Abstract
Toehold-mediated strand displacement has proven extremely powerful in programming enzyme-free DNA circuits and DNA nanomachines. To achieve multistep, autonomous, and complex behaviors, toeholds must be initially inactivated by hybridizing to inhibitor strands or domains, and then relieved from inactivation in a programmed, timed manner. Although powerful and reasonably robust, this strategy has several drawbacks that limit the architecture of DNA circuits. For example, the combination between toeholds and branch migration (BM) domains is ‘hard-wired’ during DNA synthesis, thus cannot be created or changed during the execution of DNA circuits. To solve this problem, I propose a strategy called ‘associative toehold activation’, where the toeholds and BM domains are connected via hybridization of auxiliary domains during the execution of DNA circuits. Bulged thymidines that stabilize DNA 3-way junctions substantially accelerate strand displacement reactions in this scheme, allowing fast strand displacement initiated by reversible toehold binding. To demonstrate the versatility of the scheme, I show (1) run-time combination of toeholds and BM domains, (2) run-time recombination of toeholds and BM domains, which results in a novel operation ‘toehold switching’, and (3) the design of a simple conformational self-replicator.
Introduction
One of the chief goals of molecular programming is to imbue molecular systems with human-designed programs that guide the self-assembly, self-regulation, and/or self-organization of such systems in order to achieve functions dictated by the designer. Nucleic acids (i.e., DNA and RNA) have emerged as the most prevalent programmable molecules for several theoretical and practical reasons.1-2 The simple rules of Watson-Crick base-pairing allow researchers to design orthogonal interactions (i.e., hybridizations), which can be followed by subsequent, hybridization-dependent events. This simple strategy yielded various applications in biological research such as the polymerase chain reaction (PCR), Southern blotting, Northern blotting, DNA microarrays, molecular beacons, and fluorescent in situ hybridization (FISH). In these applications, sequence-specific hybridization leads to the elongation of a DNA chain, localization of a radiolabeled or fluorescent-labeled DNA, or conformation-change of another DNA. However, in all of these applications the sequence-specific hybridization is essentially irreversible. Therefore, the program that can be embedded in these operations is usually limited to a simple choice between ‘to hybridize’ and ‘not to hybridize’. Complex, dynamic, autonomous behaviors are almost impossible to program using one-step hybridization.
Using nucleic acid-dependent and/or nucleic acid-modifying enzymes such as restriction endonucleases, DNA ligases, and DNA/RNA polymerases, it is possible to make one hybridization event dependent on another hybridization event, either directly or indirectly. By ingeniously exploiting the principle of cascaded hybridization, a number of DNA-programmed molecular automata have been designed and implemented.3-6 It is also possible to exploit this principle using ribozymes or deoxyribozymes.7 All of these systems require the formation of a complex 3D structure at the active center of an enzyme or (deoxy)ribozyme, which exerts catalytic activity to elongate, ligate, or cleave substrate nucleic acids.
It was surprising that hybridization itself can support complex behaviors without the help of any enzyme or (deoxy)ribozyme. Using the scheme called toehold-mediated strand displacement, Yurke et al. first demonstrated the control of a simple DNA nanomachine, a pair of DNA tweezers, by oligonucleotides in a sequence specific manner.8 The general scheme of toehold-mediated strand displacement can be described by Figure S1. Essentially, a short segment of single-stranded DNA (typically 4- to 10-nt) called a toehold binds to its complementary strand and, in doing so, juxtaposes a longer segment of single-stranded DNA (typically ~20 nt) called the branch migration (BM) domain, with a duplex. One strand of the duplex has the same sequence as the BM domain and is eventually displaced by the BM domain.
This scheme proved surprisingly versatile and robust. By cascading the strand displacement events in a manner called ‘toehold exchange’, moderate-scale DNA logic circuits and arbitrary chemical reaction networks can be constructed.9-10 An elegant design of reversible toehold exchange gave rise to entropy-driven catalytic circuits,11 which allows signal amplification and, ultimately, the construction of exceedingly large-scale digital and analog DNA circuits.12-13 The same scheme was also used in designing human-controlled14 and autonomous15 DNA nanomachines, as well as in programming isothermal biomolecular assembly pathways16. Here I use the term ‘DNA circuits’ to specifically refer to enzyme-free DNA circuits driven by hybridization and toehold-mediated strand displacement.
