Abstract
The invariant chain (Ii) is the critical third chain required for the MHC class II heterodimer to be properly guided through the cell, loaded with peptide, and expressed on the surface of antigen presenting cells. Here, we report the isolation of the nurse shark Ii gene, and the comparative analysis of Ii splice variants, expression, genomic organization, predicted structure, and function throughout vertebrate evolution. Alternative splicing to yield Ii with and without the putative protease-protective, thyroglobulin-like domain is as ancient as the MHC-based adaptive immune system, as our analyses in shark and lizard further show conservation of this mechanism in all vertebrate classes except bony fish. Remarkable coordinate expression of Ii and class II was found in shark tissues. Conserved Ii residues and cathepsin L orthologs suggest their long co-evolution in the antigen presentation pathway, and genomic analyses suggest 450 million years of conserved Ii exon/intron structure. Other than an extended linker preceding the thyroglobulin-like domain in cartilaginous fish, the Ii gene and protein are predicted to have largely similar physiology from shark to man. Duplicated Ii genes found only in teleosts appear to have become sub-functionalized, as one form is predicted to play the same role as that mediated by Ii mRNA alternative splicing in all other vertebrate classes. No Ii homologs or potential ancestors of any of the functional Ii domains were found in the jawless fish or lower chordates.
Keywords: MHC, antigen processing, invariant chain, evolution, shark
1. Introduction
The hallmark molecular components of the adaptive immune response have been found in the oldest group of living jawed vertebrates, the cartilaginous fish. Multiple IgH (Flajnik, 2002), IgL (Criscitiello and Flajnik, 2007), and TCR (Criscitiello et al., 2010; Rast et al., 1997) are diversified by RAG (Bernstein et al., 1994) and AID (Conticello et al., 2005) in sharks and rays. Furthermore, these animals are in the oldest extant group of vertebrates having a polymorphic, polygenic major histocompatibility complex (MHC) (Kasahara et al., 1992).
MHC class II glycoproteins present peptides to CD4+ T cells. Newly synthesized class II α and β chains assemble in the endoplasmic reticulum (ER) together with a Type II glycoprotein called the invariant chain (Ii) (Jones et al., 1979). Ii is a chaperone ensuring the correct folding and trafficking of MHC class II proteins (Bikoff et al., 1993). Ii first trimerizes before the sequential addition of three class II α/β dimers (Lamb and Cresswell, 1992). In this nine-chain complex, each Ii blocks the peptide binding groove of one of the three class II heterodimers with a peptide called CLIP (class II-associated invariant chain peptide) (Freisewinkel et al., 1993), which prevents loading of class II with ER-derived proteins and peptides and provides the groove occupancy required for the stability of class II heterodimers (Zhong et al., 1996). In the trans-Golgi the αβIi complex is diverted from the secretory pathway to the endocytic pathway via a conserved motif in the Ii cytoplasmic tail. The complex is transported to an acidified post-Golgi vesicle where first the membrane distal portion of Ii is proteolytically degraded to leave the LIP (leupeptin-induced peptide), which still blocks the peptide binding cleft and retains the Ii transmembrane and cytoplasmic segments that continue to target the complex to the endosomal MHC class II compartment (MIIC) (Blum and Cresswell, 1988). The low pH of MIIC activates proteases to further cleave the membrane-proximal portion of Ii, leaving only CLIP in the peptide binding cleft. Blockade of this progressive cleavage of the Ii results in accumulation of Ii intermediates and reduced class II surface expression (Neefjes and Ploegh, 1992). DM can then bind class II and release LIP or CLIP, facilitating the exchange for endosomal peptides before transport to the cell surface (Schafer et al., 1996). Maturation of phagosomes containing non-self cargo (versus apoptotic self-cargo) may be enhanced by toll-like receptor-mediated vesicular signaling, marking those phagosomes with pathogenic contents for fusion into the MIIC compartment (Blander and Medzhitov, 2006).
