Abstract
Epigenetic variations have been widely described to occur during the aging process. To verify if these modifications are correlated with the inter-individual phenotypic variability of elderly people, we searched for a correlation between global DNA methylation levels and frailty. We found that the global DNA methylation levels were correlated to the frailty status in middle/advanced-aged subjects but not with age. A 7-year follow-up study also revealed that a worsening in the frailty status was associated to a significant decrease in the global DNA methylation levels. These results suggest that the relaxation of the epigenetic control in aging is specifically associated with the functional decline rather than with the chronological age of individuals. Thus, the modifications of DNA methylation, representing a drawbridge between the genetic and the environmental factors affecting the age-related decay of the organism, may play an important role in determining physiological changes over old age.
Keywords: Frailty, Global DNA methylation, Epigenetics, Aging
Introduction
Frailty represents a clinical syndrome which is prevalent in the elderly population. It is characterized by the decrease of total physiological reserves and a progressive deregulation in the homeostatic network underlying maintenance and repair in the aging body (Lang et al. 2009; Bandinelli et al. 2010). A wide range of geriatric phenotypes are commonly observed to be associated with frailty including weight loss and sarcopenia, muscle weakness with low grip strength, low activity level, exhaustion, low body mass index, blood pressure instability and balance and gait abnormalities (Fried et al. 2004; Bergman et al. 2007; Topinková 2008; Davis et al. 2010). In addition, in the characterization of the clinical phenotype of frailty, it has been very important the demonstration of the role in this syndrome of multi-system impairments including immune/inflammatory, multiple hormonal and neuromuscular deregulations, as well as metabolic and vascular alterations and oxidative stress that commonly occur with aging (Ferrucci et al. 1999; Leng et al. 2002; Schalk et al. 2004; Walston et al. 2006; Barzilay et al. 2007; Landi et al. 2008; Hubbard et al. 2009; Blaum et al. 2009; Serviddio et al. 2009; Desai et al. 2010; Lustosa et al. 2010; Evans et al. 2010; Hyde et al. 2010; Maggio et al. 2010; Hubbard and Woodhouse 2010). As a consequence, frailty increases the individual vulnerability to age-related disorders, such as myocardial infarction, rheumatoid arthritis, diabetes and hypertension (Hubbard et al. 2010). Moreover, an association between frailty and the risk of mild cognitive impairment and a rapid rate of cognitive decline was described (Buchman et al. 2007; Boyle et al. 2010).
In recent years, many operational definitions of frailty have been proposed. Montesanto et al. (2010), by using a hierarchical cluster analysis that availed of specific geriatric parameters, identified a specific approach to define the frailty status. The diagnostic and predictive soundness of this classification was confirmed by a detailed survival analysis which showed higher survival chance for subjects characterized by lower degree of frailty.
Frailty is commonly considered as the resultant of a multidimensional interplay of genetic, biological, psychosocial and environmental factors (Fried et al. 2001; Rockwood et al. 2004; Rockwood and Mitnitski 2007; Bergman et al. 2007; Lang et al. 2009; Fulop et al. 2010; Landi et al. 2010). Several lines of evidence suggest that epigenetic reprogramming of gene expression occurs during aging (Fraga 2009; Kawakami et al. 2009; Gravina and Vijg 2010; Rando 2010). A correlation between epigenetic DNA modifications and quality of aging has been shown by Fraga et al. (2005), who found that global and locus-specific differences in DNA methylation in identical twins of various ages are influenced by different environmental factors and lifestyle. In vitro models and studies carried out in tissues and blood DNA samples demonstrated a decrease in global cytosine methylation during aging mostly due to the demethylation in transposable repetitive elements (Castro et al. 2003; Bjornsson et al. 2008; Bollati et al. 2009). On the other hand, additional studies reported an age-related hypermethylation in promoter regions of specific genes, with a consequent decrease of correspondent mRNA levels. In humans, this hypermethylation has been observed for genes involved in cell cycle regulation, tumor-cell invasion, DNA repair, apoptosis, metabolism and cell signaling, implicating their potential role in age-related diseases (Wilson et al. 1987; Oakes et al. 2003; Richardson 2003; Fuke et al. 2004; Fraga and Esteller 2007; Ling et al. 2008; Arai et al. 2010; Lee et al. 2010).
In the present study, we investigated whether the frailty status in elderly people was associated to global DNA methylation levels. Thus, we searched a correlation between the age-related functional decline and epigenetic modifications. In addition, a longitudinal study carried out in a subgroup of the original sample allowed us to determine whether the methylation levels were subject to changes over time in agreement with changes in the frailty status. Global DNA methylation was measured by applying the assay developed by Anisowicz et al. (2008) to peripheral blood DNAs collected from 318 subjects classified for frailty phenotype according to Montesanto et al. (2010). In the longitudinal study, both frailty evaluation and DNA methylation analysis were repeated after 7 years.
Materials and methods
Population sample
A total of 318 unrelated individuals (144 men and 174 women), 65–105 years old (median age 79.5 years), participated in the present study. All the subjects lived in Calabria (southern Italy) and their origin in the area had been ascertained up to the grandparents generation. A more detailed sample description can be found elsewhere (De Rango et al. 2010). Health status was ascertained by medical visit carried out by a geriatrician who also conducted a structured interview including questions on common diseases occurred in the past. At the same time of the visit, peripheral venous blood samples were also obtained. Before the interview, each subject consented to her/his phenotypic and genetic data to be used anonymously for genetic studies on aging (informed consent).