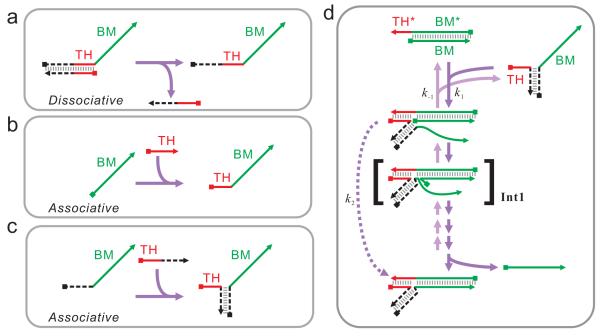
The key consideration in designing DNA circuits is programmed and timed activation of a toehold. This is typically done by first hiding the toehold in an intermolecular or intramolecular duplex, thereby preventing the toehold from hybridizing to its intended target. Upon the occurrence of certain events during the execution of a DNA circuit, the toehold can dissociate from its blocker, and is thus activated (Figure 1a). Therefore, I term this strategy ‘dissociative toehold activation’. Although dissociative toehold activation alone seems sufficient in designing a number of complex molecular systems as shown in the examples cited above, additional levels of control and new schemes of toehold activation are still in need for the design of a wider variety of DNA circuits. For example, the Turberfield group recently developed a concept called remote toehold,17 which decouples toehold binding and strand displacement, and allows additional regulation of strand displacement kinetics via controlling the stiffness of the linker region.
Figure 1.
Scheme of dissociative and associative toehold activation. (a) Dissociative toehold activation. (b) General concept of associative toehold activation. (c) Hybridization-based associative toehold activation. (d) Toehold-mediated strand displacement across a 3-way junction. See text for the discussion on the intermediate Int1 and see Text S3 for discussions on rate constants k1, k−1, and k2. In all panels, toeholds (THs) are shown as red lines. Branch-migration domains (BMs) are shown as green lines. 5′ and 3′ termni are shown as squares and arrows, respectively. Auxiliary domains are shown as dashed back lines. Complementary domains are denoted with asterisks (*).
One drawback of dissociative toehold activation that limits the possible architecture of DNA circuits is the fact that the linkage between the toehold and the BM domain is ‘hard-wired’, i.e., pre-defined during DNA synthesis. Therefore, although a toehold specifies the reactivity of the BM domain, this specification cannot be changed during the execution of DNA circuits. Such run-time toehold assignment can be potentially achieved by attaching the toehold to the BM domain by the DNA circuit itself, a scheme I call ‘associative toehold activation’ (Figure 1b). However, DNA ligation is a complex chemical reaction and likely requires enzymes or (deoxy)ribozymes. To achieve associative toehold activation in DNA circuits completely driven by hybridization and strand displacement, I asked whether the linkage between the toehold and the BM domain can be made by simple hybridization (Figure 1c). I found that although direct hybridization results in inefficient toehold-mediated strand displacement, two bulged bases at the 3-way junction substantially accelerate strand displacement. This rate acceleration enables fast strand displacement followed by reversible toehold binding, which is required to achieve multiple-turnover catalysis in several types of DNA circuits. Encouraged by this result, I investigated and achieved run-time association between toeholds and BM. Combination of dissociative and associative toehold activation should allow an expansion of the architecture of DNA circuits. To illustrate these possibilities, I first demonstrated a novel operation called ‘toehold switching’, in which the combination between toeholds and BM domains is changed during the execution of DNA circuits. I further designed and implemented a simple conformational self-replicator that consists of only 3 DNA strands. Taken together, these results not only demonstrate the possibility of associative toehold activation but also exemplify vast opportunities made possible by this scheme.
Materials and methods
Oligonucleotides
All unmodified and 5′-FAM-labeled oligonucleotides were ordered from Integrated DNA Technology (IDT, Coralville, IA) in desalted grade. Some oligonucleotides (see Figure S2 to S5 of the Supporting Information) were further purified in-house using 8% denaturing PAGE (denaturing agent: 7M urea; gel-making and gel-running buffer: 1x TBE) before use in experiments or further purification. Other fluorophore- and quencher-labeled oligonucleotides were ordered from IDT in RF-HPLC-purified grade.
General procedures for the execution of DNA circuits
All reactions were carried out in 1x TNaKMg buffer (20 mM Tris, pH 7.5; 140 mM NaCl; 5 mM KCl; 5 mM MgCl2; 0.5 mM EDTA) supplemented with 1 μM (dT)21 at 37 °C, in 384 well plates (shallow well, black, polypropylene, Nalge NUNC International, Rochester, NY). The volume of all reactions was 18 μL. The relative fluorescence units (RFUs) of the reporter constructs were monitored in real time by a TECAN Safire plate reader. All fluorescent reporters (including F:Q, S3-F:S3-Q, SA-F:SA-Q, SB-F:SB-Q, and S-F:S-Q) were prepared by hybridization of the two strands at a molar ratio of 2:1 with the quencher-bearing strand in excess to achieve efficient quenching. The procedures to calibrate fluorescence signals and to process the kinetic data are described in Text S1 of the Supporting Information.