The enzymes that cleave Ii are related to papain and known as cathepsins, which are the same proteases that degrade lysosomal contents for antigen loading (Riese and Chapman, 2000). The activation of cathepsins requires an acidic environment, and they can be divided into four categories depending on the critical component of their active sites: cysteine, aspartate, serine, or metal ions (Turk et al., 2001). Cysteine cathepsins are primarily involved in antigen processing, specifically cathepsins L and S are dedicated to this function. Cathepsin activity is regulated by several protein inhibitors, including cystatins, thyropins and even one domain of Ii itself. The thyroglobulin-like (Tg) domain of longer Ii isoforms is a strong inhibitor of cathepsin L but not cathepsin S (Bevec et al., 1996).
The mammalian Ii gene has an exon organization that largely corresponds to its structural protein domains. The first exon encodes the amino-terminal cytoplasmic tail including the endosomal targeting motifs, the second exon encodes the transmembrane domain, and the third exon encodes a linker between the membrane and the trimerization domain that includes CLIP. The fourth, fifth and sixth exons contribute to the three alpha helices and connecting strands of the trimerization domain. The seventh exon, alternatively spliced out in short isoforms, encodes the Tg domain that presumably inhibits cathepsin proteolytic action. The eighth exon nearly encodes the entire carboxy-terminal end, which is often rich in charged residues but has unclear function. The ninth protein-coding exon encodes the final amino acid or two and contains the stop codon.
Unlike MHC genes, Ii genes do not display high allelic polymorphism, but four variants of the protein are found in human as p33, p35, p41 and p43 (O’Sullivan et al., 1987). Use of an alternative start codon accounts for the small molecular weight differences between the (predominant) p33 and p41, and p35 and p43 forms. The p35 and p43 forms contain an ER-retention motif lacking in the shorter forms from the alternative initiation site; this signal is concealed upon αβ binding and allows transport of the nonamer to the Golgi (Schutze et al., 1994). The 10 kDa distinction between the p33/p35 and p41/p43 forms results from the alternatively spliced Tg domain exon, mentioned above. The Tg domain is a structural motif found in several functionally unrelated proteins (e.g. testican, equistatin, thyroglobulin) and sometimes functions as an inhibitor of cysteine proteases, often with higher target specificity than the better studied cystatins (Mihelic and Turk, 2007).
Ii knockout mice show impeded class II transport and surface expression. Class II found on the surface of Ii-deficient cells has an unstable conformation due to the lack of endogenously processed peptide, but the dimers can bind peptide added to the medium. Accordingly, cells from these mice do not present whole exogenous antigen well and the animals have greatly reduced numbers of thymic and peripheral CD4+ T cells (Bikoff et al., 1993; Viville et al., 1993). These transgenic mice studies suggest that Ii prevents class II from binding floppy, incompletely folded proteins in the ER (rather than preventing the binding of peptides transported into the ER by TAP) and stabilize the heterodimer. Ii knockout mice with restoration of either p31 or p41 (containing the Tg domain) have shown that both forms participate in class II folding and assembly, can reconstitute the CD4+ T cell population, and rescue immune responses to protein antigen (Shachar et al., 1995; Takaesu et al., 1995). The complete functions of each isoform are not known; however p41 has been shown to be necessary for airway hyper-responsiveness and IgE responses in the lung (Ye et al., 2003a).
Although crucial to class II antigen presentation, Ii and cathepsins are encoded outside of the MHC (Long et al., 1983). However, cathepsins S and L are found in MHC paralogous regions (Flajnik and Kasahara, 2010), one of many linkages that contribute to hypotheses of an ancestral “pre-adaptive immune complex” encoding antigen receptors, NK receptors and antigen processing and presentation components (Ohta et al., 2011). Ii and homologous genes have been identified in several divergent vertebrate model species, although such reports are few in comparison to class IIα/β chains. Amongst poikilothermic vertebrates, annotated Ii sequences have been submitted to public databases from reptiles and amphibians and studies have been conducted on Ii from bony fish species. The cloning of the first Ii from lower vertebrates was done in zebrafish, and this work confirmed that, like in mammals, fish Ii-like transcripts exist in multiple forms (Yoder et al., 1999). Work in rainbow trout also found two Ii products (Dijkstra et al., 2003) that are encoded by two paralogous genes (Fujiki et al., 2003) as opposed to alternative splicing. Structure of sea bass Ii was modeled more recently with analysis of potential interactions with class II and cathepsins (Silva et al., 2007). Here, we report the first description of Ii from the cartilaginous fish, the oldest vertebrate group with MHC-based adaptive immunity. We set out to determine whether the gene and its expression were phylogenetically conserved, and attempted to find genes related to precursors of Ii and cathepsins in jawless vertebrates and lower deuterosotmes.