In a previous work, by using a hierarchical cluster analysis (HCA) which availed of specific geriatric parameters, this sample was used to identify specific aging phenotypes (Montesanto et al. 2010). In particular, the sample was analyzed considering two different age groups. The first (S1) included 217 subjects (94 males and 123 females) 65–89 years old (median age 75 years); the second (S2) included 101 subjects (50 males and 51 females) older than 90 years (median age 99 years). By using this approach in the S1 sample, three clusters were identified: nonfrail (the cluster with subjects showing the best scores for the classification variables), frail (the clusters with subjects showing the worst scores for the classification variables), and prefrail (the cluster with subjects showing intermediate scores for the classification variables). In the S2 sample, two clusters were identified. Similar to the first classification, the two clusters obtained were defined as frail (the cluster with subjects showing the best scores for the classification variables) and very frail (the cluster with subjects showing the worst scores for the same variables).
The diagnostic and predictive soundness of these classifications were confirmed by a 3-year longitudinal study. In fact, a detailed survival analysis showed higher survival chance for subjects characterized by lower frailty in these classifications.
In order to better evaluate the relationship between DNA methylation levels and degree of frailty, a randomly selected sample consisting of 98 prefrail and nonfrail subjects of S1 group were reconsidered after 7 years from the baseline visit. During the follow-up period, 55 subjects (56%) died. Of the remaining 43 alive subjects, 6 refused the visit while for the remaining others (37), a new multidimensional geriatric evaluation was performed and a new sample of blood for the evaluation of DNA methylation was provided.
In vitro DNA methylation of control lambda DNA
Five hundred nanograms of lambda DNA (Sigma) were completely methylated by using 2.5 U of M.HpaII methylase (New England Biolabs). The mix was incubated at 37°C for 3 h and successively at 65°C for 15 min to inactivate the enzyme.
Restriction analysis of control lambda DNAs, control human genomic DNAs and population sample DNAs
One hundred nanograms of methylated and unmethylated lambda DNA were separately incubated with 5 U of HpaII and MspI restriction endonucleases (New England Biolabs) at 37°C overnight and successively at 65°C for 20 min to inactivate the endonucleases. The samples were loaded on a 1.4% agarose gel, electrophoresed in TAE (Tris/Acetate/EDTA) buffer and stained with ethidium bromide.
One hundred nanograms of CpGenome™ Universal Methylated DNA (Chemicon), 100 nanograms of CpGenome™ Universal Unmethylated DNA (Chemicon) and 100 ng of a mixture obtained combining 50 ng of CpGenome Universal Methylated and 50 ng of Unmethylated DNA were separately incubated with 5 U HpaII and MspI restriction endonucleases at 37°C overnight and successively at 65°C for 20 min to inactivate the endonucleases. The samples were loaded on to 1.4% agarose gel, electrophoresed in TAE (Tris/Acetate/EDTA) buffer and stained with ethidium bromide.
One hundred nanograms of population sample DNAs, extracted from venous blood buffy coats following standard procedures, were digested with HpaII and MspI by using the above experimental conditions.
Measurement of global DNA methylation levels
The global DNA methylation levels of control lambda DNAs, control human DNAs, and population sample DNAs were estimated by using the CpGlobal assay, designed and developed by Anisowicz et al. (2008). Briefly, 2 μM of both Biotin-11-dCTP and Biotin-11-dGTP (Perkin Elmer) were added to digested DNA samples in an end-fill reaction of 20 μl carried out in presence of biotinylation buffer (40 mM Tris–HCl pH 7.5, 20 mM Tris–HCl, 50 mM NaCl), and 2 U of Sequenase (USB Corporation). After incubation at 37°C for 30 min, 100 μl of Reacti-BindTM DNA Coating Solution (Pierce) were added and the samples were shaken in an orbital platform at room temperature overnight. The solution was removed and the samples were washed three times with Dulbecco’s Phosphate-Buffered Saline (Sigma). Then, 200 μl of the detector block solution (KPL) were added and the mixtures were incubated at room temperature for 30 min. After the removal of the solution, 150 μl of the Detector Block Solution containing 0.5 μg/ml of HRP Streptavidin (KPL) were added and the samples were incubated at room temperature for 30 min. After the incubation, the Detector Block Solution was removed and the samples were washed four times with Biotin Wash Solution 1X (KPL). Then, 150 μl of LumiGlo Chemiluminescence substrate (KPL) were added and after 2 min the chemiluminescence emitted from each sample was quantified in a Lumat LB9507 luminometer (EG&G Bertold). Each sample was analyzed three independent times in triplicates. In order to determine the possible “background noise” and, thus, to calculate the net luminescence for each sample a control lacking of enzyme was also analyzed. The data were calculated as Global DNA Methylation Index (GDMI) by dividing the mean net luminescence values for the HpaII enzyme to the mean net luminescence values for the MspI enzyme. Thus, the GDMI values inversely correlate to the global DNA methylation levels.
Statistical analysis
Descriptive statistics for continuous and categorical variables were used to describe socio-demographic characteristics, the frailty phenotypes and the GDMI values of the study sample. For continuous variables, measures of central tendency and dispersion, including mean, median and standard deviations were reported. Categorical variables were examined by analyzing the relevant frequency distributions. ANOVA test followed by multiple comparison test (Dunnett’s test) was used to compare the values of GDMI among the frailty groups defined by the HCA approach. Linear regression analysis was performed for testing the dependence between GDMI and age.