Test of single-turnover strand displacement across a 3-way junction
The sequences of oligonucleotides used in this section of the study are shown in Figure S2. D:P duplex was prepared by hybridizing strand D and strand P at a molar ratio of 1.5:1 (D in excess) . The reaction mixtures contained 10 nM of D:P duplex, 200 nM of F:Q duplex, 25 nM Cix (i = 1, 2, or 3; x = 8, 10, or 14), and 40 nM of C30. The function of C30 is to hybridize with excess strand D. It should be noted that D:C30 could act as a competitor of D:P for Cix. However, since the concentration of D:C30 was only ~5 nM and Cix was in large excess, the competition by D:C30 should not significantly affect the estimation of the kinetics of strand displacement reaction between D:P and Cix. The reactions were initiated by the addition of Cix.
Test of multiple-turnover reactions and toehold-switching
The sequences of oligonucleotides used in this section of the study are shown in Figure S3 and S4. Immediately before experiments, the reactant hairpins were refolded separately in 1x TNaK buffer (20 mM Tris, pH 7.5; 140 mM NaCl; 5 mM KCl) by heating to 90 °C for 1 min followed by cooling down to 23 °C at a rate of 0.1 °C/s on a thermocycler. The reactions were initiated by the addition of the hairpin M3 or the mixture of H1A and H1B.
Test of conformational self-replicator
The sequences of oligonucleotides used in this section of the study are shown in Figure S5. The Top:BotO duplex was prepared by first hybridizing ~300 pmole of Top and ~1 nmole of BotO, followed by purification of the duplex using native PAGE (gel-making and gel-running buffer: 0.25x TBE). The gel was stained with SYBR Gold (Invitrogen, Carlsbad, CA) and imaged with a STORM scanner (GE healthcare, Piscataway, NJ). The band corresponding to Top:BotO was excised and the gel slice was crushed. The DNA was then eluted into 1 mL 1x TNaK at 37 °C overnight, and concentrated using an Amicon Ultra 3K concentrator (Millipore, Bellerica, MA) to ~40 μL. This Top:BotO stock was termed DN-pure Top:BotO. To further remove mis-synthesized and/or mis-hybridized Top:BotO, 130 pmole DN-pure Top:BotO was incubated with 300 pmole of BotS in a 37 μL reaction in 1x TNaKMg for 4 hr, followed by native gel-purification, elution, and concentration as described above. The reactions were initiated by the addition of Top:BotO.
Results and discussion
Strand displacement across a 3-way junction
The question to be answered at the outset is whether a toehold joint to the BM domain across a DNA duplex can efficiently initiate strand displacement; or in other words, whether strand displacement across a 3-way junction (Figure 1d) can proceed fast enough. Some hints can be found in previous studies. For example, the Pierce group has reported the catalyzed formation of 3-way junctions from 3 hairpin substrates 16. The last step of this reaction pathway is, in fact, the displacement of the catalyst strand by a portion of the third hairpin, across a 3-way junction, using the hybridization between the second and the third hairpins as a long toehold. The study on remote toehold 17 also suggests that the requirement on the linkage between the toehold and the BM domain is not strict. Single-stranded DNA and chemical linkers (such as polyethylene glycol) of various lengths can all support toehold mediated strand displacement. Therefore the qualitative answer to this question seems to be ‘yes’: when the toehold binding is sufficiently strong, strand displacement across a 3-way junction is likely to happen efficiently.
However, very strong, nearly irreversible toehold interactions are often undesirable. For example, in many catalytic circuits such as the dual-output entropy-driven circuits 11, Seesaw circuits 12, and some types of catalyzed hairpin assembly circuits 18, the catalyst strand needs to dissociate from the product spontaneously for regeneration of the catalyst. Very strong toehold interactions will limit the steady state rate of catalysis by limiting the rate of catalyst regeneration. Practically, for a circuit to execute on a biologically relevant time scale (minutes to hours), the dissociation rate constant of toehold unbinding should be at least on the order of 1 min−1. This means the dissociation constant (Kd) of toehold binding should be no lower than mid-nanomolar range. Therefore, I set out to determine whether reversible toehold binding can mediate efficient strand displacement across a 3-way junction.
A simple DNA circuit (Figure 2a) was designed to measure the rate of strand displacement in real time. In this circuit, an invader DNA C1x, (x = 8, 10, or 14, see below) can potentially displace strand P from duplex D:P (Figure 2a, reaction i). Once displaced, strand P can in turn displace the quencher-bearing strand Q from the fluorescent reporter duplex F:Q, (Figure 2a, reaction ii), leading to an increase of fluorescence signal. It was confirmed that by using a high concentration (>100 nM) of the reporter F:Q, the generation of strand P can be reported immediately (within seconds, data not shown).
