Abstract
Background
Cutaneous leishmaniasis (CL) is a vector-borne parasitic disease characterized by the presence of one or more lesions on the skin that usually heal spontaneously after a few months. Most cases of CL worldwide occur in Southwest Asia, Africa and South America, and a number of cases have been reported among troops deployed to Afghanistan. No vaccines are available against this disease, and its treatment relies on chemotherapy. The aim of this study was to characterize parasites isolated from Canadian soldiers at the molecular level and to determine their susceptibility profile against a panel of antileishmanials to identify appropriate therapies.
Methodology/Principal Findings
Parasites were isolated from skin lesions and characterized as Leishmania tropica based on their pulsed field gel electrophoresis profiles and pteridine reductase 1 (PTR1) sequences. Unusually high allelic polymorphisms were observed at several genetic loci for the L. tropica isolates that were characterized. The drug susceptibility profile of intracellular amastigote parasites was determined using an established macrophage assay. All isolates were sensitive to miltefosine, amphotericin B, sodium stibogluconate (Pentostam) and paromomycin, but were not susceptible to fluconazole. Variable levels of susceptibility were observed for the antimalarial agent atovaquone/proguanil (Malarone). Three Canadian soldiers from this study were successfully treated with miltefosine.
Conclusions/Significance
This study shows high heterogeneity between the two L. tropica allelic versions of a gene but despite this, L. tropica isolated from Afghanistan are susceptible to several of the antileishmanial drugs available.
Author Summary
Cutaneous leishmaniasis (CL) is a vector-borne parasitic disease transmitted by the bite of sandflies, resulting in sores on the skin. No vaccines are available, and treatment relies on chemotherapy. CL has been frequently diagnosed in military personnel deployed to Afghanistan and returning from duty. The parasites isolated from Canadian soldiers were characterized by pulsed field gels and by sequencing conserved genes and were identified as Leishmania tropica. In contrast to other Leishmania species, high allelic polymorphisms were observed at several genetic loci for the L. tropica isolates that were characterized. In vitro susceptibility testing in macrophages showed that all isolates, despite their genetic heterogeneity, were sensitive to most antileishmanial drugs (antimonials, miltefosine, amphotericin B, paromomycin) but were insensitive to fluconazole. This study suggests a number of therapeutic regimens for treating cutaneous leishmaniasis caused by L. tropica among patients and soldiers returning from Afghanistan. Canadian soldiers from this study were successfully treated with miltefosine.
Introduction
Cutaneous leishmaniasis (CL) is a vector-borne parasitic disease characterized by one or more sores or nodules on the skin that often heal spontaneously after a few months, resulting in scar formation. This disease has been frequently diagnosed in military personnel who were returning from duty in Southwest Asia [1], [2], with several outbreaks observed in troops deployed to Iraq [3] and Afghanistan [2], [4]. Currently, Kabul is believed to be the largest focus of CL worldwide, having an estimated incidence of 67,500 new cases per annum [5]. Whereas CL in Iraq has been mostly caused by Leishmania major, CL in Afghanistan can either be due to Leishmania tropica or Leishmania major [6], and differences in clinical features have been observed between the two species. Notably, L. tropica tends to cause more chronic infections and may rarely progress to a systemic form of the disease termed viscerotropic leishmaniasis, a situation requiring special attention [7].
There is a lack of consensus about the best therapeutic options for the treatment of CL, mainly due to the lack of properly controlled clinical trials [8]. Because of the self-healing nature of the illness, the treatment of CL depends on several factors such as the site and number of lesions, the aetiology of the disease, and personal preferences. One of the main therapeutic options that has been used for the treatment of CL for many years relied on the local or systemic administration of pentavalent antimony [9]. Because Leishmania species are susceptible to heat, the local application of radio frequency to generate heat at the site of the lesions was also shown to yield cure rates equivalent to systemic pentavalent antimony [10], [11]. Nonetheless, the availability of effective oral treatments would constitute attractive therapeutic options against CL, and there is evidence of benefit for the use of oral triazoles like itraconazole and fluconazole against L. tropica and L. major, respectively [8]. Miltefosine, another orally administered drug, was shown to be an effective treatment against visceral leishmaniasis in India [12] and cutaneous leishmaniasis in South America [13], but there is only limited data about its efficacy against CL in Southwest Asia [14]–[16].
In this report, we describe the molecular characterization and in vitro drug susceptibility profiles of Leishmania parasites isolated from four Canadian soldiers suffering from CL after returning from Afghanistan. Primary treatment based on oral fluconazole failed to improve the appearance of lesions in three of them. We show that L. tropica was responsible for the lesions in every patient and that the parasites are highly heterogeneous but nonetheless remained sensitive to most known antileishmanials.
Methods
Ethics Statement
The skin biopsies were taken after appropriate informed consent was obtained, and as part of the routine patient care. Leishmania parasite isolates were submitted for susceptibility testing in order to assist in the clinical management of individuals with suboptimal response to fluconazole. No additional samples or procedures were done.