After the follow-up period, in order to determine the actual frailty status of the 37 subjects (re-classification) a classification tree (CT) model has been used (Breiman et al. 1984). In fact, the main HCA drawback is that classification of new cases is usually not affordable, without repeating the whole HCA analysis. Thus, we used a supervised classification approach in order to obtain an easy-to-understand model that can be used for classifying the revisited subjects. In brief, we applied the CT algorithm to the baseline data by considering the classification provided by HCA analysis in S1 group as dependent variable, while the geriatric parameters (Hand Grip, Mini Mental State Examination, Activities of Daily Living and Self Reported Health Status) as independent variables. Adjusted values (i.e., with normalization) were used for all variables (Montesanto et al. 2010). The performance of the CT model was evaluated in terms of accuracy using a tenfold cross-validation strategy. The obtained model was then used to re-classify the revisited subjects.
Statistical analyses were performed using SPSS 15.0 (SPSS Inc., Chicago, IL). A significance level (α) of 0.05 was chosen in all the tests.
Results
We carried out a series of control experiments in order to validate the reproducibility of the CpGlobal assay described in the “Materials and methods” section (Anisowicz et al. 2008). To this purpose, we analyzed two samples of lambda DNAs and three human genomic DNAs as quality control.
Global DNA methylation levels in control samples
Control lambda DNA
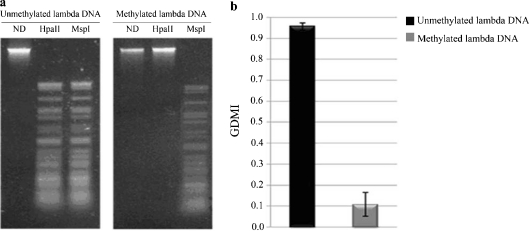
Two samples of lambda DNA were analyzed: one was unmethylated; the second one was in vitro methylated by using the bacterial M. HpaII methylase. Firstly, to verify the restriction conditions and the fully methylation we treated the two samples with the methyl-sensitive HpaII and the methyl-insensitive MspI restriction endonucleases. As shown in Fig. 1a, in contrast to unmethylated DNA, no HpaII restriction fragments were observed in the methylated lambda DNA. In fact, HpaII activity was greatly inhibited by the methylation on the internal cytosine residue in its recognition sequence. As expected, the MspI activity was not affected by the absence/presence of methylated cytosines. Then, the CpGlobal assay was applied to determine the GDMI values in the two lambda DNA samples. As expected, the GDMI value was significantly lower (about 0.1) in the methylated DNA sample than in the unmethylated DNA (about 1.0; Fig. 1b).
Fig. 1.
Control lambda DNA analysis. a Ethidium bromide-staining agarose gel showing unmethylated and methylated lambda DNAs digested with HpaII and MspI. ND: lambda DNA not digested. b GDMI values of unmethylated and methylated lambda DNAs. The values represent the main of three independent triplicate experiments with standard error mean
Control human genomic DNAs
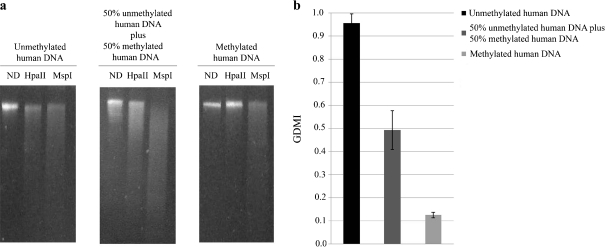
CpGlobal assay was subsequently applied to three control samples of human genomic DNA: one fully unmethylated, one fully methylated, and another obtained by mixing an equal ratio of unmethylated and methylated DNA. We restricted the samples as described above. We observed that when fully methylated human DNA was digested with HpaII, no restriction fragments were observed (Fig. 2a). Conversely, when fully unmethylated human DNA was digested with the same enzyme, several restriction fragments were observed, indicating the complete digestion of the sample. Finally, the DNA sample obtained by mixing an equal ratio of unmethylated and methylated human DNA, showed an intermediate result. In fact, the intensity of the lane of undigested DNA approximates the 50% of that untreated with HpaII, as from densitometric tracings (data not shown). As expected, the GDMI values in the fully methylated, in the mix of methylated and unmethylated DNA, and in the fully unmethylated DNA were about 0.1, 0.5, and 1.0, respectively (Fig. 2b).
Fig. 2.
Control human DNA analysis. a Ethidium bromide-staining agarose gel showing unmethylated and methylated human DNAs and a mixture of equal amount of unmethylated and methylated human DNAs digested with HpaII and MspI. ND human DNA not digested. b GDMI values of unmethylated, methylated and the mixture of unmethylated and methylated human DNAs. The values represent the main of three independent triplicate experiments with standard error mean
On the whole, the results obtained by the above control experiments demonstrated the accuracy and an overall high reproducibility of the CpGlobal assay in measuring the global DNA methylation levels of the different sample analyzed.
Global DNA methylation levels in the population sample
After the validation procedure, the CpGlobal assay was applied to measure the GDMI values of the DNA samples extracted from peripheral venous blood collected from 318 subjects (65- to 105-year-old subjects).
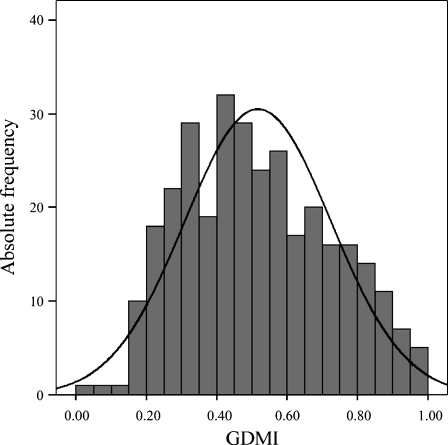
The distribution frequency of the GDMI values in the population sample is reported in Fig. 3. The distribution of these values in the sample was quite normal with a mean value of about 0.517. In addition, they were quite similar for men and women (p value = 0.424) and were not correlated with age (r = −0.040, p value = 0.474). These results suggested that global DNA methylation levels do not correlate neither with the age nor with the gender of sample analyzed.
