Abstract
Background
The functional polymorphism (rs1800566) in the NQO1 gene, a 609C>T substitution, leading to proline-to-serine amino-acid and enzyme activity changes, has been implicated in cancer risk, but individually published studies showed inconclusive results.
Methodology/Principal Findings
We performed a meta-analysis of 20 publications with a total of 5,491 cases and 5,917 controls, mainly on gastrointestinal (GI) cancers. We summarized the data on the association between the NQO1 609C>T polymorphism and risk of GI cancers and performed subgroup analyses by ethnicity, cancer site, and study quality. We found that the variant CT heterozygous and CT/TT genotypes of the NQO1 609 C>T polymorphism were associated with a modestly increased risk of GI cancers (CT vs. CC: OR = 1.10, 95% CI = 1.01 – 1.19, P heterogeneity = 0.27, I 2 = 0.15; CT/TT vs. CC: OR = 1.11, 95%CI = 1.02 – 1.20, P heterogeneity = 0.14; I 2 = 0.27). Following further stratified analyses, the increased risk was only observed in subgroups of Caucasians, colorectal cancer in Caucasians, and high quality studies.
Conclusions
This meta-analysis suggests that the NQO1 609T allele is a low-penetrance risk factor for GI cancers. Although the effect on GI cancers may be modified by ethnicity and cancer sites, small sample seizes of the subgroup analyses suggest that further larger studies are needed, especially for non-colorectal GI cancers in Caucasians and GI cancers in Asians.
Introduction
Gastrointestinal (GI) cancers are the common malignant tumors in the world [1], [2], of which colorectal cancer is the third most common cancer in males and the second in females, with over 1.2 millions of new cases and 608,700 deaths occurred in 2008 [2]. It was estimated that cancers of the esophagus, stomach, colorectum, and liver accounted for 26.4% (3.4 millions) of the total new cancer cases and 32.8% (2.5 millions) of the total cancer deaths in 2008 worldwide [2]. Although the causes of these cancers are complex and heterogeneous, chronic inflammation, cigarette smoking, heavy alcohol drinking, and poor dietary pattern are generally considered possible risk factors for these cancers [3], [4], [5], [6], [7], [8], [9], [10], [11], [12], [13]. In addition, numerous case-control, family-based and twin studies have shown that inherited genetic factors have played an important role in susceptibility to these diseases [12], [13], [14], [15], [16], [17]. Recent genome-wide association studies have also identified some susceptible loci harboring common single nucleotide polymorphisms (SNPs) for risk of GI cancers, suggesting that the low-penetrance genes are also involved in the etiology of these diseases [18], [19], [20], [21], [22].
NAD(P)H:quinone oxidoreductase 1 (NQO1) is an obligate two-electron reductase, which reduces reactive quinones to less reactive and less toxic hydroquinones. The quinones are mainly derived from endogenous quinones, such as vitamin E quinone and ubiquinone, and exogenous quinones, such as exhaust gas, cigarette smoke or diet [23], [24]. This two-electron reduction prevents the formation of semiquinones and highly reactive oxygen species (ROS), thus protecting cells against oxidative stress, cytotoxicity, and mutagenicity [25]. In addition to its catalytic role in quinones, NQO1 has been reported to show superoxide scavenging activity and protective activity against procarcinogenic benzenes [26], [27]. Notably, both in vivo and in vitro studies have demonstrated that NQO1 regulates the stability of the tumor suppressors p53 and p73, protecting them from 20S proteasomal degradation, which is important for eliminating damaged cells that are prone to cancer development [28], [29], [30], [31]. Therefore, NQO1 is considered an important defense against cancer [25], [31].
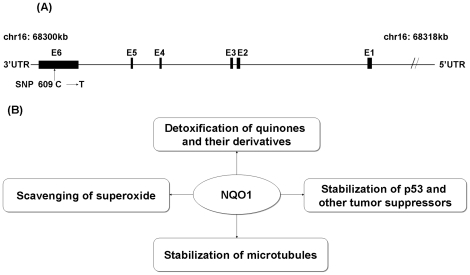
The NQO1 gene is located on chromosome 16q22.1, spanning ∼17.2 kb and consisting of 6 exons and 5 introns [32]. To date, there have been 270 SNPs identified in the NQO1 gene (http://www.ncbi.nlm.nih.gov/SNP). The most extensively studied SNP of NQO1 is a C-to-T transition at nucleotide position 609 in exon 6 (dbSNP ID: rs1800566, 609C>T; Figure 1 ), which results in a proline-to-serine amino-acid substitution at codon 187 (Pro187Ser) in the protein. Genotype-phenotype studies of the NQO1 609C>T polymorphism showed that the variant T allele was associated with reduced NQO1 enzymatic activity in both human cell lines and primary human tissues [24], [33], [34], [35]. Furthermore, there is a clear allele dosage effect of the NQO1 609T genotypes on NQO1 enzymatic activity, with the homozygotes (TT) having the lowest, the heterozygotes (CT) having the intermediate, and the wild-type homozygotes (CC) having the highest NQO1 enzyme activity [33], [36], [37], [38]. Decreased NQO1 enzymatic activity is caused by increased polyubiquination and proteosomal degradation of the mutant NQO1 protein [39]. Altered expression of NQO1 protein has been observed in liver, colon, esophagus, stomach, and pancreas cancers [40], [41], [42], [43], [44], [45]. Furthermore, the TT genotype of the NQO1 609C>T polymorphism was associated with reduced NQO1 protein expression in tumor tissues from a subset of GI cancer patients (cardiac carcinoma, gastric adenocarcinoma, esophageal adenocarcinoma, and esophageal squamous cell carcinoma) [43], [45]. Because of this SNP's functional consequence, many epidemiological studies have examined the effect of the NQO1 609C>T polymorphism on risk of GI cancers, including cancers of the esophagus, stomach, colorectum, pancreas, and liver. However, the reported genetic effects varied across the published studies, and a clear impact of this SNP on cancer risk is also limited by the insufficient statistical power of these individual studies with a relatively small sample size. Therefore, we performed a meta-analysis of published data to evaluate the influence of the NQO1 609C>T polymorphism on the risk of GI cancers.
Figure 1. NQO1 gene structure and its function.
A. NQO1 gene structure and NQO1 609C>T polymorphism location. B. The multiple functions of NQO1. As depicted, NQO1 performs multiple functions within the cell, including two-electron reduction of quinones and their derivatives, stabilization of p53 and other tumor suppressors against proteasomal degradation, and scavenging of superoxide. NQO1 has also been implicated in maintaining microtubule integrity.
Materials and Methods
Identification and eligibility of relevant studies
Using the PubMed search engine, we searched Medline databases, on the association of NQO1 609C>T polymorphism (rs1800566) with the risk of GI cancers (defined as cancers of the esophagus, stomach, colorectum, pancreas, gallbladder, liver and small/larger bowel cancer), which had been published up to October 6, 2011 with a limit to human studies in English language. The following keywords were used: ‘NAD(P)H Dehydrogenase (Quinone)’ or ‘NQO1’, ‘polymorphism’, ‘variant’, and in combination with ‘gastrointestinal/aerodigestive tract cancers’, or ‘esophageal cancer’, ‘gastric/stomach’ cancer, ‘colorectal/colon/rectum cancer’, ‘ pancreatic cancer’, ‘liver cancer’, ‘hepatocellular carcinoma’, ‘gallbladder cancer’, and ‘small/larger bowel cancer’. In addition, the references cited in the retrieved studies were also reviewed manually to identify publications on the same cancer type. If studies from the same study group had overlapped subjects, the most recent or largest study was included in the final analysis. Human population-based or hospital-based association studies were included in this meta-analysis, if they met all the following criteria: (1) an independent, unrelated case-control, nested case-control, or cohort study, (2) the NQO1 609C>T polymorphism was determined, (3) the outcome was GI cancers, (4) there were sufficient data for calculating an odds ratio (OR) with 95% confidence interval (CI), and (5) the study was reported in English. Exclusion criteria were: (1) duplicate data, (2) abstract, case report, comment, review and editorial, (3) no sufficient genotyping data were provided, (4) the outcome was benign tumors, precancerous lesions, and adenomas, and (5) family-based study.