2. Methods
2.1 Cloning of nurse shark Ii chain and cathepsins
A Ginglymostoma cirratum (nurse shark) spleen/pancreas cDNA library was constructed in the pDONR222 vector using the Gateway cloning system (Invitrogen). From this an ~8000 clone expressed sequence tag (EST) database was created after removing known housekeeping, MHC, immunoglobulin and TCR clones by subtractive colony hybridization using 137mm Magna membranes (Osmonics) for probing and high stringency washing techniques described previously (Criscitiello et al., 2004). DNA was prepared with 96-Turbo plasmid miniprep kits (Qiagen) or TempliPhi rolling circle DNA amplification (GE Healthcare) and single dye-terminator based sequencing runs were performed at the University of Maryland Biopolymer Core Facility using the universal M13 reverse primer. ESTs were used as queries against the non-redundant protein sequence database with blastx (NCBI). Gene-specific primers (Supplemental Table 1) were designed to complete sequencing of clones with high identity to Ii and cathepsins. Additional cDNA libraries from shark lymphoid tissues were assayed by 5′ and 3′ rapid amplification of cDNA ends (RACE) PCR with gene-specific Ii primers to identify all expressed splice variants. These were cloned and sequenced as above or with Zyppy plasmid DNA miniprep kit (Zymo Research), extended with BigDye XTerminator (Applied Biosystems), purified and sequenced by the Texas A&M DNA Technologies Core Laboratory.
2.2 Blotting
Total RNA was prepared for northern blotting as described (Bartl et al., 1997), and 10 μg was loaded in each lane. The nurse shark nucleotide diphosphate kinase (NDPK) probe used as a loading control was amplified with primers NDPKF and NDPKR (Kasahara et al., 1992) (Supplemental Table 1). A probe for nurse shark Ii was amplified from primers NSIiF1 and NSIiR1 which generate a probe from cDNA encoding the endosomal targeting sequence of the cytoplasmic tail to CLIP. Northern blotting and probing for nurse shark class IIA has been described previously (Kasahara et al., 1992; Ohta et al., 2004). A putatively single exon probe amplified from primers NSIiF2 and NSIiF8 was used in genomic Southern blotting of DNA from shark erythrocytes as previously described (Criscitiello et al., 2006). Blots were probed from five related sharks digested with five different enzymes as well as single enzyme blots of families (mother and pups) of analyzed MHC paternities by restriction fragment length polymorphism (RFLP) (Ohta et al., 2002).
2.3 Database mining and structural prediction
Portions of the nurse shark Ii sequences described here were used to query the database of the elephant shark genome project (http://esharkgenome.imcb.a-star.edu.sg/) by blastn and tblastn. Two scaffolds were identified in this cartilaginous fish containing exons predicted to encode Ii by visual inspection for GT/AG intron boundaries and comparison of predicted protein sequence with other vertebrates.
CD74 (nomenclature for surface expressed Ii (Koch et al., 1991)) is annotated on scaffold 29 of the Anolis carolinus genome (AnoCar1.0) but only two exons are marked. Identification of the entire Ii locus in this reptile was accomplished by first finding anole ESTs (e.g., FG760756 and FG750983) with homology to caiman and other vertebrate Ii sequences, these were used to identify the remaining exons with the exception of the first. The first exon was predicted based on visual scrutiny of ten kilobases 5 of the second exon.