Figure 2.
Kinetics of toehold-mediated strand displacement across a 3-way junction. (a) Scheme of the assay. The sequence near the 3-way junction of the invader strand is shown in the insets. For C2x, the CTTG/Y motif (see Text S2) is shown in bold. T: TYE665; rQ: IowaBlack RQ. (b and c) Kinetics of toehold-mediated strand displacement when the toehold and BM domain are directly linked via direct hybridization (C1x, b) or with two bulged thymidines (C2x, c). Real-time fluorescence measurements are shown in blue (for C18 and C28), green (for C110 and C210), and red (for C114 and C214). The results of two independent measurements were shown in the darker and lighter variations of the same color. The average fluorescence values from the 2 measurements were used to fit the time course into a single-exponential equation (fitted time course shown as dotted lines). See Text S1 for the estimation of the second-order rate constants shown in (d). The concentrations of reaction components are listed in the insets. s.f.: structure-free. The sequences of oligonucleotides used in this study are shown in Figure S2.
In DNA C1x, the toehold (shown in red, Figure 2a) is linked to the BM domain via a hairpin stem that mimics the duplex formed between the toehold-carrying strand and the BM domain-carrying strand in the scheme of hybridization-based associative toehold activation (Figure 1c). 8 nt, 10 nt, and 14 nt were chosen as the lengths of the toehold, which were predicted to result in Kd values of 40 μM, 90 nM, and 100 pM, respectively, given the particular sequences (see Figure S2) and the experimental conditions (145 mM Na+ + K+, 4.5 mM Mg2+, 37°C, near neutral pH; ΔG of hybridization: −6.2, −10.0, −14.0 kcal/mole, respectively, predicted by the DINAMelt server19). Thus, the 8-nt and 10-nt toeholds lead to reversible toehold binding whereas the 14-nt toehold leads to nearly irreversible toehold binding. These three invader DNA strands were termed C18, C110, and C114, respectively. A conventional, structure-free invader (C38) in which an 8-nt toehold is linked to the BM domain via a phosphodiester bond was also used as a positive control. Here, only 8 nt was used as the length of toehold since theoretical analysis20 and preliminary experimental data suggest that for structure-free invaders 8-nt toehold is strong enough to promote efficient strand displacement and further increasing toehold length does not significantly accelerate the reaction.
Unfortunately, although the structure-free invader C38 efficiently displaced strand P with an apparent second order rate constant (kapp, see Text S1 for its calculation) of 2 × 105 /M/s, the strand displacement reaction initiated by C18 was nearly 3 orders of magnitude slower (kapp = 6 × 102 /M/s, Figure 2b and 2d). Strand displacement initiated by C110 was also very slow (kapp = 7 × 103 /M/s), although C114, through nearly irreversible toehold binding, led to efficient strand displacement (kapp = 9 × 104 /M/s). As a conclusion, reversible toehold binding cannot initiate efficient strand displacement when the toehold and the BM domain are joined by a duplex.
The bulged thymidines accelerate strand displacement across a 3-way junction
The rate limiting step of the strand displacement by C1x after toehold binding is likely the formation of intermediate Int1, where only the first base pair is formed between the BM domain of C1x and strand D (Figure 1d). If Int1 is unstable, the activation energy is high, hence the reaction is slow. Thus, stabilization of this intermediate may accelerate strand displacement.
It has been long known that one or two bulged bases may stabilize 3-way junctions by allowing coaxial stacking of two of the three duplexes.21 Therefore, I hypothesized that such bulged bases can stabilize intermediate Int1, and in consequence accelerate the overall strand displacement reaction. To test this hypothesis, I chose two well-studied 3-way junction motifs each containing 2 bulged thymidines (see Text S2 and Figure S6) 22-23. These two motifs promote the coaxial stacking of two stems while leaving the other stem projecting outward (See Text S2).
To test whether the bulged thymidines can promote strand displacement reactions initiated by C1x, two thymidines were inserted between the toehold and the hairpin, forming molecules C2x (Figure 2a). Strikingly, the bulged thymidines substantially accelerated the strand displacement. The strand displacement reaction initiated by C28 showed a kapp value of 3 × 103 /M/s, 5 times as high as that of C18. Similarly, C210 showed a kapp value of 5 × 104 /M/s, a 7-fold improvement over C110. Even in the regime of nearly irreversible toehold binding, bulged thymidines still exhibited slight improvement: C214 showed a kapp value of 1.8 × 105 /M/s, roughly 2-fold higher than that of C114. Further discussion on these rate constants can be found in Text S3.
