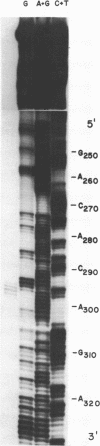
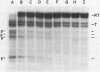
Abstract
We have determined the nucleotide sequences of four termination sites recognized by Bacillus subtilis RNA polymerase. These sites are located in the terminally repeated segment of the bacteriophage SP01 genome, where most early phage transcription occurs. The SP01 terminators have structures that are similar to those recognized by Escherichia coli RNA polymerase, containing a region of dyad symmetry followed by a stretch of HMU residues in the noncoding DNA strand (HMU is substituted for T in SP01 DNA). We note that in a terminator that is only 60% efficient in vitro, there is a greater distance between these two conserved elements that exists in more efficient terminators. We also find that RNA polymerase molecules which elongate a transcript through a partial terminator often pause at this site.
Full text
PDF


















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M. Control of transcription termination. Annu Rev Biochem. 1978;47:967–996. doi: 10.1146/annurev.bi.47.070178.004535. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Brutlag D., Atkinson M. R., Setlow P., Kornberg A. An active fragment of DNA polymerase produced by proteolytic cleavage. Biochem Biophys Res Commun. 1969 Dec 4;37(6):982–989. doi: 10.1016/0006-291x(69)90228-9. [DOI] [PubMed] [Google Scholar]
- Casey J., Davidson N. Rates of formation and thermal stabilities of RNA:DNA and DNA:DNA duplexes at high concentrations of formamide. Nucleic Acids Res. 1977;4(5):1539–1552. doi: 10.1093/nar/4.5.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelm B. K., Beard C., Geiduschek E. P. Changes in the association between Bacillus subtilis RNA polymerase core and two specificity-determining subunits during transcription. Biochemistry. 1981 Nov 10;20(23):6564–6569. doi: 10.1021/bi00526a007. [DOI] [PubMed] [Google Scholar]
- Chelm B. K., Romeo J. M., Brennan S. M., Geiduschek E. P. A transcriptional map of the bacteriophage SPO1 genome. III. A region of early and middle promoters (the gene 28 region). Virology. 1981 Jul 30;112(2):572–588. doi: 10.1016/0042-6822(81)90303-2. [DOI] [PubMed] [Google Scholar]
- Englund P. T. Analysis of nucleotide sequences at 3' termini of duplex deoxyribonucleic acid with the use of the T4 deoxyribonucleic acid polymerase. J Biol Chem. 1971 May 25;246(10):3269–3276. [PubMed] [Google Scholar]
- Friedmann T., Brown D. M. Base-specific reactions useful for DNA sequencing: methylene blue--sensitized photooxidation of guanine and osmium tetraoxide modification of thymine. Nucleic Acids Res. 1978 Feb;5(2):615–622. doi: 10.1093/nar/5.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage L. P., Geiduschek E. P. RNA synthesis during bacteriophage SPO1 development: six classes of SPO1 RNA. J Mol Biol. 1971 Apr 28;57(2):279–297. doi: 10.1016/0022-2836(71)90346-9. [DOI] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Gryczan T. J., Grandi G., Hahn J., Grandi R., Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 1980 Dec 20;8(24):6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen U., Tenen D. G., Livingston D. M., Sharp P. A. T antigen repression of SV40 early transcription from two promoters. Cell. 1981 Dec;27(3 Pt 2):603–613. doi: 10.1016/0092-8674(81)90402-5. [DOI] [PubMed] [Google Scholar]
- Lee F., Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G., Pero J. Conserved nucleotide sequences in temporally controlled bacteriophage promoters. J Mol Biol. 1981 Oct 25;152(2):247–265. doi: 10.1016/0022-2836(81)90242-4. [DOI] [PubMed] [Google Scholar]
- Mandecki W., Reznikoff W. S. A lac promoter with a changed distance between -10 and -35 regions. Nucleic Acids Res. 1982 Feb 11;10(3):903–912. doi: 10.1093/nar/10.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F. H., Tinoco I., Jr DNA-RNA hybrid duplexes containing oligo(dA:rU) sequences are exceptionally unstable and may facilitate termination of transcription. Nucleic Acids Res. 1980 May 24;8(10):2295–2299. doi: 10.1093/nar/8.10.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller K. G., Sollner-Webb B. Transcription of mouse rRNA genes by RNA polymerase I: in vitro and in vivo initiation and processing sites. Cell. 1981 Nov;27(1 Pt 2):165–174. doi: 10.1016/0092-8674(81)90370-6. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H., Kutter E., Nakanishi M. A restriction map of the bacteriophage T4 genome. Mol Gen Genet. 1980;179(2):421–435. doi: 10.1007/BF00425473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Termination of transcription and its regulation in the tryptophan operon of E. coli. Cell. 1981 Apr;24(1):10–23. doi: 10.1016/0092-8674(81)90496-7. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Rubin C. M., Schmid C. W. Pyrimidine-specific chemical reactions useful for DNA sequencing. Nucleic Acids Res. 1980 Oct 24;8(20):4613–4619. doi: 10.1093/nar/8.20.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogin M. L., Pace N. R. Nucleotide sequence of 5 S ribosomal RNA precursor from Bacillus subtilis. J Biol Chem. 1976 Jun 10;251(11):3480–3488. [PubMed] [Google Scholar]
- Stefano J. E., Gralla J. D. Spacer mutations in the lac ps promoter. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1069–1072. doi: 10.1073/pnas.79.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkington C., Pero J. Distinctive nucleotide sequences of promoters recognized by RNA polymerase containing a phage-coded "sigma-like" protein. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5465–5469. doi: 10.1073/pnas.76.11.5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., White R. L., Davis R. W. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. L., Hogness D. S. R loop mapping of the 18S and 28S sequences in the long and short repeating units of Drosophila melanogaster rDNA. Cell. 1977 Feb;10(2):177–192. doi: 10.1016/0092-8674(77)90213-6. [DOI] [PubMed] [Google Scholar]