Data extraction
Two reviewers (HY and HL) independently reviewed the articles and extracted the data from all eligible publications according to the criteria listed above. The following information was recorded for each study: first author, year of publication, country or region of origin, ethnicity, cancer type, number of cases and controls, number of cases and controls by genotype, source of control group (population-based or hospital-based), genotyping methods, minor allele frequency in controls, method for matching controls to cases. Any discrepancies between the two investigators were resolved by discussion and consultation with a third reviewer (LW).
Quality score assessment
The quality of included studies was independently assessed by the same two reviewers using the quality assessment criteria, which was modified from previously published meta-analysis of molecular association studies [46], [47]. We included the following factors related to both traditional epidemiological considerations and cancer genetic issues in terms of quality of the studies: representativeness of the cases, representativeness of the controls, ascertainment of GI cancers, control selection, genotyping examination, response rate, and total sample size. The criteria are described in detail in Table S1, and the scores were defined as 1 to 3 points given to each component or 0 if absent or the study with a sample size of less than 200. A final quality score was obtained by summation of each component giving a range from 0 (the lowest) to 15 (the highest). Studies scoring<8 were classified as low quality, and those ≥8 as high quality. Disagreements were resolved by consultation with the third reviewer.
Statistical analysis
Deviation of genotype frequencies of the NQO1 609C>T polymorphism in control subjects from Hardy-Weinberg equilibrium (HWE) was tested by using the Chi-square goodness of fit, and a P value<0.05 was considered significant. Odds ratio (OR) and corresponding 95% confidence interval (95% CI) were used to estimate the association between the NQO1 609C>T polymorphism and cancer risk. We estimated the risk for the variant homozygous TT and heterogeneous CT genotypes, compared with the wild-type homozygous CC genotype, and then for CT/TT vs. CC and TT vs. CC/CT, assuming both dominant and recessive effect models, respectively. The heterogeneity across studies was assessed with the Q test, and the heterogeneity was considered significant when a P-value<0.1 for the Q statistic [48]. If the heterogeneity was not significant, the fixed-effects model was used to estimate the summary OR and 95% CI; Otherwise, the random-effects model was used [49]. We also calculated the I 2 index, which can quantify the degree of heterogeneity in a the meta-analysis [50]. The potential source of heterogeneity across studies was explored by stratification and meta-regression analyses. Stratified analyses were conducted by several study characteristics, such as ethnicity, type of cancers (if one cancer type contains less than two studies, it was merged into the ‘other cancers’ group), and quality score of studies (quality score<8 and ≥8). In addition, the studies investigating multiple types of cancers or multiple ethnicities were separated into groups for the subgroup analysis. Both Begg's and Egger's tests [51], [52] were used to test for publication bias. A P-value<0.1 was used as an indication for the presence of potential publication bias. Sensitivity analyses were conducted by including and excluding studies not in HWE, and by removing one study at a time to assess the influence of individual studies on the pooled ORs, respectively. All analyses were performed by using Review Manager (v.5.0; Oxford, England) and Stata software (version 8.2; Stata Corp LP, College Station, TX, USA). In addition, for each statistically significant association, we estimated the false positive report probability (FPRP) using the method described by Wacholder et al [53] to evaluate the robustness of the findings. Wacholder et al suggested that estimating statistical power based on the ability to detect an OR of 1.5 (or 0.67 = 1/1.5 for an OR less than 1.0), with an alpha level equal to the observed P-value [53]. Because a single nucleotide polymorphism usually shows a relatively small effect size (i.e., OR<1.5), we presented results for an OR of 1.2. An FPRP less than 0.2 was considered as a noteworthy association [53].
Results
Characteristics of all included studies
As of October 6, 2011, we had identified 29 potentially eligible studies that have investigated the association between the NQO1 609C>T polymorphism and risk of GI cancers. After retrieving the full text of these 29 articles, we excluded 9 articles because of the following reasons: one reported the association between the NQO1 609C>T SNP and H. pylori seropositivity [54]; one did not focus on the NQO1 609C>T but on NQO1 R139W SNP (rs4986998) [55]; three were for the association between the NQO1 609C>T SNP and colorectal adenoma [56], [57], [58]; two were for the correlation between the NQO1 609C>T genotypes and NQO1 activity [59] or telomere length [60]; two were for review or meta-analysis articles [61], [62]. In addition, the Caucasian control group (252 Caucasian controls) in the study by Zhang et al. [45] had overlapped subjects used in the study by Sarbia et al. [43], and the esophageal cancer patients (193 cases) in the study by Zhang et al. [63] were also overlapped with those in the same author's study [45]. Therefore, these 252 Caucasian controls and 193 esophageal cancer patients were excluded to avoid double counting in our meta-analysis. The flow chart in Figure 2 summarizes this literature review process.
Figure 2. The flow chart of the included studies in the meta-analysis.
Overall, data from 20 publications with 5,491 cases and 5,917 controls were available for our meta-analysis. Main characteristics of the included publications are presented in Table 1 . Among the 20 publications, four studies were for esophageal cancer [45], [64], [65], [66], one for gastric cancer [67], nine for colorectal cancer [55], [59], [68], [69], [70], [71], [72], [73], [74], [75], [76], two for pancreatic cancer [77], [78], one for liver cancer [79], and three for multiple types of GI cancers [43], [63], [80]. Of all studies, 11 studies were conducted in Caucasian populations [43], [65], [66], [68], [71], [72], [73], [74], [75], [76], [77], [78], seven in Asian populations [63], [64], [67], [69], [70], [79], [80], and two in multiple populations [45], [76]. Polymerase chain reaction (PCR)-restriction fragment length polymorphism (RFLP) method was used to determine the genotype in all the included studies except for one by Mohelnikova-Duchonova et al. [78], in which the TaqMan Assay was used. The genotype frequency distributions of the NQO1 609C>T polymorphism in controls in 19 of 20 included studies were in agreement with HWE. The HWE test in the study by Lafuente et al. was not mentioned [72]; we also could not perform the HWE test for the subjects (either cases or controls) in that study, because only the total number of the combined genotypes (TT vs. CT/CC) was available. Therefore, this study was included in the analysis for the recessive model but not for other genetic models. Quality scores for the individual studies ranged from 4 to 13, with 60.0% (12 of 20) of the studies being classified as high quality (≥8).
Table 1. Characteristics of studies included in the meta-analysis.
| First author (Reference No.) | Year | Country | Cancer type | Ethnicity | No. of cases/controls | Type of case-control study | Genotyping method | Quality Score | P-valuea |
| Marjani (64) | 2010 | Iran | esophagus | Asian | 93/50 | Hospital-based | PCR-RFLP | 5 | 0.47 |
| Martino (65) | 2007 | United Kingdom | esophagus | Caucasian | 141/93 | Hospital-based | PCR-RFLP | 5 | 0.99 |
| Rahden (66) | 2004 | German | esophagus | Caucasian | 140/260 | Hospital-based | PCR-RFLP | 4 | 0.17 |
| Sarbia (43) | 2003 | German | esophagus, stomach, etc | Caucasian | 384/252 | Hospital-based | PCR-RFLP | 6 | 0.60 |
| Zhang (45) | 2003 | German | esophagus | Mixed | 450/393 | Hospital-based | PCR-RFLP | 9 | 0.77 |
| Zhang (63) | 2003 | China | stomach, esophagus | Asian | 124/165 | Hospital-based | PCR-RFLP | 7 | 0.39 |
| Hamajima (80) | 2002 | Japan | esophagus, colorectum, stomach, etc. | Asian | 391/640 | Hospital-based | PCR-RFLP | 7 | 0.17 |
| Malik (67) | 2010 | India | stomach | Asian | 108/195 | Hospital-based | PCR-RFLP | 5 | 0.31 |
| Sachse (75) | 2002 | United Kingdom | colorectum | Caucasian | 490/593 | Population-based | PCR-RFLP | 11 | 0.56 |
| Hlavata (68) | 2010 | Czech | colorectum | Caucasian | 495/495 | Hospital-based | PCR-RFLP | 10 | 0.85 |
| Sameer (69) | 2010 | India | colorectum | Asian | 86/160 | Hospital-based | PCR-RFLP | 8 | 0.45 |
| Nisa (70) | 2010 | Japan | colorectum | Asian | 684/777 | Hospital-based | PCR-RFLP | 13 | 0.07 |
| Begleiter (76) | 2006 | Canada | colorectum | Mixed | 280/327 | Hospital-based | PCR-RFLP | 9 | 0.29 |
| van der Logt (71) | 2006 | New Zealand | colorectum | Caucasian | 369/415 | Population-based | PCR-RFLP | 8 | 0.95 |
| Harth (73) | 2000 | German | colorectum | Caucasian | 323/205 | Population-based | PCR-RFLP | 9 | 0.79 |
| Mitrou (57) | 2002 | United Kingdom | colorectum | Caucasian | 206/345 | Hospital-based | PCR-RFLP | 9 | 0.96 |
| Lafuente (72)b | 2000 | Spain | colorectum | Caucasian | 247/296 | Hospital-based | PCR-RFLP | 8 | - |
| Mohelnikova-Duchonova (78) | 2010 | Czech | pancreas | Caucasian | 235/265 | Hospital-based | TaqMan assay | 8 | 0.80 |
| Bartsch (77) | 1998 | German | pancreas | Caucasian | 81/76 | Hospital-based | PCR-RFLP | 5 | 0.27 |
| Akkiz (79) | 2010 | Turkey | liver | Asian | 167/167 | Hospital-based | PCR-RFLP | 8 | 0.81 |
P-value of the chi-square goodness of fit test for Hardy-Weinberg equilibrium (HWE) in controls.