Multiple-turnover catalysis
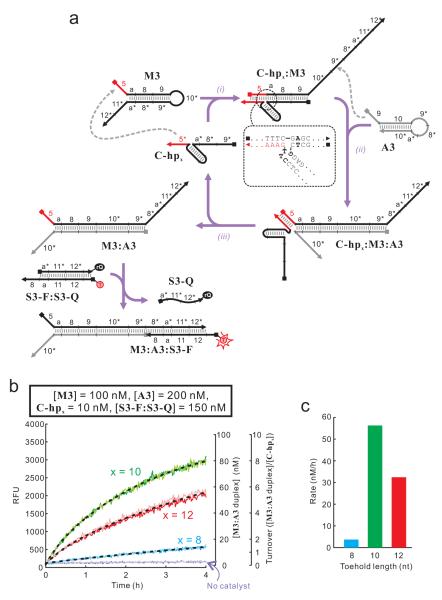
As discussed above, one of the reasons why efficient strand displacement mediated by reversible toehold binding is critical is that it allows fast regeneration of the catalyst in DNA circuits where spontaneous toehold unbinding is necessary for catalyst regeneration. Therefore, the ultimate test of the utility of the bulged thymidines seems to be multiple-turnover catalysis in such circuits, an example of which is shown in Figure 3a. In this circuit, two hairpins named M3 and A3 can potentially form duplex M3:A3. However the reaction is kinetically hindered. A hairpin-containing oligonucleotide C-hpx (where x is the length of toehold) can potentially catalyze this reaction. During the catalysis, C-hpx first uses domain 5* as a toehold to bind domain 5 of M3, potentially initiating a strand-displacement reaction across a 3-way junction to open M3, leading to the formation of C-hpx:M3 complex where the domain 9* of M3 is open (Figure 3a, reaction i). Then domain 9 of A3 can hybridize to the newly opened domain 9* of M3, initiating a conventional strand displacement to form C-hpx:M3:A3 complex (Figure 3a, reaction ii). The last step of the catalytic cycle is the spontaneous dissociation of C-hpx from M3:A3 duplex (Figure 3a, reaction iii). This circuit was used to test whether strands C-hpx, whose toehold and BM domain are linked by a duplex with two bulged thymidines, can perform multiple-turnover catalysis on biologically relevant time scale (minutes to hours).
Figure 3.
A multiple-turnover reaction catalyzed by a strand whose toehold and BM domain are linked via a hairpin stem with two bulged thymidines. (a) Scheme of the catalytic reaction and the fluorescent reporter to monitor the progression of the reaction in real time. The sequence near the 3-way junction in the intermediate C-hpx:M3 is shown in the insets. The CTTG/Y motif (see Text S2) is shown in bold. Note that the intermediates C-hpx:M3 and C-hpx:M3:A3 can also react with the fluorescent reporter in way similar to M3:A3. However these side reactions do not affect the determination of the rates and turnovers of the circuit. T: TYE665; rQ: IowaBlack RQ. (b) Real-time fluorescence readouts. The concentrations of reaction components are listed in the inset. The results of two independent measurements were shown in the darker and lighter variations of the same color. The kinetic data from the 2 measurements were averaged and used to fit into a double-exponential equation, as shown by dotted lines. (c) The initial rates of the reactions shown in a bar graph. s.f.: structure-free. The sequences of oligonucleotides used in this study are shown in Figure S3.
The formation of M3:A3 was monitored in real-time using a fluorescent reporter similar to that shown in Figure 2a. Again, three different toehold lengths (8 nt, 10 nt, and 12 nt) were chosen for C-hpx, resulting in molecules C-hp8, C-hp10, and C-hp12, respectively. As shown in Figure 4b, when 10 nM C-hp10 was mixed with 100 nM M3 and 200 nM A3, multiple-turnover catalysis with an initial rate of ~55 nM/h was observed. C-hp8 and C-hp12 also exhibited multiple-turnover catalysis, albeit with much lower rates. The slower catalysis by C-hp12 was likely caused by slower toehold dissociation. In contrast, when the two bulged thymidines of C-hp8 were deleted, no catalysis was observed (data not shown).
Figure 4.