Parasites and culture
Fresh tissue samples obtained through biopsy of the skin lesions were collected from three Canadian soldiers who returned from duty in southern Afghanistan with suspected CL lesions at the Department of Medical Microbiology and Infectious Diseases of the University of Manitoba in Winnipeg. Samples were submitted to culture, pathological examination, and PCR analyses. The histology revealed the presence of granulotomatous inflammation. The isolates identified as 017102, 431462, and 072218 underwent routine clinical laboratory studies at the National Reference Center for Parasitology in Montreal, QC. An additional skin lesion sample (identified as 18693) was collected from a Canadian soldier also returning from Afghanistan and suspected of suffering from CL at the CHUQ in Quebec, QC. Parasites were isolated from the biopsy in SDM-79 medium supplemented with 20% heat-inactivated fetal calf serum, 5 µg/ml hemin and 10 µM biopterin at pH 7.0 and 25°C. The molecular characterization of parasites was done at the Centre de Recherche en Infectiologie du Centre de Recherche du CHUL, Quebec, QC. The L. tropica strains 175 and 482, isolated from Iranian patients [17], and L. tropica MHOM/SU/74/K27, obtained from the ATCC, were used as reference isolates.
Pulsed field gel electrophoresis karyotyping
Agarose blocks containing Leishmania cells were prepared as described [18]. Briefly, cells were resuspended in HEPES buffer at a density of 5×108 cells/ml and mixed with low-melting-point agarose. Cells were lysed in the presence of 0.5 M EDTA (pH 9.5), 1% SLS, and proteinase K (500 µg/ml). Their chromosomes were electrophoresed by a BioRad (Hercules, California, United States) contour-clamped homogeneous electric field (CHEF) mapper for separating 0.1–1.0 Mbp DNAs over a period of 27 h. Chromosomes were visualized after ethidium bromide staining.
Sequence analysis
Species identification and heterogeneity were studied by sequencing the pteridine reductase 1 (PTR1), glucose-6-phosphate isomerase (GPI), nucleoside hydrolase 1 (NH1), dihydrofolate reductase-thymidylate synthase (DHFR-TS), stearic acid desaturase (SAD), mannose phosphate isomerase (MPI), aspartate aminotransferase (ASAT), 6-phosphogluconate dehydrogenase (PGD), glucose-6-phosphate dehydrogenase (G6PDH) and cytochrome B (CYTB) genes. Genomic DNA was extracted from mid-log phase parasites using the DNAzol reagent (Invitrogen) as described by the manufacturer. PCR reactions were performed in 50 µl using the primers listed in Table S1 and contained 100 ng of total gDNA, 50 pmol of each primer, 0.2 mM of dNTPs, 1.5 mM of MgCl2 and 5 U of Taq polymerase. Amplification was performed in 30 cycles, each cycle using the following conditions: denaturation at 94°C for 1 min, annealing at 58°C for 1 min and extension at 72°C for 1–2 min (depending on the size of PCR products). A final extension was performed at 72°C for 5 min. PCR products were migrated on agarose gel, purified with the QIAquick Gel Extraction Kit (Qiagen) and sequenced with an ABI Prism 3100 DNA sequencer. The PTR1 sequences obtained were compared to those of eight Leishmania reference isolates using the Lasergene Software (DNASTAR, Inc.).
Phylogenetic analysis
Multiple sequence alignments were performed on the amino acid sequence of the PTR1 coding region using ClustalW [19] with the default settings. The resulting multiple alignments were subjected to phylogenetic analysis using the neighbor-joining method [20] with the Poisson correction distance method as implemented in the MEGA3.1 software [21]. The reliabilities of each branch point were assessed by the analysis of 1000 bootstrap replicates.
Viability test
The 50% inhibitory concentrations (IC50) of drugs on macrophages were established by using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Briefly, THP-1 cells were differentiated in 96-well flat-bottom microtiter plates in a volume of 100 µl of complete RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum and 20 ng/ml phorbol myristate acetate. Plates were incubated at 37°C in the presence of 5% CO2 for 3 days. Drugs were added at 1/10 of the final concentration in a volume of 10 µl in duplicate. After 96 h of incubation, 10 µl of MTT (10 mg/ml) was added to each well and plates were further incubated for 4 h. The enzymatic reaction was stopped by the addition of 100 µl of 50% ethanol-10% sodium dodecyl sulfate. The plates were incubated for an additional 30 minutes under agitation at room temperature before reading the optical density at 570 nm with a 96-well scanner. The viability assays were performed in duplicates. As a control, the cytotoxicity of reagents used to solubilize the drugs was determined and no substantial toxicity was found.
Drug susceptibility assays
L. tropica promastigote parasites were transfected with the firefly luciferase-containing vector pSP1.2 LUC αHYGα as previously described [22]. THP-1 cells were differentiated by incubation at 37°C in the presence of 5% CO2 for 3 days in complete RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum and 20 ng/ml phorbol myristate acetate. The cells were washed with pre-warmed medium and subsequently infected with L. tropica promastigotes at a parasite/macrophage ratio of 15∶1 for 3 h. Non-internalized parasites were removed by several washes. Luciferase activity was measured after 4 days of exposure to fluconazole, Pentostam, amphotericin B, miltefosine, paromomycin or Malarone as described elsewhere [23].