the HWE test can not be conducted because only the total number of genotypes (TT vs. CT/CC) was available, and the HWE test was not mentioned in this study.
Frequency of the NQO1 609 C>T polymorphism in control populations
Of 5,917 control subjects included in this meta-analysis, 3622 were Caucasians and 2295 were Asians. The frequency distributions of the genotypes of the NQO1 609 C>T polymorphism were different between these two ethnic groups. The frequencies of the TT, CT, and CC genotypes were 3.1%, 28.2%, and 68.7%, respectively, in Caucasians and 13.1%, 44.7%, and 42.2% in Asians, respectively ( Table 2 ).
Table 2. The genotype frequencies of the NQO1 609C >T polymorphism in controls in different ethnic groups.
| Ethnic group | Number of controls | Genotype (%) | ||
| CC | CT | TT | ||
| Caucasians a | 3326 | 2286 (68.7) | 937 (28.2) | 103 (3.1) |
| Asians | 2295 | 968 (42.2) | 1027 (44.7) | 300 (13.1) |
| P-value b | 0.007 | 0.005 | 0.007 | |
The study by Lafuente et al was excluded when calculating the genotype frequency because the numbers for the CC and TT genotypes were not provided in this study.
Two-side Student's t test within the stratum.
Association between the NQO1 609C>T polymorphism and the risk of GI cancers
Overall, as shown in Table 3 , compared to the wild-type CC homozygous genotype, the CT heterozygous genotype was significantly associated with a modestly increased risk for GI cancers (CT vs. CC: OR = 1.10, 95% CI = 1.01 – 1.19). A main effect also was significant in the dominant model (CT/TT vs. CC: OR = 1.11, 95% CI = 1.02 – 1.20) ( Figure 3 ). There was no significant heterogeneity among the studies (P heterogeneity = 0.27 and I2 = 0.15 for CT vs. CC; P heterogeneity = 0.14 and I2 = 0.27 for CT/TT vs. CC). We found similar effects in the homozygous comparison (TT vs.CC: OR = 1.20, 95% CI: 0.96 – 1.50) and in the recessive model comparison (TT vs. CT/CC: OR = 1.22, 95% CI: 0.98 to 1.51). However, these effects did not reach statistical significance. A modest heterogeneity among the studies was observed (P heterogeneity = 0.09 and I2 = 0.32 for TT vs. CC; P heterogeneity = 0.06 and I2 = 0.36 for TT vs. CT/CC). Subsequent sensitivity analyses were performed by removing the individual study sequentially, and we found that all but one Japanese study by Hamajima et al. [80] slightly influenced the overall pooled ORs. After exclusion of this study, a significant increased risk was found in the homozygous comparison (TT vs.CC: OR = 1.27, 95% CI: 1.03 – 1.47) or in the recessive model comparison (TT vs. CT/CC: OR = 1.29, 95% CI: 1.05 – 1.59), and the heterogeneity among the studies was not significant (P heterogeneity = 0.18 and I2 = 0.23 for TT vs. CC; P heterogeneity = 0.20 and I2 = 0.20 for TT vs. CT/CC), suggesting that this study may contribute to the observed heterogeneity across studies.
Table 3. Meta-analysis for the association between the NQO1 609C>T polymorphism and cancer risk.
| Variables | No. of subjects Cases/Controls | na | CT vs. CC | TT vs.CC | CT/TT vs. CCb | TT vs. CT/CCc | ||||||||
| OR (95% CI) | I2 | P d | OR (95% CI) | I2 | P d | OR (95% CI) | I2 | P d | OR (95% CI) | I2 | P d | |||
| Overall | 5491/5917 | 20 | 1.10 (1.01 – 1.19) | 0.15 | 0.27 | 1.20 (0.96 – 1.50) | 0.32 | 0.09 | 1.11 (1.02 – 1.20) | 0.27 | 0.14 | 1.22 (0.98 – 1.51) | 0.36 | 0.06 |
| Caucasians | 3645/3622 | 12 | 1.13 (1.01 – 1.26) | 0.22 | 0.24 | 1.20 (0.91 – 1.58) | 0.19 | 0.27 | 1.14 (1.02 – 1.26) | 0.27 | 0.19 | 1.25 (0.96 – 1.62) | 0.25 | 0.20 |
| esophagus | 599/605 | 3 | 1.10 (0.85 – 1.43) | 0.20 | 0.29 | 1.27 (0.30 – 5.42) | 0.76 | 0.01 | 1.11 (0.75 – 1.66) | 0.59 | 0.09 | 1.26 (0.31 – 5.05) | 0.75 | 0.02 |
| stomach | 320/252 | 1 | 1.62 (1.12 – 2.33) | - | - | 2.31 (0.71 – 7.50) | - | - | 1.66 (1.16 – 2.37) | - | - | 2.00 (0.62 – 6.45) | - | - |
| colorectum | 2410/2676 | 7 | 1.13 (0.99 – 1.29) | 0.33 | 0.19 | 1.11 (0.79 – 1.56) | 0.0 | 0.48 | 1.13 (1.00 – 1.28) | 0.11 | 0.34 | 1.20 (0.88 – 1.64) | 0.27 | 0.23 |
| pancreas | 316/341 | 2 | 0.96 (0.68 – 1.35) | 0.0 | 0.46 | 1.08 (0.49 – 2.37) | 0.0 | 0.88 | 0.97 (0.70 – 1.35) | 0.0 | 0.52 | 1.12 (0.51 – 2.43) | 0.0 | 0.97 |
| Asians | 1846/2295 | 8 | 1.07 (0.94 – 1.23) | 0.43 | 1.25 (0.90 – 1.73) | 0.05 | 1.07 (0.94 – 1.21) | 0.17 | 1.22 (0.90 – 1.67) | 0.04 | ||||
| esophagus | 388/831 | 3 | 1.07 (0.79 – 1.45) | 0.23 | 0.28 | 1.12 (0.52 – 2.39) | 0.62 | 0.07 | 1.09 (0.81 – 1.45) | 0.43 | 0.17 | 1.08 (0.53 – 2.20) | 0.64 | 0.06 |
| stomach | 375/1000 | 3 | 1.12 (0.85 – 1.47) | 0.0 | 0.20 | 1.41 (0.98 – 2.03) | 0.46 | 0.22 | 1.21 (0.93 – 1.56) | 0.0 | 0.12 | 1.44 (0.84 – 2.46) | 0.59 | 0.24 |
| colorectum | 916/1577 | 3 | 0.99 (0.82 – 1.18) | 0.37 | 0.20 | 0.86 (0.65 – 1.13) | 0.33 | 0.22 | 0.96 (0.81 – 1.15) | 0.52 | 0.12 | 0.88 (0.68 – 1.14) | 0.30 | 0.24 |
| liver | 167/167 | 1 | 1.28 (0.82 – 2.00) | - | - | 1.24 (0.48 – 3.20) | - | - | 1.27 (0.83 – 1.96) | - | - | 1.12 (0.44 – 2.83) | - | - |
| Quality of study | ||||||||||||||
| high | 4358/4610 | 12 | 1.10 (1.00 – 1.22) | 0.18 | 0.27 | 1.13 (0.92 – 1.39) | 0.0 | 0.48 | 1.11 (1.01 – 1.22) | 0.15 | 0.30 | 1.17 (0.96 – 1.41) | 0.08 | 0.36 |
| low | 1133/1307 | 8 | 1.03 (0.86 – 1.24) | 0.0 | 0.45 | 1.28 (0.77 – 2.13) | 0.56 | 0.03 | 1.09 (0.92 – 1.30) | 0.31 | 0.18 | 1.29 (0.79 – 2.10) | 0.56 | 0.03 |
Number of studies. The studies investigating multiple types of cancers or multiple ethnicities were separated into groups for the subgroup analysis.