Annotated and partially annotated genomic sequences of Ii loci of human, chicken and the frog Xenopus tropicalis as well as ICLP-1 and ICLP-2 loci of zebrafish genomic sequences were checked against available cDNA data to tabulate and compare exon/intron sizes and phases. These loci were studied using genome browsers at NCBI, Ensemble and UCSC websites where open reading frames 5 and 3 to the Ii ortholog were analyzed for conserved synteny of the region amongst vertebrates.
Similar BLAST approaches were used to identify additional lower vertebrate Ii and cartilaginous fish cathepsin EST sequences using nurse shark and other vertebrate sequences as query (Supplemental Table 2) and to search for orthologs of Ii in lower chordates.
2.4 Phylogenetic analysis
Amino acid alignments of Ii homologs were initially made in Bioedit with ClustalW employing gap opening penalties of 10 and gap extension penalties of 0.1 for pairwise alignments then 0.2 for multiple alignments and the protein-weighting matrix of Gonnett or Blossum (Figure 2 and Supplementary Figure 1) (Hall, 1999; Tamura et al., 2007). These alignments were then heavily modified by hand. MEGA was used to infer the phylogenetic relationships of Ii homologs. Evolutionary distances were computed using the Dayhoff matrix (Schwarz and Dayhoff, 1979) and 387 column positions in the 37 selected sequences. A Neighbor-Joining tree was made from 1000 bootstrap replicates, using pairwise deletion.
Figure 2. Amino acid alignment identifies nurse shark and other chondrichthian Ii when compared to other vertebrate sequences.
Yellow highlighting (spanning two residues) indicates phase zero, green highlighting indicates phase one and blue highlighting indicates phase two intron positioning. Genbank accession numbers are shown in Supplemental Table 2. Endosomal targeting motifs (L-L/I/V) are underlined, as are the six conserved cysteines of Tg-like domains and (in the human sequences) the three alpha helices of the trimerization domain. Asparagine-linked glycosylation motifs (N-X-S/T, X≠P) are in italics. Also in italics are polar residues corresponding to the hydrophilic patch of the transmembrane domain implicated in Ii trimerization.
3. Results
3.1 Isolation and identification of nurse shark Ii chain
Generation of a nurse shark EST database from spleen and pancreas yielded an Ii clone (clone 104D3, comprising 1802 bp, Figure 1) that showed high identity to many Ii sequences of bony fish and tetrapods (highest protein match to the pike Esox lucius, expect 6e-19, 35% amino acid identity over 196 positions). This sequence was used to design primers (Supplemental Table 1) for 5′ and 3′ RACE PCR. Many clones were sequenced from several primer combinations amplified from peripheral blood and spleen to obtain all expressed genes, but only three other reproducible coding sequence variants were isolated, all exemplified by clone D2 (Figure 1). D2 is a long splice variant of EST clone 104D3 with a 195 bp insertion and also contains two single nucleotide polymorphisms (SNP) and two small deletions (indel). The indel creating serine 35 (104D3 aa numbering) and the point mutation exchanging glutamic acid for glutamine at 211 both appear to be allelic polymorphisms, since each occur independently in both the long and short splice isoforms.
Figure 1. Nurse shark cDNAs with homology to Ii chain.
Nucleotide and putative amino acid sequence of nurse shark cDNA clones 104D3 (top) and D2 (bottom). Gaps introduced for alignment are shown as dashes. Single base point mutations and insertion/deletions are highlighted. Amino acids of the endosomal targeting motif are highlighted in pink, the transmembrane domain in green, CLIP yellow, trimerization domain blue and Tg domain red. NCBI has given these sequences accession numbers JF507710 and JF507711.