Results
Molecular characterization of Leishmania isolates
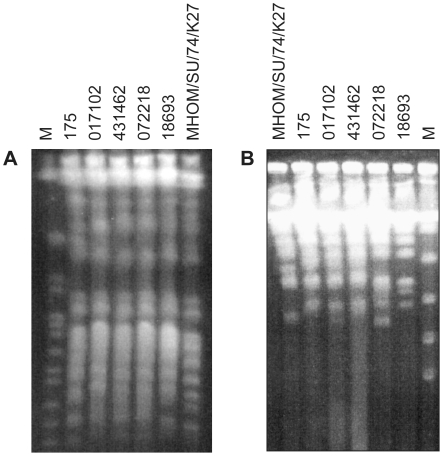
Parasites recovered from biopsy samples of four Canadian military personnel who returned from deployment in Kandahar, Afghanistan, with clinical manifestations of CL were characterized by pulsed field gel electrophoresis (PFGE). PFGE conditions optimized for the analysis of larger chromosomes did not show any major differences in the chromosome numbers and sizes between our isolates (Fig. 1A) and revealed that they were genetically closely related to the ATCC L. tropica strain MHOM/SU/74/K27 and to a L. tropica isolate recovered from a patient suffering from CL in Iran (L. tropica 175) [17], [24]. The analysis of smaller chromosomes revealed considerable karyotype differences, however (Fig. 1B).
Figure 1. Karyotypes of Afghan and Iranian Leishmania tropica isolates as characterized by PFGE.
Cells were embedded and lyzed in agarose and their chromosomes were electrophoresed and stained with ethidium bromide. A. 600–1300 kb electrophoresis B. 100–500 kb electrophoresis. The field isolates 017102, 431462, 072218 and 18693 from Afghanistan have closely related karyotype to the L. tropica reference strains 175 and MHOM/SU/74/K27. M, yeast chromosomes molecular weight marker (BioRad).
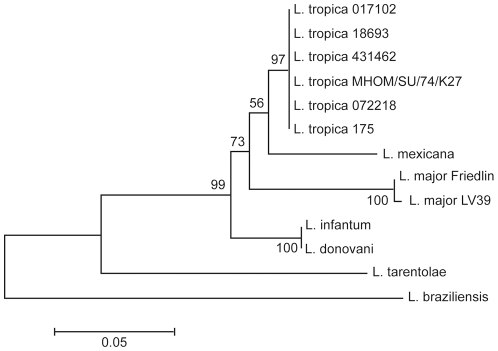
The isolates were further characterized on the basis of the pteridine reductase 1 (PTR1) sequence [17]. PCR fragments of the coding region of PTR1 were amplified from genomic DNA extracted from the clinical isolates and sequenced. The sequences generated were compared to those of eight Leishmania reference strains and were shown to be closely related to L. tropica sequences (data not shown). A neighbor-joining phylogenetic analysis generated from the translated PTR1 sequences further confirmed that the four CL strains derived from Canadian soldiers were L. tropica parasites (Fig. 2).
Figure 2. Phylogenetic analysis of the PTR1 sequences.
Amino acid sequences were aligned using the ClustalW algorithm. The resulting multiple alignment was subjected to phylogenetic analysis by using the neighbor-joining method with Poisson correction as implemented in the MEGA3.1 software. The field isolates 017102, 431462, 072218, and 18693 from Afghanistan are clustering with the L. tropica reference strains 175 and MHOM/SU/74/K27. The reliabilities of each branch point were assessed by the analysis of 1000 bootstrap replicates.
Leishmania tropica genetic heterozygosity
The PTR1 nucleotide sequences of the four L. tropica isolated from Canadian soldiers revealed the presence of single nucleotide polymorphisms (SNPs) at five different positions (Table 1). The changes in nucleotide were conservative (Table 1), and the same polymorphisms were also observed in two other strains of L. tropica (strains 482 and MHOM/SU/74/K27) that we have analyzed (Table 2). The heterozygous gene sequences were detected as split peaks in the chromatogram generated by the sequencing of the PTR1 locus in both directions, using DNA extracted from populations of parasites. This type of polymorphism was not observed when sequencing the PTR1 gene of L. infantum and L. major (Table 2). To assess whether these polymorphisms corresponded to population heterogeneity or to parasite heterozygocity, the PTR1 sequence of cloned parasites from three distinct L. tropica strains (MHOM/SU/74/K27, 072218 and 482) was determined (3 clones for each strain). Again, the same PTR1 polymorphisms were detected in every clone tested. Each allele was detected at a frequency of 50%, which suggests that parasites were harbouring two distinct alleles. The same two alleles were detected in every L. tropica strain studied (Table 1), which is reflected by the homogenous clustering of the L. tropica isolates in the neighbour-joining phylogenetic analysis (Fig. 2).