Fig. 3.
Frequency distribution of GDMI values in the total population sample
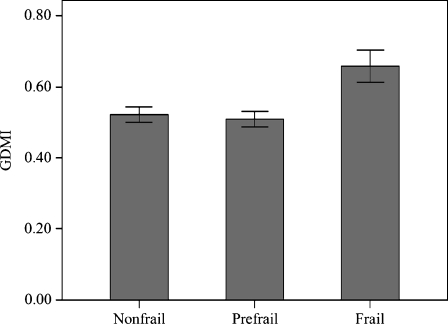
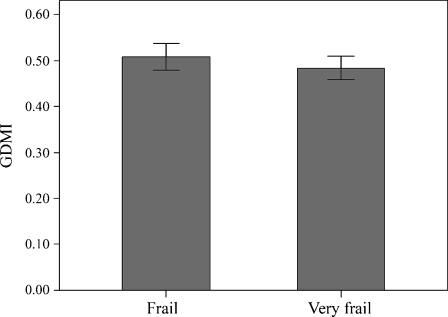
Subsequently, we wondered whether the GDMI values were correlated to the frailty status rather than to chronologic age. To answer this question, we availed of the HCA classifications reported in Montesanto et al. (2010), that allowed to classify this sample in different aging phenotypes (see “Materials and methods”). The mean GDMI values across the S1 and S2 groups are shown in Figs. 4 and 5. We can observe that frail subjects of S1 group exhibit GDMI values significantly higher than those prefrail (0.658 ± 0.201 vs 0.508 ± 0.223, respectively, p value = 0.006) and nonfrail subjects (0.658 ± 0.201 vs 0.521 ± 0.196, respectively, p value = 0.006). In S2 group no difference in GDMI values was detected across the frailty phenotypes (0.484 ± 0.191 and 0.509 ± 0.197 for very frail and frail, respectively). Moreover, GDMI values were quite similar for men and women in both groups (in S1 sample 0.534 ± 0.220 vs 0.522 ± 0.210, respectively; p value = 0.668; in S2 sample 0.514 ± 0.217 vs 0.477 ± 0.167, respectively; p value = 0.335). These results indicated that a correlation between the global DNA methylation levels and the frailty phenotype exists in middle-aged subjects, but not in ultranonagenarians.
Fig. 4.
Mean GDMI values across the groups defined by cluster analysis in S1 sample
Fig. 5.
Mean GDMI values across the groups defined by cluster analysis in S2 sample
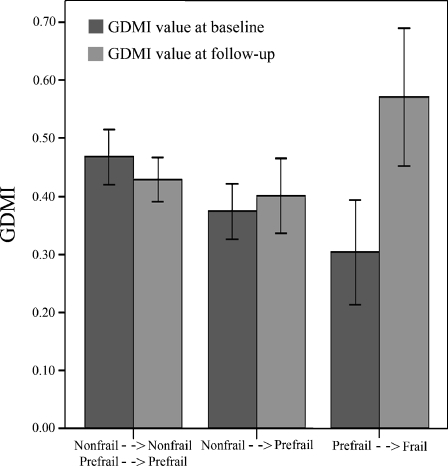
Then, in order to better evaluate the relationship between DNA methylation levels and degree of frailty, 37 prefrail and nonfrail subjects of S1 sample were revisited after 7 years from the baseline visit. Figure 6 shows the GDMI values at baseline (black bars) and after the follow-up period (gray bars) with respect to the changes in the frailty status after this period. We can observe that in subjects who, after the follow-up period, have maintained their nonfrail or prefail frailty status or have changed their frailty status from nonfrail to prefrail, mean GDMI value did not show significant changes over time (about 0.4). On the contrary, in subjects who became frail, mean GDMI value was significantly increased (about 0.6) over time compared to the first measurement.
Fig. 6.
Variations of GDMI values with respect to the variations of the frailty status after the follow-up period
Discussion
Understanding the mechanisms that modulate the quality of aging remains one the most challenging research topics. Several lines of evidence have demonstrated how the characterization of frailty, that represents a state of vulnerability for adverse health outcomes, may contribute to disentangle the molecular mechanisms influencing the functional decline of elderly people and thus to characterize and to better define the aging process (Fried et al. 2004). The impact of genetic variants of both nuclear and mitochondrial DNA on the inter-individual susceptibility to functional decline and vulnerability to diseases in the elderly people has been largely demonstrated (Maggio et al. 2006; Moore et al. 2010; Matteini et al. 2010). Similarly, different reports have shown the influence of environmental and social factors on frailty. A “drawbridge” across genetic factors and environment may be represented by epigenetic variations which in turn depend on hereditary, environmental and stochastic factors and might explain the inter-individual variability in the frailty status (Sutherland and Costa 2003; Fraga 2009; Schneider et al. 2010). Indeed, most studies on DNA methylation demonstrated that aging is associated with a relaxation of epigenetic control, thus contributing to some age-related phenotypes such as functional and cognitive decline and the development of pathologies.