3.2 Mining of other Ii sequences from poikilothermic vertebrates
Isolation of the nurse shark Ii sequence prompted database searches for Ii in other species. We found ESTs of complete and partial Ii from dogfish shark (Squalus acanthus) and Pacific electric ray (Torpedo californica) and performed partial genomic analysis of Ii from the more primitive Holocephalin, the elephant shark (Callorhinchus milii). Other bony fish Ii ESTs were found, full genomic annotation was completed for the green anole lizard (Anolis carolina) and the Ii exon/intron structure was manually acquired for the two zebrafish ICLPs, Xenopus tropicalis, chicken and human Ii genes. Accession numbers are shown in Supplemental Table 2 and all sequences analyzed are aligned in Supplemental Figure 1.
3.3 Sequence analysis of Ii from cartilaginous fish
The primary functional domains that have been described in Ii of higher vertebrates (transmembrane, trimerization, CLIP, Tg; all colored domains in Figure 1) are present in Ii of both major radiations of elasmobranchs (modern sharks (Selachi) and skates and rays (Batoidea), Figure 2). Additionally, the elephant shark Ii confirms that at least the trimerization domain and Tg are present in the more primitive holocephalians. Protein identity to the complete nurse shark long isoform (D2 adding N-terminal MSADEQQNALL) in pairwise alignments range from 33% with trout S25-7 Ii, to 29% with caiman to 26% with chicken and human. The long connecting linker domain between the trimerization domain and Tg seems to be a common feature in the cartilaginous fish lineage.
3.3.1 Cytoplasmic tail and transmembrane domain
As a Type II transmembrane protein, Ii has an amino terminal cytoplasmic tail. No evidence was found by 5′RACE for use of an alternative start codon in nurse shark or in the databases for any other ectothermic vertebrate. Database searches only found clear evidence from the rhesus macaque (XM_001099491) and a new world marmoset (XR_087115) of alternative start codon use similar to human, and the giant panda Ii has a putative 15aa alternative amino terminal peptide similar in sequence to the primate sequences. The Arg-Arg-Ser-Arg ER localization signal in this longer alternative initiation product of human Ii is how such signals were discovered (Bakke and Dobberstein, 1990; Schutze et al., 1994), yet this signal cannot be detected in the cytoplasmic tails of other vertebrate Ii. Di-leucine like motifs have been identified in the mammalian Ii cytoplasmic region which serve as endosomal targeting motifs (Pieters et al., 1993) and mediate Ii interaction with the clathrin adaptor proteins AP1 and AP2 (Kongsvik et al., 2002). One such candidate sequence is seen in the nurse shark Ii at position 21–22 (Figure 2). Acidic residues preceding the di-leucine are conserved in the shark motif and have proven necessary for sorting to large endosomes (Pond et al., 1995). The EST included in Figure 2 for Ii of the electric ray has the longest cytoplasmic domain of any Ii studied that does not use an alternative start methionine.
The transmembrane region of Ii is much more conserved among vertebrates than any other part of the molecule, typically rich in hydrophobic residues. The transmembrane region of cartilaginous fish Ii maintains two of three polar residues that in human form a hydrophilic patch important for trimerization (Ashman and Miller, 1999). The nurse shark sequence has QvAS at positions 75–78 where the human employs QaTT.
3.3.2 CLIP (Class II associated peptide)
CLIP has been shown in mammals to be crucial for class II folding, transport, and peptide groove occupancy (Romagnoli and Germain, 1994), yet has proven difficult to compare amongst vertebrates as evidenced by very different published alignments that include sequences from bony fish (Dijkstra et al., 2003; Fujiki et al., 2003; Silva et al., 2007). Indeed, assigning different gap penalties and inclusion or exclusion of different sequences can drastically change alignments of this region performed by programs such as Clustal and Muscle. Silva et al. noted striking conservation of large and small CLIP side chains in a teleost Ii that bind in conserved pockets of human MHC (Ghosh et al., 1995), although this required inserting a three amino acid gap in the mammalian CLIP core region that occupies the MHC class II groove (Silva et al., 2007). When cartilaginous fish, amphibian and reptile sequences are included in the alignment such conservation patterns are not as evident. The regions between the transmembrane domain and CLIP, and CLIP and the carboxy-proximal trimerization domain, show similarly poor conservation between shark and other vertebrate groups. Similarly, the cytoplasmic region showed some divergence between the bony fish and other vertebrate sequences, CLIP and the region linking the CLIP with the trimerization domain show significant differences in teleost Ii in comparison to other vertebrates.