Table 1. Polymorphisms in L. tropica isolatesa , b.
| Genesc | Nucleotidesd | Amino acids |
| PTR1 | G 243 A | Ala 81 Ala |
| G 561 T | Pro 187 Pro | |
| T 624 A | Ala 208 Ala | |
| A 642 G | Pro 214 Pro | |
| T 690 C | Ala 230 Ala | |
| NH1 | T 243 A | Ile 81 Ile |
| A 698 G | His 233 Arg | |
| C 752 G | Pro 251 Arg | |
| G 828 T | Ala 276 Ala | |
| DHFRTS | A 911 G | Gln 304 Arg |
| G 1095 A | Gln 365 Gln | |
| A 1149 G | Leu 383 Leu | |
| A 1469 C | Glu 490 Ala | |
| SAD | C 189 T | Ser 63 Ser |
| C 1251 T | Gly 417 Gly | |
| T 1335 A | Asp 445 Glu | |
| GPI | G 109 T | Ala 37 Ser |
| C 135 G | Ala 45 Ala | |
| G 230 A | Ser 77 Asn | |
| C 383 T | Ala 128 Val | |
| C 699 T | Asn 233 Asn | |
| C 888 T | Val 296 Val | |
| T 1290 C | Ile 430 Ile | |
| PGD | G 54 C | Ala 18 Ala |
| A 114 G | Thr 38 Thr | |
| G 249 T | Thr 83 Thr | |
| A 447 G | Pro 149 Pro | |
| A 489 G | Ala 163 Ala | |
| G 623 T | Arg 208 Leu | |
| G 666 T | Glu 222 Asp | |
| A 679 G | Asn 227 Asp | |
| G 876 C | Met 292 Ile | |
| A 884 G | Tyr 295 Cys | |
| G 903 C | Ala 301 Ala | |
| G 1056 C | Leu 352 Leu | |
| A 1228 G | Asn 410 Lys | |
| C1293 T | Ala 431 Ala | |
| G 1383 C | Gly 461 Gly | |
| ASAT | A 55 C | Lys 19 Gln |
| A 978 C | Ala 326 Ala | |
| C 1141 T | Pro 381 Ser | |
| G6PDH | A 178 G | Asn 60 Asp |
| A 198 A | Glu 66 Glu | |
| C 294 T | Gly 98 Gly | |
| A 463 G | Asn 155 Asp | |
| T 669 C | Gly 223 Gly | |
| A 1038 G | Pro 346 Pro | |
| A 1039 G | Ile 347 Val | |
| G 1083 C | Ala 361 Ala | |
| C 1191 T | Gly 397 Gly | |
| A 1542 G | Pro 514 Pro |
Nucleotide polymorphisms and corresponding amino acid changes are shown.
The polymorphisms in bold cause an amino acid substitution.
The polymorphisms indicated were common to every L. tropica isolates analyzed, except for PGD, G6PDH, and SAD, which were not polymorphic in L. tropica MHOM/SU/74/K27.
For nucleotide numbering, +1 corresponds to the A of the ATG translation initiation codon.
PTR1 – pteridine reductase 1; NH1 – nucleoside hydrolase 1; DHFRTS – dihydrofolate reductase-thymidylate synthase; SAD – stearic acid desaturase; GPI – glucose-6-phosphate isomerase; PGD – 6-phosphogluconate dehydrogenase; ASAT – aspartate aminotransferase; G6PDH – glucose-6-phosphate dehydrogenase.
Table 2. Single nucleotide polymorphisms within Leishmania species for 10 genesa.
| Gene | GPI | PTR1 | NH1 | PGD | ASAT | G6PDH | DHFR-TS | SAD | MPI | CYTB |
| Chromosome | 12 | 23 | 29 | 35 | 24 | 34 | 6 | 14 | 32 | * |
| L. majorFriedlin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L. infantumJPCM5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| L. infantumLEM 3843 | 0 | 0 | 0 | ND | ND | ND | ND | ND | ND | ND |
| L. donovani infantum | 0 | 0 | 0 | ND | ND | ND | ND | ND | ND | ND |
| L. tropicaMHOM/SU/74/K27 | 7 | 5 | 4 | 0 | 3 | 0 | 4 | 0 | 0 | 0 |
| L. tropicaIRAN 482 | 7 | 5 | 4 | 15 | 6 | 10 | 4 | 3 | 0 | 0 |
| L. tropica072218 | 7 | 5 | 4 | 15 | 6 | 10 | 4 | 3 | 0 | 0 |
ND – not done.
For each gene, the number of heterozygous sites is indicated.
*: The CYTB gene is located on kinetoplast DNA.
Abreviations: PTR1 – pteridine reductase 1; NH1 – nucleoside hydrolase 1; DHFR-TS – dihydrofolate reductase-thymidylate synthase; SAD – stearic acid desaturase; GPI – glucose-6-phosphate isomerise; PGD – 6-phosphogluconate dehydrogenase; ASAT – aspartate aminotransferase; G6PDH – glucose-6-phosphate dehydrogenase; MPI – mannose phosphate isomerise; CYTB – Cytochrome B.