In the present study, we looked at epigenetic changes occurring in blood collected from individuals classified by a precise frailty phenotype based on the individual degree of cognitive, functional, and psychological status (Montesanto et al. 2010). We availed of a method that, although could not provide precise quantitative measurements of the DNA methylation levels, is highly effective in comparative analyses of these levels among different samples. In addition, as discussed by Anisowicz et al. (2008), this method is an excellent alternative to the different “gold standard technologies” such as the methods based on chromatography, the high-performance capillary electrophoresis, the radio-labeling of CpG sites using M.SssI methyltransferase, methyl-C antibody, and pyrosequencing. In fact, this method requires small amount of genomic DNA, it is not radioactive, it is highly accurate and reproducible and allow to measure global DNA methylation in many samples simultaneously. It is also worth mentioning that the use of DNA from peripheral blood can be effective in global DNA methylation studies, although this analysis misses the important effects of tissue specific methylation (Thompson et al. 2010).
As the study deals with 65–85-year-old subjects, we found no correlation between global DNA methylation levels and age but frail individuals (with high vulnerability) exhibit lower global methylation levels than those that are in a better condition (prefrail and nonfrail). On the contrary, no difference was observed in these levels among ultranonagenarians, despite their differential classification in frail and very frail subjects. In this paper, we reported also the results obtained in a longitudinal study where DNA methylation levels and frailty status were reanalyzed after 7 years of follow-up. Methylation levels were unchanged in subjects who after the follow-up period did not became frail. On the contrary, in subjects who were prefrail at baseline and became frail after the follow-up period, these levels were significantly decreased and were comparable to those observed in frail subjects at baseline.
To our knowledge, this is the first study demonstrating as a correlation between global methylation levels and frailty occurs in aging. Therefore, the loss of global DNA methylation seems to be specifically associated to the functional decline and not to chronological age, at least after 65 years of age. It might be worth noticing that frailty is associated with age, and then it is possible that in some cases an association between age and methylation is observed as a consequence of the association between frailty and methylation. This result is particularly interesting because it could provide a re-interpretation of some studies in which variations in methylation levels were not observed in samples of middle age individuals (Tra et al. 2002; Siegmund et al. 2007; Schneider et al. 2010; Agrawal et al. 2010).
We did not observe a link between DNA methylation levels and gender, although several studies found higher global DNA methylation levels in males regardless to age of the subjects analyzed (Fuke et al. 2004; Shimabukuro et al. 2006). However, as in the case of the association between age and global methylation, gender-associated differences in DNA methylation were restricted to specific loci and often to mitotic cells (El-Maarri et al. 2007). It might also be worth mentioning that no correlation between the use of drugs and methylation has been observed (data not shown).
Since methylation is associated to gene silencing, it is possible that in old frail subjects, a reduction in DNA methylation could activate genes being in a repressed status. Some of these genes might be needed to maintain the reserve capacity and the efficiency in the response to adverse stressors at levels ensuring the survival (Coneyworth et al. 2009). Likely, the hypomethylation observed in frail subjects can be associated to an accentuated deficiency to achieve the remethylation after DNA replication that, as reported in Gravina and Vijg (2010), occurs during aging. In addition, the above hypomethylation could derepress silenced retrotransposons, such as L1 or IAP, thus inducing genome instability that might account for the decline typical of frailty status (Barbot et al. 2002; Menendez et al. 2004; Maslov and Vijg 2009). Moreover, longitudinal data provide an important contribution to studies which consider frailty as a dynamic process. In fact, our findings are in line with those reported by Bjornsson et al. (2008) who observed time-dependent changes in global DNA methylation within the same individual in two separate populations (one from Iceland and one from Utah) exhibiting both decrease and increase in methylation greater than 20% over an 11- to 16-year span. A progressive loss of DNA methylation over time in repetitive elements disperses through the genome, particularly in Alu elements, was also reported by Bollati et al. (2009).
On the whole, our results contribute to understand how changes in methylation levels are associated with the aging process. We are not able to know whether the changes in frailty status induce the changes in methylation levels or, vice versa, whether these levels determine the frailty status. In any case, our results suggest, in agreement with a series of previous evidences, that specific age-related phenotypes are modulated not only by genetic and environmental factors but also by the effects of environmental factors (dietary, life style) on the individual genome.
Acknowledgments
The work was supported by Fondi di Ateneo of the University of Calabria. AM was supported financially by a fellowship (assegno di ricerca) of the University of Calabria.
Footnotes
Dina Bellizzi and Patrizia D’Aquila equally contributed to this paper.