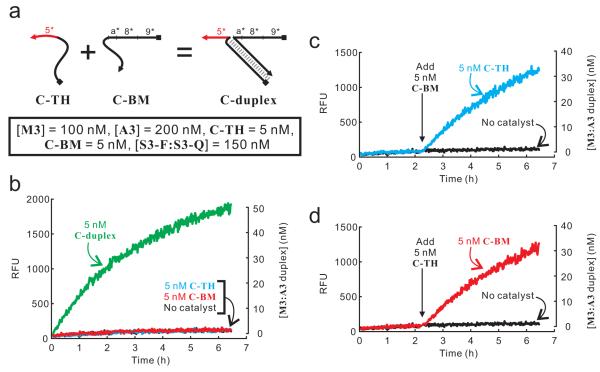
Scheme (a) and performance (b to d) of hybridization-based associative toehold activation. The substrates and reporter in these reactions are the same as those shown in Figure 3. (b) Comparison among reactions in the presence of 5 nM toehold-carrying strand C-TH alone, 5 nM BM domain-carrying strand C-BM alone, and 5 nM of the annealed product of the two strands. (c and d) Addition of 5 nM C-BM to a reaction mixture containing 5 nM C-TH (c), or vice versa (d). All reactions contained M3, A3, and S3-F:S3-Q, whose concentrations are listed in the inset of (a). The identity of each trace is labeled on the figures. The sequences of oligonucleotides used in this study are shown in Figure S3.
Associative toehold activation
Replacing the hairpin stem of C-hpx with a duplex formed by hybridization of two strands is expected to cause little (if any) difference in its catalytic activity. If this is the case, the scheme of hybridization-based associative toehold activation shown in Figure 1c can be validated. To directly test this prediction, C-hp10 was split into two strands: the toehold-carrying strand C-TH and the BM domain-carrying strand C-BM (Figure 4a). These two strands can hybridize to form C-duplex, which is expected to act similar to C-hp10. Indeed, neither 5 nM C-TH nor 5 nM C-BM alone was able to catalyze the formation of M3:A3 from M3 and A3, whereas 5 nM of the hybridization product of the two strands exhibited similar catalytic activity as C-hp10 (Figure 4b). Moreover, when 5 nM C-TH was added to the reaction mixture that contains 5 nM C-BM (or vice versa), the formation of active catalyst was observed immediately, demonstrating the feasibility of associative toehold activation during the execution of a DNA circuit (Figure 4c and 4d).
Expanded architecture of DNA circuits
The mechanism of toehold activation determines the mechanism of information processing in DNA circuits, and ultimately determines the architecture of DNA circuits. Associative toehold activation may potentially enable many new ways to design DNA circuits. As examples of these possibilities, I designed two circuits to achieve functions that are non-trivial to engineer with previous design principles.
Examples 1: Toehold-switching
Since associative toehold activation does not require hard-wiring (i.e., through covalent bonds) between the toehold and the BM domain, the combination between the two domains can be changed during the execution of a DNA circuit. I call this operation ‘toehold switching’ (Figure 5a). Toehold switching allows simultaneous inactivation of one catalyst and activation of another catalyst. More importantly, during the switching process, the total activity of the two catalysts (as defined by the total concentration of the BM domain that is linked to a toehold) remains constant.
Figure 5.
Toehold switching. (a) The scheme of toehold switching. The catalyst, duplex C-THA:C-BM2, is converted into C-THB:C-BM2 upon the addition of C-THB. C-THA:C-BM2 and C-THB:C-BM2 catalyze two hairpin assembly reactions shown as Circuit 1 and Circuit 2, respectively. The mechanism of catalysis is similar to that shown in Figure 3. Intermediates of the reactions are now shown. The products of the two reactions are monitored by fluorescent reporters SA-F:SA-Q and SB-F:SB-Q, simultaneously. F: FAM; fQ: IowaBlack FQ; T: TYE665; rQ: IowaBlack RQ. (b) Performance of toehold switching. The identity of each trace is labeled on the figure. Whenever present, the concentrations of reactants are listed in the inset. The sequences of oligonucleotides used in this study are shown in Figure S4.
To demonstrate this process, I designed two hairpin assembly reactions, H1A + H2A → H1A:H2A and H1B + H2B → H1B:H2B, that can be catalyzed by the same BM domain (carried by the strand C-BM2, where the subscript ‘2’ is used to distinguish this strand from C-BM described above) linked to two different toeholds (carried by strands C-THA and C-THB, respectively). As before, the formation of H1A:H2A and H1B:H2B was monitored in real time using two fluorescent reporters of different colors (FAM and TYE665, respectively). Initially, 100 nM H1A and H2A and 200 nM H1B and H2B were mixed with 10 nM duplex C-THA:C-BM2. As expected, only the formation of H1A:H2A was catalyzed (Figure 5b). Notably, when 20 nM C-THB was added to the reaction, the catalyzed formation of H1B:H2B was quickly observed whereas the concentration of H1A:H2A ceased increasing (Figure 5b). This observation indicated that C-THB quickly displaced C-THA to form C-THB:C-BM2, and depleted C-THA:C-BM2 at the same time.