To assess the extent of the genetic polymorphism in our panel of L. tropica strains, nine additional genes located on distinct chromosomes, i.e. glucose-6-phosphate isomerase (GPI), nucleoside hydrolase 1 (NH1), dihydrofolate reductase-thymidylate synthase (DHFR-TS), stearic acid desaturase (SAD), mannose phosphate isomerase (MPI), aspartate aminotransferase (ASAT), 6-phosphogluconate dehydrogenase (PGD), glucose-6-phosphate dehydrogenase (G6PDH), and cytochrome B (CYTB), were also sequenced in clones of strains 482, 072218 and MHOM/SU/74/K27. Heterozygous sites were observed at every locus (Table 1) except for MPI and CYTB (Table 2). In addition, the same alleles were detected in every L. tropica strains studied, except for the ASAT gene, which had three additional polymorphic sites common to L. tropica 482 and 072218 that were absent from L. tropica MHOM/SU/74/K27 (Table 2), and the PGD, G6PDH, and SAD genes, which were not polymorphic in L. tropica MHOM/SU/74/K27 (Table 2). In contrast to the PTR1 locus, however, the polymorphisms observed at the NH1, DHFR-TS, SAD, PGD, G6PDH, ASAT, and GPI loci were non-conservative for at least one position, with some genes having several non-conservative heterozygous sites (Table 1). These polymorphisms were not observed in other species outside L. tropica (Table 2). To rule out polymerase errors, the sequencing of at least three independent PCR products was done for each particular nucleotide position, as it would be quite rare that the presence of the same SNPs in different PCR reaction comes from polymerase errors. Moreover, for some genes (for example PTR1), PCR products were TA cloned and independent clones were sequenced for the confirmation of the SNPs using primers within the pGEM-T easy vector, hence ruling out the possibility of polymerase errors.
Drug susceptibility profiles of L. tropica clinical isolates
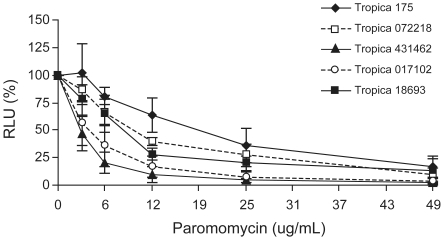
Three of the four Canadian soldiers were initially treated with oral fluconazole without any clinical improvements. To establish whether the therapeutic failure was due to parasites unresponsive to fluconazole and to test whether these parasites were sensitive to classical antileishmanials, in vitro susceptibility testing was performed with the four L. tropica isolates using the human monocyte cell line THP-1 and recombinant parasites transfected with the firefly luciferase gene. The latter system is a convenient and rapid quantitative method to monitor the growth of intracellular parasites [23]. Growth was compared between mock-treated parasites and parasites exposed to fluconazole, Pentostam, amphotericin B, miltefosine, and paromomycin. We observed that L. tropica intracellular amastigotes were insensitive to fluconazole at concentrations that were achievable in vitro (Table 3). Fluconazole was highly active when we tested it against C. albicans (results not shown). In contrast, when compared to our L. tropica reference strain 175, all L. tropica isolates tested were sensitive to amphotericin B, miltefosine, Pentostam, and paromomycin as intracellular parasites (Table 3 and Figure 3). The only exception was L. tropica 18693, for which a slightly higher miltefosine EC50 was observed in comparison to the other L. tropica strains (Table 3).
Table 3. Susceptibility of L. tropica clinical isolates to antileishmanial drugs.
| Strains | EC50 a | |||||
| AmphotericinB (µg/ml) | Paromomycin (µg/ml) | Miltefosine (µg/ml) | Pentostam (µg/ml) | Malarone (µg/ml) | Fluconazole(µg/ml) | |
| 175 | 0.51±0.08 | 19.0±8.1 | 0.19±0.07 | 300±32 | >8 | >275 |
| 017102 | 0.59±0.29 | 7.4±1.7 | 0.11±0.004 | 188±39 | 6.6±0.1 | >275 |
| 431462 | 0.32±0.18 | 3.2±0.9 | 0.09±0.004 | 142±4 | 3.6±0.6 | >275 |
| 072218 | 0.51±0.07 | 9.8±1.8 | 0.15±0.07 | 142±12 | 4.4±1.1 | >275 |
| 18693 | 0.72±0.33 | 5.8±4.8 | 0.65±0.33 | ND | >8 | >275 |
Determined as amastigotes in THP-1 cells.
Figure 3. Paromomycin susceptibility of Leishmania tropica field isolates recovered from soldiers suffering from CL.
The four L. tropica isolates from Afghanistan are sensitive to paromomycin as intracellular amastigotes. Intracellular parasites were incubated for 4 days with the indicated concentrations of paromomycin. The mean of two independent experiments done in duplicate is shown. Values are represented as numbers of relative light units (RLU).
One soldier infected by CL had to travel to Central Africa and was on Malarone anti-malaria prophylaxis. Intriguingly, his cutaneous lesions cicatrized while being on Malarone prophylaxis, so we tested whether Malarone had any activity against L. tropica isolates using the intracellular amastigote assay. The toxicity of Malarone to the THP-1 cells was first established by MTT viability assay, and this cell line was found to display a Malarone IC50 of 32 ug/ml. No THP-1 cytotoxicity was observed for Malarone concentrations up to 10 ug/ml. Using drug concentrations below 10 ug/ml, we found that three L. tropica strains were sensitive to Malarone as intracellular amastigotes (Table 3), including the strain that was isolated from the patient whose CL regressed during Malarone prophylaxis.