3.3.3 Trimerization domain and linker region
Carboxy-proximal to CLIP is a trimerization domain containing three conserved alpha helices (underlined in Figure 2). Many residues in these helices of human Ii that are important for nonamer formation are evolutionarily conserved (Jasanoff et al., 1998). Four leucines (165 and 166 of helix a, 195 and 198 of helix b) are well conserved, and several other residues (Trp210, Phe213, Glu214, Trp216, Trp220, and Phe223) are nearly invariant. Amino acids making the crucial bonds required for packing of the third (c) alpha helix in the Ii trimer (Bijlmakers et al., 1994) are conserved, and three residues (Phe208, Trp211, Trp215) are invariant over evolutionary time (see also Supplemental Figure 1). In cartilaginous fish Ii, one (elephant shark and electric ray) or two (nurse and dogfish shark) putative N-linked glycosylation sites are found in or flanking alpha helix A, as has been seen in other vertebrates.
3.3.4 Tg domain and carboxy terminus
The longer isoform (D2) of nurse shark Ii includes a Tg domain. This isoform has been shown in mammals to be generated by insertion of a 195bp exon (Strubin et al., 1986). Tg domains display inhibitory activity against cysteine- or cation-dependent proteases and are called thyropins (Lenarcic and Bevec, 1998). The Tg domain of Ii from shark also is a likely thyropin. This cysteine-rich domain in mammals interacts with the active site of some cathepsins in an inhibitory fashion, and can discriminate cathepsins L from S (Guncar et al., 1999a). All six cysteines that in human Ii form three disulfide bonds that stabilize the tertiary structure of the domain are conserved in shark Ii, suggesting that the shark Ii Tg domain assumes a two sub-domain structure seen in human Ii. Indeed, these six cysteines (and a tryptophan between the fourth and fifth cysteine) are found in all vertebrate Ii having a Tg domain (Figure 2 and Supplemental Figure 1).
Like in most vertebrates, some short teleost Ii clones were found. These Tg-less fish Ii were first identified from the trout (INVX) but this Ii homolog was encoded by a gene distinct from the Tg-containing trout Ii chains, rather than the products of alternative splicing (Fujiki et al., 2003). Sequences with a predicted domain structure like INVX have also been submitted from Atlantic salmon (Leong et al., 2010). This may be due to the teleost-specific genome duplication, generating two Ii loci.
The Ii carboxy terminal portion adjacent to the Tg domain is much shorter in nurse shark than other vertebrates, except the cyprinid ICLP-1’s in which it is also short. Little is known about this portion of Ii but it could be involved in CD74 signaling of macrophage inhibitory factor with CD44 (Shi et al., 2006). This region is rich in charged residues in bony vertebrates, and teleost Ii (except the ICLP-1’s) have a distinctive stretch of aliphatic and other hydrophobic residues preceding the region of glutamic and aspartic acids, lysines and arginines (Figure 2 and Supplemental Figure 1).
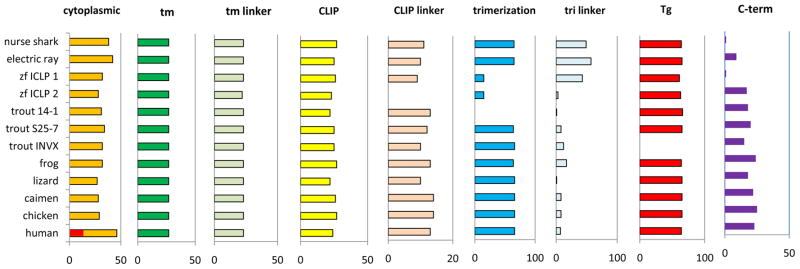
3.4 Comparative domain structure
Measurement of predicted domains and intervening protein linker sequence showed general conservation between Ii of different classes of jawed vertebrates (Figure 3). The most conspicuous deviation is shared by Ii from cartilaginous fish and ICLP-1 of bony fish, where an extended linker between the trimerization domain (or A alpha helix of trimerization domain in the case of ICLP-1) and Tg domain is coincident with a shorter carboxy terminal tail than is found in tetrapod and other bony fish Ii forms. As the ICLP-1 and nurse shark linkers are dissimilar to each other (and anything else in the databases) it is doubtful that there is rescue in this region of any function missing in their short carboxy terminus.