References
- Agrawal A, Tay J, Yang GE, Agrawal S, Gupta S. Age-associated epigenetic modifications in human DNA increase its immunogenicity. Aging. 2010;20:93–100. doi: 10.18632/aging.100121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisowicz A, Huang H, Braunschweiger KI, Liu Z, Giese H, Wang H, Mamaev S, Olejnik J, Massion PP, Mastro RG. A high-throughput and sensitive method to measure global DNA methylation: application in lung cancer. BMC Cancer. 2008;8:222. doi: 10.1186/1471-2407-8-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Kasahara I, Sawabe M, Honma N, Aida J, Tabubo K. Role of methylation of the hMLH1 gene promoter in the development of gastric and colorectal carcinoma in the elderly. Geriatr Gerontol Int Suppl. 2010;1:S207–S212. doi: 10.1111/j.1447-0594.2010.00590.x. [DOI] [PubMed] [Google Scholar]
- Bandinelli S, Corsi AM, Milaneschi Y, Vazzana R. Frailty and the homeostatic network. Acta Biomed. 2010;81(Suppl 1):15–18. [PubMed] [Google Scholar]
- Barbot W, Dupressoir A, Lazar V, Heidmann T. Epigenetic regulation of an IAP retrotransposon in the aging mouse: progressive demethylation and de-silencing of the element by its repetitive induction. Nucleic Acids Res. 2002;30:2365–2373. doi: 10.1093/nar/30.11.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzilay JI, Blaum C, Moore T, Xue QL, Hirsch CH, Walston JD, Fried LP. Insulin resistance and inflammation as precursors of frailty: the Cardiovascular Health Study. Arch Intern Med. 2007;167:635–641. doi: 10.1001/archinte.167.7.635. [DOI] [PubMed] [Google Scholar]
- Bergman H, Ferrucci L, Guralnik J, Hogan DB, Hummel S, Karunananthan S, Wolfson C. Frailty: an emerging research and clinical paradigm—issues and controversies. J Gerontol A Biol Sci Med Sci. 2007;62:731–737. doi: 10.1093/gerona/62.7.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, Yu W, Rongione MA, Ekström TJ, Harris TB, Launer LJ, Eiriksdottir G, Leppert MF, Sapienza C, Gudnason V, Feinberg AP. Intra-individual change over time in DNA methylation with familial clustering. JAMA. 2008;299:2877–2883. doi: 10.1001/jama.299.24.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaum CS, Xue QL, Tian J, Semba RD, Fried LP, Walston J. Is hyperglycemia associated with frailty status in older women? J Am Geriatr Soc. 2009;57:840–847. doi: 10.1111/j.1532-5415.2009.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Schwartz J, Wright R, Litonjua A, Tarantini L, Suh H, Sparrow D, Vokonas P, Baccarelli A. Decline in genomic DNA methylation through aging in a cohort of elderly subjects. Mech Ageing Dev. 2009;130:234–239. doi: 10.1016/j.mad.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle PA, Buchman AS, Wilson RS, Leurgans SE, Bennett DA. Physical frailty is associated with incident mild cognitive impairment in community-based older persons. J Am Geriatr Soc. 2010;58:248–255. doi: 10.1111/j.1532-5415.2009.02671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and Regression Trees. Wadsworth Statistics/Probability Series. Belmont, CA: Wadsworth; 1984. [Google Scholar]
- Buchman AS, Boyle PA, Wilson RS, Tang Y, Bennett DA. Frailty is associated with incident Alzheimer's disease and cognitive decline in the elderly. Psychosom Med. 2007;69:483–489. doi: 10.1097/psy.0b013e318068de1d. [DOI] [PubMed] [Google Scholar]
- Castro R, Rivera I, Struys EA, Jansen EE, Ravasco P, Camilo ME, Blom HJ, Jakobs C, Tavares de Almeida I. Increased homocysteine and S-adenosylhomocysteine concentrations and DNA hypomethylation in vascular disease. Clin Chem. 2003;49:1292–1296. doi: 10.1373/49.8.1292. [DOI] [PubMed] [Google Scholar]
- Coneyworth LJ, Mathers JC, Ford D. Does promoter methylation of the SLC30A5 (ZnT5) zinc transporter gene contribute to the ageing-related decline in zinc status? Proc Nutr Soc. 2009;68:142–147. doi: 10.1017/S0029665109001104. [DOI] [PubMed] [Google Scholar]
- Davis DH, Rockwood MR, Mitnitski AB, Rockwood K. Impairments in mobility and balance in relation to frailty. Arch Gerontol Geriatr. 2010 doi: 10.1016/j.archger.2010.06.013. [DOI] [PubMed] [Google Scholar]
- Rango F, Montesanto A, Berardelli M, Mazzei B, Mari V, Lattanzio F, Corsonello A, Passarino G. To grow old in southern Italy: a comprehensive description of the old and oldest old in Calabria. Gerontology. 2010 doi: 10.1159/000316941. [DOI] [PubMed] [Google Scholar]
- Desai A, Grolleau-Julius A, Yung R. Leukocyte function in the aging immune system. J Leukoc Biol. 2010;87:1001–1009. doi: 10.1189/jlb.0809542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarri O, Becker T, Junen J, Manzoor SS, Diaz-Lacava A, Schwaab R, Wienker T, Oldenburg J. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum Genet. 2007;122:505–514. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- Evans WJ, Paolisso G, Abbatecola AM, Corsonello A, Bustacchini S, Strollo F, Lattanzio F. Frailty and muscle metabolism dysregulation in the elderly. Biogerontology. 2010;11:527–536. doi: 10.1007/s10522-010-9297-0. [DOI] [PubMed] [Google Scholar]
- Ferrucci L, Harris TB, Guralnik JM, Tracy RP, Corti MC, Cohen HJ, Penninx B, Pahor M, Wallace R, Havlik RJ. Serum IL-6 level and the development of disability in older persons. J Am Geriatr Soc. 1999;47:639–646. doi: 10.1111/j.1532-5415.1999.tb01583.x. [DOI] [PubMed] [Google Scholar]