Discussion
We describe here the drug susceptibility and molecular characterization of L. tropica isolates derived from Canadian soldiers returning from Afghanistan. The isolates were identified as L. tropica by phylogenetic studies based on the PTR1 sequence, an approach proven to be useful for the molecular identification of Leishmania species [17]. Moreover, the PFGE karyotypes of the recovered Leishmania parasites were similar to those of L. tropica reference strains. This is consistent with epidemiological data that showed the majority of CL cases in Afghanistan being due to this species [5], [25]. Interestingly, the sequence of PTR1 revealed several SNPs in distinct L. tropica isolates. This phenomenon appeared to be widespread across the L. tropica genome, as it was also observed at other genetic loci on different chromosomes. Most of the loci analyzed code for proteins that are part of the panel of enzymes used for the characterization of Leishmania species by multilocus enzyme electrophoresis [26]. Among these, six (GPI, NH1, ASAT, G6PDH, PGD, and MPI) were further shown to be useful markers for the molecular characterization of Leishmania strains and species [27]–[29]. CYTB was chosen as a mitochondrial gene representative, since it has also been reported to be phylogenetically informative [30], [31]. The SAD and DHFR-TS loci were randomly chosen. DNA sequencing of cloned parasites revealed a number of heterozygous sites at these loci, some of which led to non-conservative changes. Although the prevailing mode of reproduction of Leishmania appears to be clonal [32], heterozygosity at several sites within genes or at distinct loci is suggestive of genetic exchange between strains [27], and this phenomenon has previously been observed in other Leishmania species [27], [28],[32]–[35]. Most of these studies used microsatellite markers with high mutation rates as indicators of heterozygocity, however, and this is the first report about extensive heterozygocity within coding regions in L. tropica. Nonetheless, the heterozygosity of the L. tropica isolates appears to be fixed, the same alleles being found among strains for most of the loci studied except for the reference L. tropica MHOM/SU/74/K27. This is suggestive of clonal propagation within foci of endemicity and is consistent with the anthroponotic mode of transmission of L. tropica in urban and peri-urban environments of Afghanistan [5]. L. tropica parasites were known to display genetic heterogeneity at the population level [34]–[37] and to be responsible for a spectrum of clinical manifestations including cutaneous, chronic, or viscerotropic leishmaniasis [7]. Unfortunately, the small number of isolates available for analysis prevented correlating heterozygocity with clinical data or drug susceptibility. However, this seems to be unique to L. tropica since other species did not show this level of allelic polymorphism (Table 2).
Although CL is generally self-limiting, the complexity of the clinical spectrum associated with L. tropica infections emphasizes the need for treatment. Evidence suggested that the disruption of ergosterol biosynthesis by oral azoles is an effective treatment against CL [38], [39]. However, a species-specific effect was found to be important to the clinical outcome conferred by azole molecules, with itraconazole and fluconazole being more active against L. tropica and L. major, respectively [8]. Here, the failure of oral fluconazole to improve the appearance of cutaneous lesions was indeed explained by the intrinsic resistance of our L. tropica isolates, the amastigote parasites being insensitive to the highest fluconazole concentration achievable in vitro using an established intracellular assay (Table 3). All isolates were sensitive to the other drugs tested, however, with the exception of Malarone, for which variable levels of susceptibility were observed. While anecdotal, CL regressed in one soldier during Malarone prophylaxis. Although we cannot exclude spontaneous healing, it might be worthwhile to evaluate the usefulness of this drug against CL in properly controlled experiments.
Miltefosine is an orally administered antileishmanial approved for the treatment of visceral leishmaniasis in India [12], with demonstrated efficacy against CL in some regions of South America [13]. In contrast, mostly sporadic data have been reported regarding the efficacy of miltefosine against CL in Southwest Asia [14]–[16]. Based on the results of our drug susceptibility screening, soldiers were treated with miltefosine and healing of their CL lesions was observed [40]. The patient treated with Malarone received miltefosine but elected to discontinue therapy due to abdominal pain and in the face of a contracting lesion. The other soldiers tolerated medication well and lesions resolved at follow up.
Supporting Information
Primers used in this study.
(DOC)
Acknowledgments
We thank Dr. Philippe Leprohon for a critical revision of the manuscript.
Footnotes
The authors have declared that no competing interests exist.
This work was funded by Canadian Institute of Health Research (CIHR) grants to M.O. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. M.O. also holds the Canada Research Chair in Antimicrobial Resistance and is a Burroughs Wellcome Fund Scholar in Molecular Parasitology.