Figure 3. Invariant chain domain conservation in cartilaginous fish and tetrapods.
Representative predicted protein sequences were chosen to compare the entire longest splice forms of invariant chain homologs found. Xenopus laevis was used for frog. Human is shown with earlier alternative start site that gives 16 additional amino acids to the cytoplasmic tail, shown in red. Length is measured in amino acids.
As stated above, some teleost Ii forms only appear without certain domains. Splice forms are never found of either zebrafish ICLP or trout 14-1 with a complete trimerization domain and trout and salmon INVX does not contain the Tg domain (Dijkstra et al., 2003; Fujiki et al., 2003; Yoder et al., 1999).
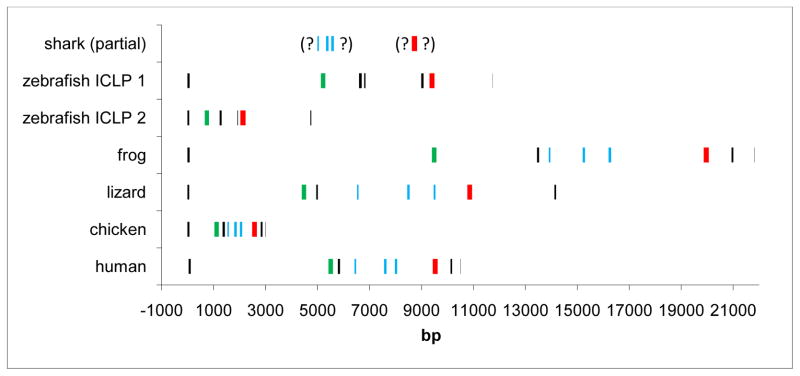
3.5 Genomic organization of the Ii locus in vertebrates
We compared protein coding exon/intron lengths and splice sites where available from shark to man (Figure 4, annotation of green anole lizard and elephant shark shown in Supplemental Figures 2 and 3, respectively). Similar intron positions and phases were found in frog, lizard, chicken and man (Figure 2). Although intron sizes were much longer in X. tropicalis and much shorter in chicken (typical for chickens), the relative size of these introns to those of other higher mammals is consistent with expansions and contractions of these loci. A separate small exon for the stop codon was not found in the anole lizard as it was in other tetrapods.
Figure 4. Exon-intron organization and relative size similar from shark to man.
Exons encoding the transmembrane domain are shown in green, the exons approximately encoding the three alpha-helices of the trimerization domain are shown in blue, and the Tg domain in red. Question marks denote missing data from elephant shark scaffolds, exon content in each set of parentheses is on one unmapped scaffold. Drawn to scale, distance in base pairs is shown at bottom.
The two zebrafish ICLP loci analyzed showed that ICLP-1 on chromosome 14 was twice as long as ICLP-2 on chromosome 12, including an additional exon for the linker between the partial trimerization domain and Tg. As might be expected considering the sequence divergence, the partial trimerization domains of the ICLPs and subsequent linker showed the most diversity in exon/intron organization. This region in shark contains a phase two intron not consistent with other vertebrates, and the ICLPs contained a phase zero intron the position of which could not be aligned with precision to the introns of other vertebrate Ii. The three domains encoding the elephant shark’s trimerization domain were found together on one scaffold, and the Tg domain was found on another.