Example 2: A simple conformational self-replicator
Self-replication is a fundamental process that has implications for both the understanding of molecular evolution and the development of biotechnology. Several artificial chemical systems capable of self-replication have been demonstrated,24-25 including those that rely on cleavage or ligation activities of ribozymes and deoxyribozymes.26-27 Although in most of these cases replication involves formation or cleavage of covalent bonds, it is known that self-replication can also occur through conformational change, with prions as a classic example.28 Here, I show that a conformational self-replicator can be easily designed by combining the schemes of associative and dissociative toehold activation.
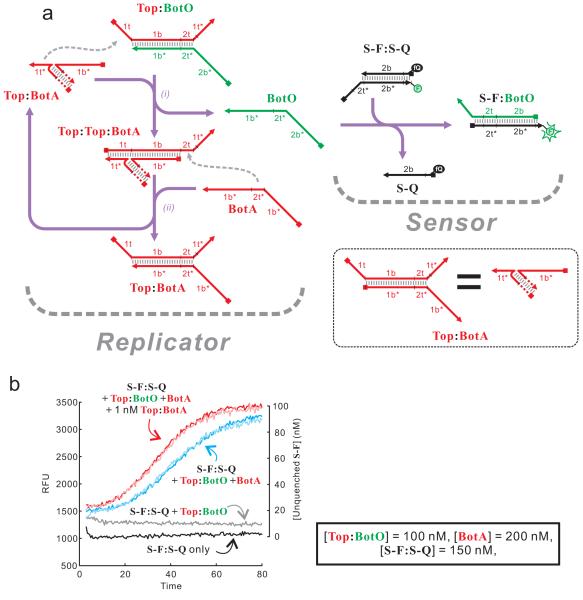
The design of a conformational replicator is shown in Figure 6a. The circuit consists of only three strands, one arbitrarily named the top strand (or Top), and two named the bottom strands. One of the bottom strands can trigger the output reporter and was thus named BotO. The other bottom strand acts as part the autocatalyst and was thus named BotA. The two initial substrates of the self-replication are the Top:BotO duplex and free BotA, whereas the catalyst and the product are both Top:BotA duplex. Top contains domains 1t and 1t* on it 5′ and 3′ termini, respectively. Therefore, the Top:BotA duplex, as the catalyst, can use its domain 1t* to bind domain 1t of a Top:BotO duplex (a substrate). This initiates a strand displacement reaction that displaces BotO, leaving domain 2t of Top unhybridized (Figure 6a, reaction i). Consequently, BotA (analogous to the fuel strand in Seesaw gates 12) can use its domain 2t* as a toehold to bind domain 2t of Top and initiate the reverse strand displacement that ultimately displaces the catalyst Top:BotA, forming another Top:BotA (Figure 6a, reaction ii). The displaced BotO can be monitored by a fluorescent reporter that recognizes domain 2t* and 2b* of BotO. The overall reaction is driven partially by the entropy increase as a result of the balanced distribution of BotO and BotA on Top, as in Seesaw gates, and partially by the enthalpically favorable sensing reaction (S-F:S-Q + BotO → S-F:BotO + S-Q). As shown in Figure 6b, exponential increase of unquenched S-F was observed when Top:BotO duplex and BotA were mixed with the fluorescent reporter. The initial trigger of the self-replication was probably circuit leakage (i.e., formation of Top:BotA in the absence of the catalyst). As expected, when exogenous catalyst was introduced to the reaction, a faster initial rate of replication was observed.
Figure 6.
A simple conformational self-replicator. (a) The scheme of the replicator and the sensor to monitor the progression of the reaction. For clarity, the names of DNA strands are written in the same color as strands themselves. Due to space limitations, when Top:BotA acts as a catalyst, only the part used to catalyze the reaction is shown in detail, as indicated in the inset. (b) Kinetics of self-replication. The identity of each trace is labeled on the figure. Whenever present, the concentrations of reaction components are listed in the inset. For self-replication reactions with (red traces) and without (blue traces) external catalyst (1 nM of Top:BotA), two independent measurements were carried out and shown in the darker and lighter variations of the same color. The sequences of oligonucleotides used in this study are shown in Figure S5.