CT/TT vs. CC: Dominant model.
TT vs. CT/CC: Recessive model.
Heterogeneity across studies.
Figure 3. Forest plot (Fixed effects model) describing the association of the NQO1 609C>T polymorphism with risk of gastrointestinal (GI) cancers.
The NQO1 609C>T polymorphism was associated with a modestly increased risk of GI cancers in a dominant model (CT/TT vs. CC).
In the stratified analysis by ethnicity, as shown in Table 3 , significantly elevated cancer risks were found among Caucasians in the heterozygous genotype comparison (CT vs. CC: OR = 1.13, 95% CI: 1.01 –1.26, P heterogeneity = 0.24 and and I2 = 0.22) and the dominant model comparison (CT/TT vs. CC: OR = 1.14, 95% CI 5 1.02 – 1.26, P heterogeneity = 0.26 and I2 = 0.27), but not in the homozygous genotype comparison (TT vs. CC: OR = 1.20, 95% CI: 0.91 – 1.58, P heterogeneity = 0.27 and I2 = 0.19) and the recessive model comparison (TT vs. CT/CC: OR = 1.25, 95% CI: 0.96 – 1.62, P heterogeneity = 0.20 and I2 = 0.25). No significant heterogeneity was observed for all the genetic mode1 comparisons. The leave-one-out sensitivity analysis found that no single study dramatically influenced the overall pooled ORs (data not shown). In Asians, no significant association between the NQO1 609C>T polymorphism and the risk of GI cancers was found for all variant genotypes (CT vs.CC: OR = 1.07, 95% CI: 0.94 – 1.23, P heterogeneity = 0.43 and I2 = 0.0; TT vs.CC: OR = 1.25, 95% CI: 0.90 – 1.73, P heterogeneity = 0.05 and I2 = 0.50), the dominant model (CT/TT vs. CC: OR = 1.07, 95% CI: 0.94 – 1.26, P heterogeneity = 0.17 and I2 = 0.32) and the recessive model (TT vs. CT/CC: OR = 1.22, 95% CI: 0.90 – 1.21, P heterogeneity = 0.27 and I2 = 0.52). However, the leave-one-out sensitivity analysis showed that after removing the study by Hamajima et al. [80], the heterogeneity among studies diminished, and a significant association was found in the recessive model (TT vs.CT/CC: OR = 1.36, 95% CI: 1.02 – 1.81, P heterogeneity = 0.23 and I2 = 0.26). In further stratification analysis by cancer site ( Table 3 ), a modestly significant increased risk was found for the colorectal cancer under the dominant model in Caucasians (CT/TT vs. CC: OR = 1.13, 95% CI: 1.00 – 1.28, P heterogeneity = 0.34 and I2 = 0.11). However, no significant association was observed for other cancer sites either in Caucasians or in Asians. The leave-one-out sensitivity analysis showed that no single study dramatically influenced the overall pooled ORs (data not shown).
We also performed subgroup analysis by quality score of studies ( Table 3 ). We found that the CT heterozygous genotype was significantly associated with a modestly increased risk for GI cancers, compared to the wild-type homozygous genotype (CC) in the studies with high quality score (≥8.0) (CT vs.CC: OR = 1.10, 95% CI: 1.00 – 1.22; P heterogeneity = 0.27 and I2 = 0.18), and such an effect was also found in the dominant genetic model (CT/TT vs.CC: OR = 1.11, 95% CI: 1.01 – 1.22; P heterogeneity = 0.30 and I2 = 0.15). Similar effects were also found for the homozygous genotype comparison (TT vs.CC: OR = 1.13, 95% CI: 0.92 – 1.39; P heterogeneity = 0.48 and I2 = 0.0) and for the recessive genetic model comparison (TT vs.CT/CC: OR = 1.17, 95% CI: 0.96 – 1.41; P heterogeneity = 0.36 and I2 = 0.08), though they did not reach statistical significance. In the subgroup of low quality studies, no significant association between the NQO1 609C>T polymorphism and the risk of GI cancers was observed. Sensitivity analyses showed that no single study influenced quantitatively the overall pooled ORs (data not shown).
Evaluation of heterogeneity
In the present study, we used the Q test and the I2 index to evaluate the heterogeneity across studies. As shown in Table 2, although the Q test showed that there was no significant heterogeneity in some overall comparisons and subgroup analyses, the I2 index suggested that a low to high heterogeneity across studies presented in most of comparisons. We assessed heterogeneity across studies by ethnicity, cancer site, and quality of studies, and found that they did not contribute the heterogeneity observed across the studies in the overall meta-analysis (TT vs.CC: t = −0.24, P = 0.815 for ethnicity, t = 0.02, P = 0.988 for cancer sites, and t = 0.39 8, P = 0.703 for quality of studies; TT vs.CT/CC: t = 0.00, P = 1.000 for ethnicity, t = −0.29, P = 0.773 for cancer sites, and t = 0.29, P = 0.777 for quality of studies). These factors were also not found to contribute to the heterogeneity across studies in some of the subgroup analysis (data not shown). Together with the results from the leave-one-out sensitivity analysis as mentioned above, the study by Hamajima et al. could be the main source of the observed heterogeneity across the studies in this meta-analysis.
Publication bias
Both Begg's and Egger's tests were performed to evaluate the publication bias of the included studies. The shape of the funnel plots did not reveal any evidence of obvious asymmetry for all genetic models in the overall meta-analysis ( Figure 4 ). The Begg's test and Egger's test did not present any significantly statistical evidence of publication bias for any of the genetic models (CT vs.CC: P Begg = 0.529 and P Egger = 0.369, TT vs.CC: P Begg = 0.726 and P Egger = 0.690, CT/TT vs.CC: P Begg = 1.000 and P Egger = 0.671, and TT vs.CT/CC: P Begg = 0.626 and P Egger = 0.700.) Neither funnel plots nor Begg's and Egger's tests detected any obvious evidence of publication bias in the subgroup analyses for all genetic models (data not shown).
Figure 4. Funnel plot analysis to detect publication bias.
Each point represents an individual study for the indicated association.
Finally, because many subgroup comparisons were conducted, we calculated false positive report probability (FPRP) for each statistically significant result. As shown in Table 4 , with the assumption of a moderate prior probability of 0.1 and the OR for the specific genotype was 1.2, the FPRP values for the significant findings in the heterozygous genotype comparison (CT vs. CC) and the dominant model (CT/TT vs. CC) in all subjects, and in the dominant model in Caucasians (CT/TT vs. CC) were 0.138, 0.074, 0.099, respectively. However, greater FPRP values were observed for other significant associations between the NQO1 609C>T polymorphism and risk of GI cancers.