References
- 1.Goldrick BA. Another hazard of war: infectious diseases: leishmaniasis, malaria threaten troops, millions worldwide. Am J Nurs. 2004;104:64–66. doi: 10.1097/00000446-200412000-00030. [DOI] [PubMed] [Google Scholar]
- 2.van Thiel PP, Leenstra T, de Vries HJ, van der Sluis A, van Gool T, et al. Cutaneous leishmaniasis (Leishmania major infection) in Dutch troops deployed in northern Afghanistan: epidemiology, clinical aspects, and treatment. Am J Trop Med Hyg. 2010;83:1295–1300. doi: 10.4269/ajtmh.2010.10-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weina PJ, Neafie RC, Wortmann G, Polhemus M, Aronson NE. Old world leishmaniasis: an emerging infection among deployed US military and civilian workers. Clin Infect Dis. 2004;39:1674–1680. doi: 10.1086/425747. [DOI] [PubMed] [Google Scholar]
- 4.Faulde M, Schrader J, Heyl G, Amirih M, Hoerauf A. Zoonotic cutaneous leishmaniasis outbreak in Mazar-e Sharif, northern Afghanistan: an epidemiological evaluation. Int J Med Microbiol. 2008;298:543–550. doi: 10.1016/j.ijmm.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 5.Reithinger R, Mohsen M, Aadil K, Sidiqi M, Erasmus P, et al. Anthroponotic cutaneous leishmaniasis, Kabul, Afghanistan. Emerg Infect Dis. 2003;9:727–729. doi: 10.3201/eid0906.030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faulde M, Schrader J, Heyl G, Amirih M. Differences in transmission seasons as an epidemiological tool for characterization of anthroponotic and zoonotic cutaneous leishmaniasis in northern Afghanistan. Acta Trop. 2008;105:131–138. doi: 10.1016/j.actatropica.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 7.Magill AJ, Grogl M, Gasser RA, Jr, Sun W, Oster CN. Visceral infection caused by Leishmania tropica in veterans of Operation Desert Storm. N Engl J Med. 1993;328:1383–1387. doi: 10.1056/NEJM199305133281904. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez U, Pinart M, Reveiz L, Alvar J. Interventions for Old World cutaneous leishmaniasis. Cochrane Database Syst Rev. 2008:CD005067. doi: 10.1002/14651858.CD005067.pub3. [DOI] [PubMed] [Google Scholar]
- 9.Wortmann G, Miller RS, Oster C, Jackson J, Aronson N. A randomized, double-blind study of the efficacy of a 10- or 20-day course of sodium stibogluconate for treatment of cutaneous leishmaniasis in United States military personnel. Clin Infect Dis. 2002;35:261–267. doi: 10.1086/341406. [DOI] [PubMed] [Google Scholar]
- 10.Aronson NE, Wortmann GW, Byrne WR, Howard RS, Bernstein WB, et al. A randomized controlled trial of local heat therapy versus intravenous sodium stibogluconate for the treatment of cutaneous Leishmania major infection. PLoS Negl Trop Dis. 4:e628. doi: 10.1371/journal.pntd.0000628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reithinger R, Aadil K, Kolaczinski J, Mohsen M, Hami S. Social impact of leishmaniasis, Afghanistan. Emerg Infect Dis. 2005;11:634–636. doi: 10.3201/eid1104.040945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sundar S, Jha TK, Thakur CP, Engel J, Sindermann H, et al. Oral miltefosine for Indian visceral leishmaniasis. N Engl J Med. 2002;347:1739–1746. doi: 10.1056/NEJMoa021556. [DOI] [PubMed] [Google Scholar]
- 13.Soto J, Arana BA, Toledo J, Rizzo N, Vega JC, et al. Miltefosine for new world cutaneous leishmaniasis. Clin Infect Dis. 2004;38:1266–1272. doi: 10.1086/383321. [DOI] [PubMed] [Google Scholar]
- 14.Stojkovic M, Junghanss T, Krause E, Davidson RN. First case of typical Old World cutaneous leishmaniasis treated with miltefosine. Int J Dermatol. 2007;46:385–387. doi: 10.1111/j.1365-4632.2007.03153.x. [DOI] [PubMed] [Google Scholar]
- 15.Mohebali M, Fotouhi A, Hooshmand B, Zarei Z, Akhoundi B, et al. Comparison of miltefosine and meglumine antimoniate for the treatment of zoonotic cutaneous leishmaniasis (ZCL) by a randomized clinical trial in Iran. Acta Trop. 2007;103:33–40. doi: 10.1016/j.actatropica.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 16.van Thiel PP, Leenstra T, Kager PA, de Vries HJ, van Vugt M, et al. Miltefosine treatment of Leishmania major infection: an observational study involving Dutch military personnel returning from northern Afghanistan. Clin Infect Dis. 50:80–83. doi: 10.1086/648726. [DOI] [PubMed] [Google Scholar]
- 17.Hadighi R, Mohebali M, Boucher P, Hajjaran H, Khamesipour A, et al. Unresponsiveness to Glucantime treatment in Iranian cutaneous leishmaniasis due to drug-resistant Leishmania tropica parasites. PLoS Med. 2006;3:e162. doi: 10.1371/journal.pmed.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grondin K, Papadopoulou B, Ouellette M. Homologous recombination between direct repeat sequences yields P-glycoprotein containing amplicons in arsenite resistant Leishmania. Nucleic Acids Res. 1993;21:1895–1901. doi: 10.1093/nar/21.8.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 22.Sereno D, Roy G, Lemesre JL, Papadopoulou B, Ouellette M. DNA transformation of Leishmania infantum axenic amastigotes and their use in drug screening. Antimicrob Agents Chemother. 2001;45:1168–1173. doi: 10.1128/AAC.45.4.1168-1173.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roy G, Dumas C, Sereno D, Wu Y, Singh AK, et al. Episomal and stable expression of the luciferase reporter gene for quantifying Leishmania spp. infections in macrophages and in animal models. Mol Biochem Parasitol. 2000;110:195–206. doi: 10.1016/s0166-6851(00)00270-x. [DOI] [PubMed] [Google Scholar]