Conclusion
In this paper I demonstrated a new scheme – hybridization-based associative toehold activation – that allows the expansion of the architecture of DNA circuits. Although direct hybridization between the toeholds and BM domains results in slow strand displacement across 3-way junctions, two bulged thymidines substantially accelerate this process so that reversible toehold binding is sufficient to initiate fast strand displacement. This new scheme allows recombination between toeholds and BM domains during the execution of DNA circuits, as shown by the demonstration of run-time toehold activation and toehold switching. A simple conformational self-replicator, enabled by this scheme, is also shown. I believe this scheme can be widely used in designing DNA circuits and DNA nanomachines, which may potentially lead to wider applications of these devices in a variety of fields from biosensing to DNA-programmed chemical synthesis.
Supplementary Material
Acknowledgement
This work was supported by National Institute of Health (R01 AI092839-01, R01 GM094933-01), National Security Science and Engineering Faculty Fellowship (FA9550-10-1-0169), and Welch Foundation (F-1654). X.C. is partially supported by a postdoctoral trainee fellowship from Cancer Prevention Research Institute of Texas (CPRIT).
Footnotes
Supporting Information A detailed description of the procedures for data processing; discussion on the stabilization of DNA 3-way junctions by bulged bases and its impact on the kinetics of DNA circuits; as well as sequences of oligonucleotides. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Seeman NC. Annual review of biochemistry. 2010;79:65. doi: 10.1146/annurev-biochem-060308-102244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Zhang DY, Seelig G. Nat Chem. 2011;3:103. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- (3).Benenson Y, Gil B, Ben-Dor U, Adar R, Shapiro E. Nature. 2004;429:423. doi: 10.1038/nature02551. [DOI] [PubMed] [Google Scholar]
- (4).Kim J, White KS, Winfree E. Mol Syst Biol. 2006;2:68. doi: 10.1038/msb4100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Kim J, Winfree E. Mol Syst Biol. 2011;7:465. doi: 10.1038/msb.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Montagne K, Plasson R, Sakai Y, Fujii T, Rondelez Y. Mol Syst Biol. 2011;7:476. doi: 10.1038/msb.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Stojanovic MN, Stefanovic D. Nat Biotechnol. 2003;21:1069. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]
- (8).Yurke B, Turberfield AJ, Mills AP, Jr., Simmel FC, Neumann JL. Nature. 2000;406:605. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- (9).Seelig G, Soloveichik D, Zhang DY, Winfree E. Science. 2006;314:1585. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- (10).Soloveichik D, Seelig G, Winfree E. Proc Natl Acad Sci U S A. 2010;107:5393. doi: 10.1073/pnas.0909380107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zhang DY, Turberfield AJ, Yurke B, Winfree E. Science. 2007;318:1121. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]
- (12).Qian L, Winfree E. J R Soc Interface. 2011 doi: 10.1098/rsif.2010.0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Qian L, Winfree E. Science. 2011;332:1196. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- (14).Gu H, Chao J, Xiao SJ, Seeman NC. Nature. 2010;465:202. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Omabegho T, Sha R, Seeman NC. Science. 2009;324:67. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Yin P, Choi HM, Calvert CR, Pierce NA. Nature. 2008;451:318. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- (17).Genot AJ, Zhang DY, Bath J, Turberfield AJ. J Am Chem Soc. 2011;133:2177. doi: 10.1021/ja1073239. [DOI] [PubMed] [Google Scholar]
- (18).Li B, Ellington AD, Chen X. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Markham NR, Zuker M. Nucleic Acids Res. 2005;33:W577. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Zhang DY, Winfree E. J Am Chem Soc. 2009;131:17303. doi: 10.1021/ja906987s. [DOI] [PubMed] [Google Scholar]
- (21).Leontis NB, Kwok W, Newman JS. Nucleic Acids Res. 1991;19:759. doi: 10.1093/nar/19.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Wu B, Girard F, van Buuren B, Schleucher J, Tessari M, Wijmenga S. Nucleic Acids Res. 2004;32:3228. doi: 10.1093/nar/gkh645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).van Buuren BN, Overmars FJ, Ippel JH, Altona C, Wijmenga SS. J Mol Biol. 2000;304:371. doi: 10.1006/jmbi.2000.4224. [DOI] [PubMed] [Google Scholar]
- (24).Sievers D, von Kiedrowski G. Nature. 1994;369:221. doi: 10.1038/369221a0. [DOI] [PubMed] [Google Scholar]
- (25).Zielinski WS, Orgel LE. Nature. 1987;327:346. doi: 10.1038/327346a0. [DOI] [PubMed] [Google Scholar]
- (26).Levy M, Ellington AD. Proc Natl Acad Sci U S A. 2003;100:6416. doi: 10.1073/pnas.1130145100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Lincoln TA, Joyce GF. Science. 2009;323:1229. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Tuite MF, Serio TR. Nat Rev Mol Cell Biol. 2010;11:823. doi: 10.1038/nrm3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.