Table 4. False positive reporting probability values for associations between the NQO1 609C>T polymorphism and the risk of GI cancers.
| Genotype | OR (95% CI) | Prior probability | ||||
| 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||
| All subjects | ||||||
| CT vs. CC | 1.10 (1.01 – 1.19) | 0.051 | 0.138 | 0.638 | 0.947 | 0.994 |
| CT/TT vs. CC | 1.11 (1.02 – 1.20) | 0.026 | 0.074 | 0.469 | 0.899 | 0.989 |
| Caucasians | ||||||
| CT vs. CC | 1.13 (1.01 – 1.26) | 0.088 | 0.225 | 0.762 | 0.970 | 0.997 |
| CT/TT vs. CC | 1.14 (1.02 – 1.26) | 0.035 | 0.099 | 0.547 | 0.924 | 0.992 |
| Colorectum cancer in Caucasians | ||||||
| CT/TT vs. CC | 1.13 (1.00 – 1.28) | 0.165 | 0.373 | 0.867 | 0.985 | 0.998 |
| High quality of study | ||||||
| CT vs. CC | 1.10 (1.00 – 1.22) | 0.184 | 0.403 | 0.881 | 0.987 | 0.999 |
| CT/TT vs. CC | 1.11 (1.01 – 1.22) | 0.088 | 0.224 | 0.761 | 0.970 | 0.997 |
Discussion
In the present meta-analysis with 5,491 cases and 5,917 controls, the variant CT heterozygous genotype and the combined CT/TT genotype of the NQO1 609 C>T polymorphism was found to be associated with a modestly increased risk of GI cancers, and no significant heterogeneity was found across studies. It was also noted that, when limiting the pooled analysis to the studies with high quality, the results were persistent and robust, with the NQO1 609 T allele being significantly associated with the increased risk of GI cancers. Publication bias was not observed in this study. These findings suggest that the NQO1 609C>T polymorphism may modify the risk of GI cancers.
Our findings have some biological plausibility, because NQO1 performs multiple functions within the cell. Conclusive evidence suggests that NQO1 has a protective function in cellular defense against the toxicity of electrophilic and oxidizing metabolites of xenobiotic quinones [81]. In addition, its induction protects cells against carcinogenesis [27], [28], [29], [30], [31], [81]. Constitutive expression of NQO1 has been found in most human tissues, where its expression is highly induced by various stimuli, including antioxidants, oxidants, xenobiotics, heavy metals, UV light, and ionizing radiation [37]. It has been shown that NQO1 is overexpressed in many human tumors, including cancers of the lung, breasts, liver, esophagus, stomach, colon, pancreas, and bladder [24], [31], [42], [43], [82], [83], [84], [85]. The NQO1 knockout mice were reported to exhibit marked increased sensitivity to 7,12-dimethylbenz(a)anthracene (DMBA)- and benzo(a)pyrene (BP)-induced skin carcinogenesis [86], [87].
Human NQO1 is polymorphic [88], of which the NQO1 609C>T polymorphism, in terms of its frequency and phenotypic consequences, is most prominent and thus intensively studied. Our results are consistent with the potentially altered biological functions of NQO1 by the 609C>T polymorphism. Although the association of the homozygous variant genotype (TT) with overall cancer risk did not achieve statistical significance, the magnitude and direction for association for GI cancers were persistent in both overall and some subgroups in our meta-analysis. Because the frequency of the TT genotype of the NQO1 609C>T polymorphism was low in the published study populations, with 3.1% and 13.1% of the controls being the TT homozygote in Caucasians and Asians, respectively, we might not have sufficient statistical power to detect the weak effect of this variant genotype on risk of GI cancers. Further studies with larger sample sizes are warranted.
GI cancers represent a heterogeneous group of malignancies. Except for some shared risk factors, different primary sites of GI cancers have different risk factors and thus different etiologies. For example, in addition to smoking and alcohol consumption, H. Pylori infection is involved in stomach cancer and HBV/HCV infection is involved in liver cancer, while dietary exposure to heterocyclic amines (HCAs), nitrosamines, polycyclic aromatic hydrocarbons (PAHs) derived from red meat and processed meat is a key risk factor for colorectum cancer. Such etiologic heterogeneity in GI cancers raises the possibility that the NQO1 polymorphism may be associated with specific types of GI cancers, because NQO1 plays an important role in detoxifying dietary carcinogenic compounds such as HCAs, PAHs and nitrosamines [89]. Therefore, the functional NQO1 609 C>T polymorphism resulting in decreased activity of NQO1 enzyme may increase risk of colorectum cancer. Indeed, in the stratification analysis by cancer site in Caucasians and Asians, significantly elevated risk associated with the NQO1 609T allele was only found for colorectal cancer among Caucasians but not in Asians. A previous meta-analysis by Chao et al. [62] found an association between the NQO1 609T allele and an increased risk for colorectal cancer (CT/TT vs. CC: OR = 1.18, 95% CI: 1.02 – 1.35) among 1637 cases and 1854 controls in the Caucasian population, and this association remained statistically significant in this expanded meta-analysis that had included additional subjects (2410 cases and 2676 controls in the Caucasian population). However, the meta-analysis for the NQO1 609C>T polymorphism and colorectal cancer risk was not performed in Asian population in Chao's study. We also did not find significant associations between this SNP and risk of other cancer sites, such as cancers of the esophagus, stomach, and pancreas either in Caucasians or in Asians. This lack of significance could be due to either no effect of this SNP on these cancer sites or limited statistical power to detect such a weak association. In our meta-analysis, only two studies with 316 cases and 341 controls for pancreatic cancer in Caucasians, three studies with 375 cases and 1,000 controls for gastric cancer and three studies with 916 cases and 1577 controls for colorectum were conducted in Asians. Therefore, our results should be interpreted with caution. Because the NQO1 609C>T polymorphism is functional and potentially to be associated with risk of cancer as shown in this meta-analysis, further larger studies are needed, especially for non-colorectal GI cancers in Caucasians and GI cancers in Asians.
Certain potential limitations exist in our meta-analysis. Firstly, although the Begg's test and Egger's test did not show any publication bias, selection bias could have occurred, because only studies published in English were included in our meta-analysis. Secondly, all the studies included in this meta-analysis were hospital-based case-control studies. In this instance, the hospital-based controls may not be representative of the general population. Thirdly, the numbers of published studies were still not sufficiently large for the analysis of the effect of the variant TT genotype on risk of GI cancers and for some subgroups. Furthermore, we were unable to perform further subgroup analyses for a particular cancer site in different ethnic populations due to a limited number of published studies available to be included. For example, only one Caucasian study and one Asian study for gastric cancer and liver cancer were available for this meta-analysis, respectively. Fourthly, the FPRP analyses showed that with the assumption of a prior probability of 0.1, the FPRP values for the significant findings in overall comparisons and the comparison in the dominant model in Caucasians were below 0.2, providing some measures of robustness for our observations. However, greater FPRP values were observed for the other significant associations between the NQO1 609C>T polymorphism and risk of GI cancers, suggesting some possible bias in the findings. Finally, due to lacking individual original data, we did not take into account the other factors such as sex, ethnicity, smoking and drinking status, that could modify the risk of estimate [57], [62], [69], when we evaluated the effect of the NQO1 609 C>T polymorphism on the risk of GI cancers. A more precise analysis could have been conducted, if individual data were available. Furthermore, gene-environment and gene-gene interactions should also be considered in further studies.
In summary, despite the above-mentioned limitations, our meta-analysis suggests that the minor allele T of the NQO1 609C>T polymorphism may be associated with a moderately increased risk of GI cancers. Although the effect on cancer risk may be modified by ethnicity and cancer sites, small sample sizes in some subgroups suggest that future large and well-designed studies in different ethnic populations and different sites of GI cancers are needed to validate our findings.
Supporting Information
Score of quality assessment.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported in part by National Institutes of Health grants R01ES011740 and R01CA131274 (Qingyi Wei). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. No additional external funding received for this study.