- Fraga MF. Genetic and epigenetic regulation of aging. Curr Opin Immunol. 2009;21:446–453. doi: 10.1016/j.coi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suñer D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005 doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried LP, Tangen CM, Walston J, Newman AB, Hirsch C, Gottdiener J, Seeman T, Tracy R, Kop WJ, Burke G, McBurnie MA, Cardiovascular Health Study Collaborative Research Group Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci. 2001;56:M146–M156. doi: 10.1093/gerona/56.3.M146. [DOI] [PubMed] [Google Scholar]
- Fried LP, Ferrucci L, Darer J, Williamson JD, Anderson G. Untangling the concepts of disability, frailty, and comorbidity: implications for improved targeting and care. J Gerontol A Biol Sci Med Sci. 2004;59:255–263. doi: 10.1093/gerona/59.3.M255. [DOI] [PubMed] [Google Scholar]
- Fuke C, Shimabukuro M, Petronis A, Sugimoto J, Oda T, Miura K, Miyazaki T, Ogura C, Okazaki Y, Jinno Y. Age related changes in 5-methylcytosine content in human peripheral leukocytes and placentas: an HPLC-based study. Ann Hum Genet. 2004;68:196–204. doi: 10.1046/j.1529-8817.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- Fulop T, Larbi A, Witkowski JM, McElhaney J, Loeb M, Mitnitski A, Pawelec G. Aging, frailty and age-related diseases. Biogerontology. 2010;11:547–563. doi: 10.1007/s10522-010-9287-2. [DOI] [PubMed] [Google Scholar]
- Gravina S, Vijg J. Epigenetic factors in aging and longevity. Pflugers Arch. 2010;459:247–258. doi: 10.1007/s00424-009-0730-7. [DOI] [PubMed] [Google Scholar]
- Hubbard RE, Woodhouse KW. Frailty, inflammation and the elderly. Biogerontology. 2010;11:635–641. doi: 10.1007/s10522-010-9292-5. [DOI] [PubMed] [Google Scholar]
- Hubbard RE, O'Mahony MS, Savva GM, Calver BL, Woodhouse KW. Inflammation and frailty measures in older people. J Cell Mol Med. 2009;13:3103–3109. doi: 10.1111/j.1582-4934.2009.00733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard RE, Andrew MK, Fallah N, Rockwood K. Comparison of the prognostic importance of diagnosed diabetes, co-morbidity and frailty in older people. Diabet Med. 2010;27:603–606. doi: 10.1111/j.1464-5491.2010.02977.x. [DOI] [PubMed] [Google Scholar]
- Hyde Z, Flicker L, Almeida OP, Hankey GJ, McCaul KA, Chubb SA, Yeap BB. Low free testosterone predicts frailty in older men: the health in men study. J Clin Endocrinol Metab. 2010;95:3165–3172. doi: 10.1210/jc.2009-2754. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Nakamura A, Ishigami A, Goto S, Takahashi R. Age-related difference of site-specific histone modifications in rat liver. Biogerontology. 2009;10:415–421. doi: 10.1007/s10522-008-9176-0. [DOI] [PubMed] [Google Scholar]
- Landi F, Russo A, Pahor M, Capoluongo E, Liperoti R, Cesari M, Bernabei R, Onder G. Serum high-density lipoprotein cholesterol levels and mortality in frail, community-living elderly. Gerontology. 2008;54:71–78. doi: 10.1159/000111381. [DOI] [PubMed] [Google Scholar]
- Landi F, Liperoti R, Russo A, Capoluongo E, Barillaro C, Pahor M, Bernabei R, Onder G. Disability, more than multimorbidity, was predictive of mortality among older persons aged 80 years and older. J Clin Epidemiol. 2010;63:752–759. doi: 10.1016/j.jclinepi.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Lang PO, Michel JP, Zekry D. Frailty syndrome: a transitional state in a dynamic process. Gerontology. 2009;55:539–549. doi: 10.1159/000211949. [DOI] [PubMed] [Google Scholar]
- Lee J, Jeong DJ, Kim J, Lee S, Park JH, Chang B, Jung SI, Yi L, Han Y, Yang Y, Kim KI, Lim JS, Yang I, Jeon S, Bae DH, Kim CJ, Lee MS. The anti-aging gene KLOTHO is a novel target for epigenetic silencing in human cervical carcinoma. Mol Cancer. 2010;9:109. doi: 10.1186/1476-4598-9-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng S, Chaves P, Koenig K, Walston J. Serum interleukin-6 and hemoglobin as physiological correlates in the geriatric syndrome of frailty: a pilot study. J Am Geriatr Soc. 2002;50:1268–1271. doi: 10.1046/j.1532-5415.2002.50315.x. [DOI] [PubMed] [Google Scholar]
- Ling C, Guerra S, Lupi R, Rönn T, Granhall C, Luthman H, Masiello P, Marchetti P, Groop L, Prato S. Epigenetic regulation of PPARGC1A in human type 2 diabetic islets and effect on insulin secretion. Diabetologia. 2008;51:615–622. doi: 10.1007/s00125-007-0916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustosa LP, Coelho FM, Silva JP, Pereira DS, Parentoni AN, Dias JM, Dias RC, Pereira LS. The effects of a muscle resistance program on the functional capacity, knee extensor muscle strength and plasma levels of IL-6 and TNF-alpha in pre-frail elderly women: a randomized crossover clinical trial—a study protoco. Trials. 2010;11:82. doi: 10.1186/1745-6215-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio M, Guralnik JM, Longo DL, Ferrucci L. Interleukin-6 in aging and chronic disease: a magnificent pathway. J Gerontol A Biol Sci Med Sci. 2006;61:575–584. doi: 10.1093/gerona/61.6.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio M, Cattabiani C, Lauretani F, Ferrucci L, Luci M, Valenti G, Ceda G. The concept of multiple hormonal dysregulation. Acta Biomed. 2010;81(Suppl 1):19–29. [PMC free article] [PubMed] [Google Scholar]
