10.1093/nar/2.10.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein L., Thomas M., Davis R. W. EcoRI endonuclease cleavage map of bacteriophage P4-DNA. Virology. 1975 Aug;66(2):420–427. doi: 10.1016/0042-6822(75)90214-7. [DOI] [PubMed] [Google Scholar]
- Jack W. E., Terry B. J., Modrich P. Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4010–4014. doi: 10.1073/pnas.79.13.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. Hairpin-loop formation by inverted repeats in supercoiled DNA is a local and transmissible property. Nucleic Acids Res. 1981 Mar 25;9(6):1271–1289. doi: 10.1093/nar/9.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M. The inverted repeat as a recognizable structural feature in supercoiled DNA molecules. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6468–6472. doi: 10.1073/pnas.77.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonossoff G. P., Butler P. J., Klug A. Sequence-dependent variation in the conformation of DNA. J Mol Biol. 1981 Jul 15;149(4):745–760. doi: 10.1016/0022-2836(81)90356-9. [DOI] [PubMed] [Google Scholar]
- Mickel S., Arena V., Jr, Bauer W. Physical properties and gel electrophoresis behavior of R12-derived plasmid DNAs. Nucleic Acids Res. 1977;4(5):1465–1482. doi: 10.1093/nar/4.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickel S., Bauer W. Isolation, by tetracycline selection, of small plasmids derived from R-factor R12 in Escherichia coli K-12. J Bacteriol. 1976 Jul;127(1):644–655. doi: 10.1128/jb.127.1.644-655.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickel S., Ohtsubo E., Bauer W. Heteroduplex mapping of small plasmids derived from R-factor R12: in vivo recombination occurs at IS1 insertion sequences. Gene. 1977;2(3-4):193–210. doi: 10.1016/0378-1119(77)90017-8. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ryder T., Inokuchi H., Ohtsubo H., Ohtsubo E. Genes and sites involved in replication and incompatibility of an R100 plasmid derivative based on nucleotide sequence analysis. Mol Gen Genet. 1980;179(3):527–537. doi: 10.1007/BF00271742. [DOI] [PubMed] [Google Scholar]
- Rubin R. A., Modrich P. Substrate dependence of the mechanism of EcoRI endonuclease. Nucleic Acids Res. 1978 Aug;5(8):2991–2997. doi: 10.1093/nar/5.8.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D. I., Blattner F. R., Davies J. The isolation and partial characterization of a new restriction endonuclease from Providencia stuartii. Nucleic Acids Res. 1976 Feb;3(2):343–353. doi: 10.1093/nar/3.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]