References
- 1.Milosavljevic T, Kostic-Milosavljevic M, Jovanovic I, Krstic M. Gastrointestinal and liver tumours and public health in Europe. Eur Rev Med Pharmacol Sci. 2010;14:259–262. [PubMed] [Google Scholar]
- 2.Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 3.Key TJ. Fruit and vegetables and cancer risk. Br J Cancer. 2011;104:6–11. doi: 10.1038/sj.bjc.6606032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Randi G, Edefonti V, Ferraroni M, La Vecchia C, Decarli A Dietary patterns and the risk of colorectal cancer and adenomas. Nutrition Reviews. 2010;68:389–408. doi: 10.1111/j.1753-4887.2010.00299.x. [DOI] [PubMed] [Google Scholar]
- 5.Compare D, Rocco A, Nardone G. Risk factors in gastric cancer. Eur Rev Med Pharmacol Sci. 2010;14:302–308. [PubMed] [Google Scholar]
- 6.Di Luzio R, Moscatiello S, Marchesini G. Role of nutrition in gastrointestinal oncological patients. Eur Rev Med Pharmacol Sci. 2010;14:277–284. [PubMed] [Google Scholar]
- 7.Menon R, Riera A, Ahmad A. A global perspective on gastrointestinal diseases. Gastroenterol Clin North Am. 2011;40:427–439, ix. doi: 10.1016/j.gtc.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 8.Ullman TA, Itzkowitz SH. Intestinal inflammation and cancer. Gastroenterology. 2011;140:1807–1816. doi: 10.1053/j.gastro.2011.01.057. [DOI] [PubMed] [Google Scholar]
- 9.Piazuelo MB, Epplein M, Correa P. Gastric cancer: an infectious disease. Infect Dis Clin North Am. 2010;24:853–869, vii. doi: 10.1016/j.idc.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wroblewski LE, Peek RM, Jr, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010;23:713–739. doi: 10.1128/CMR.00011-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouchard MJ, Navas-Martin S. Hepatitis B and C virus hepatocarcinogenesis: lessons learned and future challenges. Cancer Lett. 2011;305:123–143. doi: 10.1016/j.canlet.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dragani TA. Risk of HCC: genetic heterogeneity and complex genetics. J Hepatol. 2010;52:252–257. doi: 10.1016/j.jhep.2009.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Yaghoobi M, Bijarchi R, Narod SA. Family history and the risk of gastric cancer. Br J Cancer. 2010;102:237–242. doi: 10.1038/sj.bjc.6605380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baglietto L, Jenkins MA, Severi G, Giles GG, Bishop DT, et al. Measures of familial aggregation depend on definition of family history: meta-analysis for colorectal cancer. J Clin Epidemiol. 2006;59:114–124. doi: 10.1016/j.jclinepi.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 15.Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, et al. Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 16.Grady WM. Genetic testing for high-risk colon cancer patients. Gastroenterology. 2003;124:1574–1594. doi: 10.1016/s0016-5085(03)00376-7. [DOI] [PubMed] [Google Scholar]
- 17.Johns LE, Houlston RS. A systematic review and meta-analysis of familial colorectal cancer risk. Am J Gastroenterol. 2001;96:2992–3003. doi: 10.1111/j.1572-0241.2001.04677.x. [DOI] [PubMed] [Google Scholar]
- 18.Le Marchand L. Genome-wide association studies and colorectal cancer. Surg Oncol Clin N Am. 2009;18:663–668. doi: 10.1016/j.soc.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomlinson IP, Webb E, Carvajal-Carmona L, Broderick P, Howarth K, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet. 2008;40:623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 20.Wang LD, Zhou FY, Li XM, Sun LD, Song X, et al. Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54. Nat Genet. 2010;42:759–763. doi: 10.1038/ng.648. [DOI] [PubMed] [Google Scholar]
- 21.Haerian MS, Baum L, Haerian BS. Association of 8q24.21 loci with the risk of colorectal cancer: A systematic review and meta-analysis. J Gastroenterol Hepatol. 2011;26:1475–1484. doi: 10.1111/j.1440-1746.2011.06831.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhang H, Zhai Y, Hu Z, Wu C, Qian J, et al. Genome-wide association study identifies 1p36.22 as a new susceptibility locus for hepatocellular carcinoma in chronic hepatitis B virus carriers. Nat Genet. 2010;42:755–758. doi: 10.1038/ng.638. [DOI] [PubMed] [Google Scholar]
- 23.Ross D, Kepa JK, Winski SL, Beall HD, Anwar A, et al. NAD(P)H:quinone oxidoreductase 1 (NQO1): chemoprotection, bioactivation, gene regulation and genetic polymorphisms. Chem Biol Interact. 2000;129:77–97. doi: 10.1016/s0009-2797(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 24.Traver RD, Siegel D, Beall HD, Phillips RM, Gibson NW, et al. Characterization of a polymorphism in NAD(P)H: quinone oxidoreductase (DT-diaphorase). Br J Cancer. 1997;75:69–75. doi: 10.1038/bjc.1997.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsvetkov P, Reuven N, Shaul Y Ubiquitin-independent p53 proteasomal degradation. Cell Death Differ. 17:103–108. doi: 10.1038/cdd.2009.67. [DOI] [PubMed] [Google Scholar]
- 26.Siegel D, Gustafson DL, Dehn DL, Han JY, Boonchoong P, et al. NAD(P)H:quinone oxidoreductase 1: role as a superoxide scavenger. Mol Pharmacol. 2004;65:1238–1247. doi: 10.1124/mol.65.5.1238. [DOI] [PubMed] [Google Scholar]
- 27.Dinkova-Kostova AT, Talalay P. NAD(P)H:quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch Biochem Biophys. 2010;501:116–123. doi: 10.1016/j.abb.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asher G, Tsvetkov P, Kahana C, Shaul Y. A mechanism of ubiquitin-independent proteasomal degradation of the tumor suppressors p53 and p73. Genes Dev. 2005;19:316–321. doi: 10.1101/gad.319905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Asher G, Lotem J, Kama R, Sachs L, Shaul Y. NQO1 stabilizes p53 through a distinct pathway. Proc Natl Acad Sci U S A. 2002;99:3099–3104. doi: 10.1073/pnas.052706799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asher G, Lotem J, Sachs L, Kahana C, Shaul Y. Mdm-2 and ubiquitin-independent p53 proteasomal degradation regulated by NQO1. Proc Natl Acad Sci U S A. 2002;99:13125–13130. doi: 10.1073/pnas.202480499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nioi P, Hayes JD. Contribution of NAD(P)H:quinone oxidoreductase 1 to protection against carcinogenesis, and regulation of its gene by the Nrf2 basic-region leucine zipper and the arylhydrocarbon receptor basic helix-loop-helix transcription factors. Mutat Res. 2004;555:149–171. doi: 10.1016/j.mrfmmm.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 32.Rosvold EA, McGlynn KA, Lustbader ED, Buetow KH. Identification of an NAD(P)H:quinone oxidoreductase polymorphism and its association with lung cancer and smoking. Pharmacogenetics. 1995;5:199–206. doi: 10.1097/00008571-199508000-00003. [DOI] [PubMed] [Google Scholar]
- 33.Siegel D, McGuinness SM, Winski SL, Ross D. Genotype-phenotype relationships in studies of a polymorphism in NAD(P)H:quinone oxidoreductase 1. Pharmacogenetics. 1999;9:113–121. doi: 10.1097/00008571-199902000-00015. [DOI] [PubMed] [Google Scholar]