- Maslov AY, Vijg J. Genome instability, cancer and aging. Biochim Biophys Acta. 2009;1790:963–969. doi: 10.1016/j.bbagen.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matteini AM, Walston JD, Bandeen-Roche K, Arking DE, Allen RH, Fried LP, Chakravarti A, Stabler SP, Fallin MD. Transcobalamin-II variants, decreased vitamin B12 availability and increased risk of frailty. J Nutr Health Aging. 2010;14:73–77. doi: 10.1007/s12603-010-0013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez L, Benigno BB, McDonald JF. L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Mol Cancer. 2004;3:12. doi: 10.1186/1476-4598-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montesanto A, Lagani V, Martino C, Dato S, Rango F, Berardelli M, Corsonello A, Mazzei B, Mari V, Lattanzio F, Conforti D, Passarino G. A novel, population-specific approach to define frailty. Age. 2010;32:385–395. doi: 10.1007/s11357-010-9136-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AZ, Biggs ML, Matteini A, O’Connor A, McGuire S, Beamer BA, Fallin MD, Fried LP, Walston J, Chakravarti A, Arking DE (2010) Polymorphisms in the mitochondrial DNA control region and frailty in older adults. PLoS One 5. doi:10.1371/journalpone.0011069 [DOI] [PMC free article] [PubMed]
- Oakes CC, Smiraglia DJ, Plass C, Trasler JM, Robaire B. Aging results in hypermethylation of ribosomal DNA in sperm and liver of male rats. Proc Natl Acad Sci USA. 2003;100:1775–1780. doi: 10.1073/pnas.0437971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando TA. Epigenetics and aging. Exp Gerontol. 2010;45:253–254. doi: 10.1016/j.exger.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson B. Impact of aging on DNA methylation. Ageing Res Rev. 2003;2:245–261. doi: 10.1016/S1568-1637(03)00010-2. [DOI] [PubMed] [Google Scholar]
- Rockwood K, Mitnitski A. Frailty in relation to the accumulation of deficits. J Gerontol A Biol Sci Med Sci. 2007;62:722–727. doi: 10.1093/gerona/62.7.722. [DOI] [PubMed] [Google Scholar]
- Rockwood K, Howlett SE, MacKnight C, Beattie BL, Bergman H, Hébert R, Hogan DB, Wolfson C, McDowell I. Prevalence, attributes, and outcomes of fitness and frailty in community-dwelling older adults: report from the Canadian study of health and aging. J Gerontol A Biol Sci Med Sci. 2004;59:1310–1317. doi: 10.1093/gerona/59.12.1310. [DOI] [PubMed] [Google Scholar]
- Schalk BW, Visser M, Deeg DJ, Bouter LM. Lower levels of serum albumin and total cholesterol and future decline in functional performance in older persons: the longitudinal aging study Amsterdam. Age Ageing. 2004;33:266–272. doi: 10.1093/ageing/afh073. [DOI] [PubMed] [Google Scholar]
- Schneider E, Pliushch G, El Hajj N, Galetzka D, Puhl A, Schorsch M, Frauenknecht K, Riepert T, Tresch A, Müller AM, Coerdt W, Zechner U, Haaf T. Spatial, temporal and interindividual epigenetic variation of functionally important DNA methylation patterns. Nucleic Acids Res. 2010;38:3880–3890. doi: 10.1093/nar/gkq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serviddio G, Romano AD, Greco A, Rollo T, Bellanti F, Altomare E, Vendemiale G. Frailty syndrome is associated with altered circulating redox balance and increased markers of oxidative stress. Int J Immunopathol Pharmacol. 2009;22:819–827. doi: 10.1177/039463200902200328. [DOI] [PubMed] [Google Scholar]
- Shimabukuro M, Sasaki T, Imamura A, Tsujita T, Fuke C, Umekage T, Tochigi M, Hiramatsu K, Miyazaki T, Oda T, Sugimoto J, Jinno Y, Okazaki Y. Global hypomethylation of peripheral leukocyte DNA in male patients with schizophrenia: a potential link between epigenetics and schizophrenia. J Psychiatr Res. 2006;41:1042–1046. doi: 10.1016/j.jpsychires.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Siegmund KD, Connor CM, Campan M, Long TI, Weisenberger DJ, Biniszkiewicz D, Jaenisch R, Laird PW, Akbarian S. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland JE, Costa M. Epigenetics and the environment. Ann NY Acad Sci. 2003;983:151–160. doi: 10.1111/j.1749-6632.2003.tb05970.x. [DOI] [PubMed] [Google Scholar]
- Thompson RF, Atzmon G, Gheorghe C, Liang HQ, Lowes C, Greally JM, Barzilai N. Tissue-specific dysregulation of DNA methylation in aging. Aging Cell. 2010;9:506–518. doi: 10.1111/j.1474-9726.2010.00577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topinková E. Aging, disability and frailty. Ann Nutr Metab. 2008;52(Suppl 1):6–11. doi: 10.1159/000115340. [DOI] [PubMed] [Google Scholar]
- Tra J, Kondo T, Lu Q, Kuick R, Hanash S, Richardson B. Infrequent occurrence of age-dependent changes in CpG island methylation as detected by restriction landmark genome scanning. Mech Ageing Dev. 2002;123:1487–1503. doi: 10.1016/S0047-6374(02)00080-5. [DOI] [PubMed] [Google Scholar]
- Walston J, Hadley EC, Ferrucci L, Guralnik JM, Newman AB, Studenski SA, Ershler WB, Harris T, Fried LP. Research agenda for frailty in older adults: toward a better understanding of physiology and etiology: summary from the American Geriatrics Society/National Institute on Aging Research Conference on Frailty in Older Adults. J Am Geriatr Soc. 2006;54:991–1001. doi: 10.1111/j.1532-5415.2006.00745.x. [DOI] [PubMed] [Google Scholar]
- Wilson VL, Smith RA, Ma S, Cutler RG. Genomic 5-methyldeoxycytidine decreases with age. J Biol Chem. 1987;262:9948–9951. [PubMed] [Google Scholar]