- 24.Hadighi R, Boucher P, Khamesipour A, Meamar AR, Roy G, et al. Glucantime-resistant Leishmania tropica isolated from Iranian patients with cutaneous leishmaniasis are sensitive to alternative antileishmania drugs. Parasitol Res. 2007;101:1319–1322. doi: 10.1007/s00436-007-0638-0. [DOI] [PubMed] [Google Scholar]
- 25.Ashford RW, Kohestany KA, Karimzad MA. Cutaneous leishmaniasis in Kabul: observations on a ‘prolonged epidemic’. Ann Trop Med Parasitol. 1992;86:361–371. doi: 10.1080/00034983.1992.11812679. [DOI] [PubMed] [Google Scholar]
- 26.Rioux JA, Lanotte G, Serres E, Pratlong F, Bastien P, et al. Taxonomy of Leishmania. Use of isoenzymes. Suggestions for a new classification. Ann Parasitol Hum Comp. 1990;65:111–125. doi: 10.1051/parasite/1990653111. [DOI] [PubMed] [Google Scholar]
- 27.Mauricio IL, Yeo M, Baghaei M, Doto D, Pratlong F, et al. Towards multilocus sequence typing of the Leishmania donovani complex: resolving genotypes and haplotypes for five polymorphic metabolic enzymes (ASAT, GPI, NH1, NH2, PGD). Int J Parasitol. 2006;36:757–769. doi: 10.1016/j.ijpara.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Zemanova E, Jirku M, Mauricio IL, Horak A, Miles MA, et al. The Leishmania donovani complex: genotypes of five metabolic enzymes (ICD, ME, MPI, G6PDH, and FH), new targets for multilocus sequence typing. Int J Parasitol. 2007;37:149–160. doi: 10.1016/j.ijpara.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 29.Tsukayama P, Lucas C, Bacon DJ. Typing of four genetic loci discriminates among closely related species of New World Leishmania. Int J Parasitol. 2009;39:355–362. doi: 10.1016/j.ijpara.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Luyo-Acero GE, Uezato H, Oshiro M, Takei K, Kariya K, et al. Sequence variation of the cytochrome b gene of various human infecting members of the genus Leishmania and their phylogeny. Parasitology. 2004;128:483–491. doi: 10.1017/s0031182004004792. [DOI] [PubMed] [Google Scholar]
- 31.Asato Y, Oshiro M, Myint CK, Yamamoto Y, Kato H, et al. Phylogenic analysis of the genus Leishmania by cytochrome b gene sequencing. Exp Parasitol. 2009;121:352–361. doi: 10.1016/j.exppara.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 32.Lukes J, Mauricio IL, Schonian G, Dujardin JC, Soteriadou K, et al. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci U S A. 2007;104:9375–9380. doi: 10.1073/pnas.0703678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rougeron V, De Meeus T, Hide M, Waleckx E, Bermudez H, et al. Extreme inbreeding in Leishmania braziliensis. Proc Natl Acad Sci U S A. 2009;106:10224–10229. doi: 10.1073/pnas.0904420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwenkenbecher JM, Frohlich C, Gehre F, Schnur LF, Schonian G. Evolution and conservation of microsatellite markers for Leishmania tropica. Infect Genet Evol. 2004;4:99–105. doi: 10.1016/j.meegid.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 35.Schwenkenbecher JM, Wirth T, Schnur LF, Jaffe CL, Schallig H, et al. Microsatellite analysis reveals genetic structure of Leishmania tropica. Int J Parasitol. 2006;36:237–246. doi: 10.1016/j.ijpara.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 36.Schonian G, Schnur L, el Fari M, Oskam L, Kolesnikov AA, et al. Genetic heterogeneity in the species Leishmania tropica revealed by different PCR-based methods. Trans R Soc Trop Med Hyg. 2001;95:217–224. doi: 10.1016/s0035-9203(01)90173-7. [DOI] [PubMed] [Google Scholar]
- 37.Schnur LF, Nasereddin A, Eisenberger CL, Jaffe CL, El Fari M, et al. Multifarious characterization of leishmania tropica from a Judean desert focus, exposing intraspecific diversity and incriminating phlebotomus sergenti as its vector. Am J Trop Med Hyg. 2004;70:364–372. [PubMed] [Google Scholar]
- 38.Berman JD. Activity of imidazoles against Leishmania tropica in human macrophage cultures. Am J Trop Med Hyg. 1981;30:566–569. doi: 10.4269/ajtmh.1981.30.566. [DOI] [PubMed] [Google Scholar]
- 39.Alrajhi AA, Ibrahim EA, De Vol EB, Khairat M, Faris RM, et al. Fluconazole for the treatment of cutaneous leishmaniasis caused by Leishmania major. N Engl J Med. 2002;346:891–895. doi: 10.1056/NEJMoa011882. [DOI] [PubMed] [Google Scholar]
- 40.Keynan Y, Larios OE, Wiseman MC, Plourde M, Ouellette M, et al. Use of oral miltefosine for cutaneous leishmaniasis in Canadian soldiers returning from Afghanistan. Can J Infect Dis Med Microbiol. 2008;19:394–396. doi: 10.1155/2008/802710. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used in this study.
(DOC)