- 34.Misra V, Klamut HJ, Rauth AM. Transfection of COS-1 cells with DT-diaphorase cDNA: role of a base change at position 609. Br J Cancer. 1998;77:1236–1240. doi: 10.1038/bjc.1998.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Traver RD, Horikoshi T, Danenberg KD, Stadlbauer TH, Danenberg PV, et al. NAD(P)H:quinone oxidoreductase gene expression in human colon carcinoma cells: characterization of a mutation which modulates DT-diaphorase activity and mitomycin sensitivity. Cancer Res. 1992;52:797–802. [PubMed] [Google Scholar]
- 36.Moran JL, Siegel D, Ross D. A potential mechanism underlying the increased susceptibility of individuals with a polymorphism in NAD(P)H:quinone oxidoreductase 1 (NQO1) to benzene toxicity. Proc Natl Acad Sci U S A. 1999;96:8150–8155. doi: 10.1073/pnas.96.14.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ross D, Siegel D. NAD(P)H:quinone oxidoreductase 1 (NQO1, DT-diaphorase), functions and pharmacogenetics. Methods Enzymol. 2004;382:115–144. doi: 10.1016/S0076-6879(04)82008-1. [DOI] [PubMed] [Google Scholar]
- 38.Kuehl BL, Paterson JW, Peacock JW, Paterson MC, Rauth AM. Presence of a heterozygous substitution and its relationship to DT-diaphorase activity. Br J Cancer. 1995;72:555–561. doi: 10.1038/bjc.1995.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegel D, Anwar A, Winski SL, Kepa JK, Zolman KL, et al. Rapid polyubiquitination and proteasomal degradation of a mutant form of NAD(P)H:quinone oxidoreductase 1. Mol Pharmacol. 2001;59:263–268. doi: 10.1124/mol.59.2.263. [DOI] [PubMed] [Google Scholar]
- 40.Awadallah NS, Dehn D, Shah RJ, Russell Nash S, Chen YK, et al. NQO1 expression in pancreatic cancer and its potential use as a biomarker. Appl Immunohistochem Mol Morphol. 2008;16:24–31. doi: 10.1097/PAI.0b013e31802e91d0. [DOI] [PubMed] [Google Scholar]
- 41.Belinsky M, Jaiswal AK. NAD(P)H:quinone oxidoreductase1 (DT-diaphorase) expression in normal and tumor tissues. Cancer Metastasis Rev. 1993;12:103–117. doi: 10.1007/BF00689804. [DOI] [PubMed] [Google Scholar]
- 42.Chiu MM, Ko YJ, Tsou AP, Chau GY, Chau YP. Analysis of NQO1 polymorphisms and p53 protein expression in patients with hepatocellular carcinoma. Histol Histopathol. 2009;24:1223–1232. doi: 10.14670/HH-24.1223. [DOI] [PubMed] [Google Scholar]
- 43.Sarbia M, Bitzer M, Siegel D, Ross D, Schulz WA, et al. Association between NAD(P)H: quinone oxidoreductase 1 (NQ01) inactivating C609T polymorphism and adenocarcinoma of the upper gastrointestinal tract. Int J Cancer. 2003;107:381–386. doi: 10.1002/ijc.11430. [DOI] [PubMed] [Google Scholar]
- 44.Siegel D, Ross D. Immunodetection of NAD(P)H:quinone oxidoreductase 1 (NQO1) in human tissues. Free Radic Biol Med. 2000;29:246–253. doi: 10.1016/s0891-5849(00)00310-5. [DOI] [PubMed] [Google Scholar]
- 45.Zhang J, Schulz WA, Li Y, Wang R, Zotz R, et al. Association of NAD(P)H: quinone oxidoreductase 1 (NQO1) C609T polymorphism with esophageal squamous cell carcinoma in a German Caucasian and a northern Chinese population. Carcinogenesis. 2003;24:905–909. doi: 10.1093/carcin/bgg019. [DOI] [PubMed] [Google Scholar]
- 46.Thakkinstian A, McKay GJ, McEvoy M, Chakravarthy U, Chakrabarti S, et al. Systematic review and meta-analysis of the association between complement component 3 and age-related macular degeneration: a HuGE review and meta-analysis. Am J Epidemiol. 2011;173:1365–1379. doi: 10.1093/aje/kwr025. [DOI] [PubMed] [Google Scholar]
- 47.Thakkinstian A, D'Este C, Eisman J, Nguyen T, Attia J. Meta-analysis of molecular association studies: vitamin D receptor gene polymorphisms and BMD as a case study. J Bone Miner Res. 2004;19:419–428. doi: 10.1359/JBMR.0301265. [DOI] [PubMed] [Google Scholar]
- 48.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 49.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 50.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 52.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goto Y, Hamajima N, Honda H, Matsuo K, Yamamoto K, et al. Association between Helicobacter pylori seropositivity and NAD(P)H:quinone oxidoreductase 1 (NQO1) C609T polymorphism observed in outpatients and health checkup examinees. Gastric Cancer. 2005;8:12–17. doi: 10.1007/s10120-004-0308-1. [DOI] [PubMed] [Google Scholar]
- 55.Landi S, Gemignani F, Moreno V, Gioia-Patricola L, Chabrier A, et al. A comprehensive analysis of phase I and phase II metabolism gene polymorphisms and risk of colorectal cancer. Pharmacogenet Genomics. 2005;15:535–546. doi: 10.1097/01.fpc.0000165904.48994.3d. [DOI] [PubMed] [Google Scholar]
- 56.Tijhuis MJ, Visker MH, Aarts JM, Laan W, de Boer SY, et al. NQO1 and NFE2L2 polymorphisms, fruit and vegetable intake and smoking and the risk of colorectal adenomas in an endoscopy-based population. Int J Cancer. 2008;122:1842–1848. doi: 10.1002/ijc.23246. [DOI] [PubMed] [Google Scholar]
- 57.Mitrou PN, Watson MA, Loktionov AS, Cardwell C, Gunter MJ, et al. Role of NQO1C609T and EPHX1 gene polymorphisms in the association of smoking and alcohol with sporadic distal colorectal adenomas: results from the UKFSS Study. Carcinogenesis. 2007;28:875–882. doi: 10.1093/carcin/bgl194. [DOI] [PubMed] [Google Scholar]
- 58.Hou L, Chatterjee N, Huang WY, Baccarelli A, Yadavalli S, et al. CYP1A1 Val462 and NQO1 Ser187 polymorphisms, cigarette use, and risk for colorectal adenoma. Carcinogenesis. 2005;26:1122–1128. doi: 10.1093/carcin/bgi054. [DOI] [PubMed] [Google Scholar]
- 59.Tijhuis MJ, Boerboom AM, Visker MH, Op den Camp L, Nagengast FM, et al. The influence of fruit and vegetable consumption and genetic variation on NAD(P)H:quinone oxidoreductase (NQO1) phenotype in an endoscopy-based population. Nutr Cancer. 2008;60:204–215. doi: 10.1080/01635580701684849. [DOI] [PubMed] [Google Scholar]
- 60.Takagi S, Kinouchi Y, Hiwatashi N, Hirai M, Suzuki S, et al. Correlative polymorphism of NAD(P)H: quinone oxidoreductase (NQO1) with telomere shortening in colorectal cancer. Anticancer Res. 2002;22:2749–2752. [PubMed] [Google Scholar]
- 61.Hamajima N, Naito M, Kondo T, Goto Y. Genetic factors involved in the development of Helicobacter pylori-related gastric cancer. Cancer Sci. 2006;97:1129–1138. doi: 10.1111/j.1349-7006.2006.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chao C, Zhang ZF, Berthiller J, Boffetta P, Hashibe M. NAD(P)H:quinone oxidoreductase 1 (NQO1) Pro187Ser polymorphism and the risk of lung, bladder, and colorectal cancers: a meta-analysis. Cancer Epidemiol Biomarkers Prev. 2006;15:979–987. doi: 10.1158/1055-9965.EPI-05-0899. [DOI] [PubMed] [Google Scholar]
- 63.Zhang JH, Li Y, Wang R, Geddert H, Guo W, et al. NQO1 C609T polymorphism associated with esophageal cancer and gastric cardiac carcinoma in North China. World J Gastroenterol. 2003;9:1390–1393. doi: 10.3748/wjg.v9.i7.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marjani HA, Biramijamal F, Rakhshani N, Hossein-Nezhad A, Malekzadeh R. Investigation of NQO1 genetic polymorphism, NQO1 gene expression and PAH-DNA adducts in ESCC. A case-control study from Iran. Genet Mol Res. 2010;9:239–249. doi: 10.4238/vol9-1gmr693. [DOI] [PubMed] [Google Scholar]
- 65.di Martino E, Hardie LJ, Wild CP, Gong YY, Olliver JR, et al. The NAD(P)H:quinone oxidoreductase I C609T polymorphism modifies the risk of Barrett esophagus and esophageal adenocarcinoma. Genet Med. 2007;9:341–347. doi: 10.1097/gim.0b013e3180654ccd. [DOI] [PubMed] [Google Scholar]
- 66.von Rahden BH, Stein HJ, Langer R, von Weyhern CW, Schenk E, et al. C609T polymorphism of the NAD(P)H:quinone oxidoreductase I gene does not significantly affect susceptibility for esophageal adenocarcinoma. Int J Cancer. 2005;113:506–508. doi: 10.1002/ijc.20576. [DOI] [PubMed] [Google Scholar]
- 67.Malik MA, Zargar SA, Mittal B. Role of NQO1 609C>T and NQO2-3423G>A polymorphisms in susceptibility to gastric cancer in Kashmir valley. DNA Cell Biol. 2010;30:297–303. doi: 10.1089/dna.2010.1115. [DOI] [PubMed] [Google Scholar]
- 68.Hlavata I, Vrana D, Smerhovsky Z, Pardini B, Naccarati A, et al. Association between exposure-relevant polymorphisms in CYP1B1, EPHX1, NQO1, GSTM1, GSTP1 and GSTT1 and risk of colorectal cancer in a Czech population. Oncol Rep. 2010;24:1347–1353. doi: 10.3892/or_00000992. [DOI] [PubMed] [Google Scholar]
- 69.Sameer AS, Shah ZA, Syeed N, Rasool R, Afroze D, et al. NAD(P)H:quinone oxidoreductase 1 (NQO1) Pro187Ser polymorphism and colorectal cancer predisposition in the ethnic Kashmiri population. Asian Pac J Cancer Prev. 2010;11:209–213. [PubMed] [Google Scholar]
- 70.Nisa H, Kono S, Yin G, Toyomura K, Nagano J, et al. Cigarette smoking, genetic polymorphisms and colorectal cancer risk: the Fukuoka Colorectal Cancer Study. BMC Cancer. 2010;10:274. doi: 10.1186/1471-2407-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van der Logt EM, Bergevoet SM, Roelofs HM, Te Morsche RH, Dijk Y, et al. Role of epoxide hydrolase, NAD(P)H:quinone oxidoreductase, cytochrome P450 2E1 or alcohol dehydrogenase genotypes in susceptibility to colorectal cancer. Mutat Res. 2006;593:39–49. doi: 10.1016/j.mrfmmm.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 72.Lafuente MJ, Casterad X, Trias M, Ascaso C, Molina R, et al. NAD(P)H:quinone oxidoreductase-dependent risk for colorectal cancer and its association with the presence of K-ras mutations in tumors. Carcinogenesis. 2000;21:1813–1819. doi: 10.1093/carcin/21.10.1813. [DOI] [PubMed] [Google Scholar]
- 73.Harth V, Donat S, Ko Y, Abel J, Vetter H, et al. NAD(P)H quinone oxidoreductase 1 codon 609 polymorphism and its association to colorectal cancer. Arch Toxicol. 2000;73:528–531. doi: 10.1007/s002040050004. [DOI] [PubMed] [Google Scholar]
- 74.Mitrou P, Watson M, Bingham S, Stebbings WS, Speakman CT, et al. NQO1 and mEH exon 4 (mEH4) gene polymorphisms, smoking and colorectal cancer risk. IARC Sci Publ. 2002;156:495–497. [PubMed] [Google Scholar]
- 75.Sachse C, Smith G, Wilkie MJ, Barrett JH, Waxman R, et al. A pharmacogenetic study to investigate the role of dietary carcinogens in the etiology of colorectal cancer. Carcinogenesis. 2002;23:1839–1849. doi: 10.1093/carcin/23.11.1839. [DOI] [PubMed] [Google Scholar]
- 76.Begleiter A, Hewitt D, Maksymiuk AW, Ross DA, Bird RP. A NAD(P)H:quinone oxidoreductase 1 polymorphism is a risk factor for human colon cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:2422–2426. doi: 10.1158/1055-9965.EPI-06-0661. [DOI] [PubMed] [Google Scholar]
- 77.Bartsch H, Malaveille C, Lowenfels AB, Maisonneuve P, Hautefeuille A, et al. Genetic polymorphism of N-acetyltransferases, glutathione S-transferase M1 and NAD(P)H:quinone oxidoreductase in relation to malignant and benign pancreatic disease risk. The International Pancreatic Disease Study Group. Eur J Cancer Prev. 1998;7:215–223. doi: 10.1097/00008469-199806000-00006. [DOI] [PubMed] [Google Scholar]
- 78.Mohelnikova-Duchonova B, Marsakova L, Vrana D, Holcatova I, Ryska M, et al. Superoxide dismutase and nicotinamide adenine dinucleotide phosphate: quinone oxidoreductase polymorphisms and pancreatic cancer risk. Pancreas. 2010;40:72–78. doi: 10.1097/MPA.0b013e3181f74ad7. [DOI] [PubMed] [Google Scholar]
- 79.Akkiz H, Bayram S, Bekar A, Akgollu E, Ulger Y, et al. No association of NAD(P)H: quinone oxidoreductase 1 (NQO1) C609T polymorphism and risk of hepatocellular carcinoma development in Turkish subjects. Asian Pac J Cancer Prev. 2010;11:1051–1058. [PubMed] [Google Scholar]
- 80.Hamajima N, Matsuo K, Iwata H, Shinoda M, Yamamura Y, et al. NAD(P)H: quinone oxidoreductase 1 (NQO1) C609T polymorphism and the risk of eight cancers for Japanese. Int J Clin Oncol. 2002;7:103–108. doi: 10.1007/s101470200013. [DOI] [PubMed] [Google Scholar]
- 81.Ross D. Quinone reductases multitasking in the metabolic world. Drug Metab Rev. 2004;36:639–654. doi: 10.1081/dmr-200033465. [DOI] [PubMed] [Google Scholar]
- 82.Schlager JJ, Powis G. Cytosolic NAD(P)H:(quinone-acceptor)oxidoreductase in human normal and tumor tissue: effects of cigarette smoking and alcohol. Int J Cancer. 1990;45:403–409. doi: 10.1002/ijc.2910450304. [DOI] [PubMed] [Google Scholar]
- 83.Marin A, Lopez de Cerain A, Hamilton E, Lewis AD, Martinez-Penuela JM, et al. DT-diaphorase and cytochrome B5 reductase in human lung and breast tumours. Br J Cancer. 1997;76:923–929. doi: 10.1038/bjc.1997.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Malkinson AM, Siegel D, Forrest GL, Gazdar AF, Oie HK, et al. Elevated DT-diaphorase activity and messenger RNA content in human non-small cell lung carcinoma: relationship to the response of lung tumor xenografts to mitomycin Cl. Cancer Res. 1992;52:4752–4757. [PubMed] [Google Scholar]
- 85.Lyn-Cook BD, Yan-Sanders Y, Moore S, Taylor S, Word B, et al. Increased levels of NAD(P)H: quinone oxidoreductase 1 (NQO1) in pancreatic tissues from smokers and pancreatic adenocarcinomas: A potential biomarker of early damage in the pancreas. Cell Biol Toxicol. 2006;22:73–80. doi: 10.1007/s10565-006-0156-3. [DOI] [PubMed] [Google Scholar]
- 86.Long2nd DJ, Waikel RL, Wang XJ, Perlaky L, Roop DR, et al. NAD(P)H:quinone oxidoreductase 1 deficiency increases susceptibility to benzo(a)pyrene-induced mouse skin carcinogenesis. Cancer Res. 2000;60:5913–5915. [PubMed] [Google Scholar]
- 87.Long2nd DJ, Waikel RL, Wang XJ, Roop DR, Jaiswal AK. NAD(P)H:quinone oxidoreductase 1 deficiency and increased susceptibility to 7,12-dimethylbenz[a]-anthracene-induced carcinogenesis in mouse skin. J Natl Cancer Inst. 2001;93:1166–1170. doi: 10.1093/jnci/93.15.1166. [DOI] [PubMed] [Google Scholar]
- 88.Kelsey KT, Ross D, Traver RD, Christiani DC, Zuo ZF, et al. Ethnic variation in the prevalence of a common NAD(P)H quinone oxidoreductase polymorphism and its implications for anti-cancer chemotherapy. Br J Cancer. 1997;76:852–854. doi: 10.1038/bjc.1997.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rauth AM, Goldberg Z, Misra V. DT-diaphorase: possible roles in cancer chemotherapy and carcinogenesis. Oncol Res. 1997;9:339–349. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Score of quality assessment.
(DOC)