Southern blotting with genomic DNA of a family of nurse sharks usually resulted in two bands hybridizing to an Ii transmembrane-CLIP probe (Supplemental Figure 4) consistent with a single Ii locus in shark, rather than multiple loci as in bony fish. RFLP were compared with known patterns for MHC (Ohta et al., 2002) and shark Ii was not MHC-linked (data not shown).
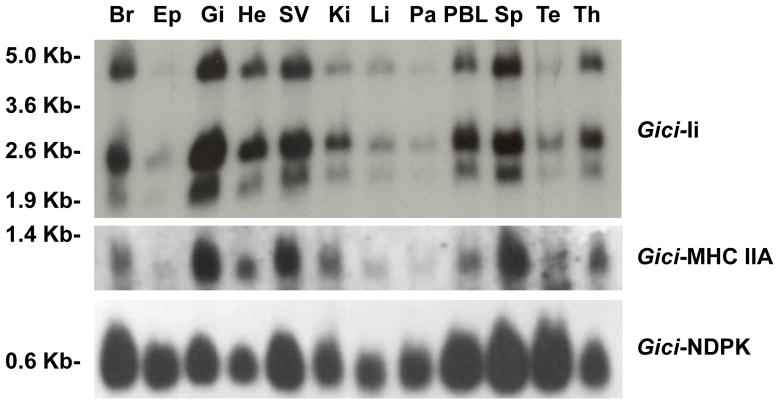
3.7 Tissue expression
Northern blotting on RNA from many nurse shark tissues demonstrated coordinate regulation of Ii transcripts with those of MHC class II (Figure 5). Expression was highest in gill, spleen and spiral valve (shark intestine), with lower but obvious expression also in peripheral blood leukocytes, thymus and brain. Consistent with what is known in mammals, such synchronized expression of these two genes suggests shared transcriptional regulation early in vertebrate adaptive immunity. In mammals both genes’ promoters (and that of HLA-DM) depend on the class II transactivator (CIITA) to coordinate upregulation of transcription in response to interferon γ ((Brown et al., 1993), reviewed in (Ting and Trowsdale, 2002)). CIITA has yet to be unambiguously identified below bony fish but it appears likely that it or an analogous system regulates their expression in shark. We did identify a candidate CIITA partially encoded on an elephant shark scaffold (AAVX01120910.1) which shares 42% identity and 66% similarity (e=4e-49) with CIITA of zebra finch (Figure 6).
Figure 5. Tissue expression of shark Ii mRNA.
Northern blot hybridization of nurse shark tissues (Br, brain; Ep, epigonal; Gi, gills; He, SV, spiral valve; Ki, kidney; Li, liver; Pa, pancreas; PBL, peripheral blood leukocytes; Sp, spleen; Te, testis; and Th, thymus) with the Gici-Ii probe demonstrates that three transcripts for Ii are expressed at similar levels relative to MHCIIA. Size in bases of the transcripts is shown on the left.
Figure 6. CIITA of cartilaginous fish.
Amino acid alignment of partial putative elephant shark (eShark) ortholog of CIITA. Yellow highlighting indicates identity with other vertebrate CIITA proteins. Predicted domains of protein shown under alignment. Other sequences included in the alignment: finch XP002195062, opossum XM001376433, zebra danio XP001343072, and human NP000237.
Multiple 5′ and 3′ RACE experiments yielded Ii cDNA clones yielding transcripts of 1.8B (D2) and 2.3B (104D3) (Figure 1) based on alternative polyadenylation sites in the 3′ untranslated region differing by 497B (assuming 200B poly-A tails (Wahle and Keller, 1992)). Each of these has an alternative splice isoform possible without the 195B exon encoding the Tg domain. The northern blotting confirmed predominant bands migrating near the 1.6–1.8B and 2.1–2.3B sequence lengths of each form with and without the Tg domain exon, but the slightly larger transcripts were higher than predicted possibly do to longer poly-A tails or longer 5′ untranslated regions. There was also a larger 4.8B band that we could not identify from cDNA. This large transcript displayed parallel tissue expression levels as the lower bands. Therefore there is likely an additional polyadenylation variant or exon splice isoform that has yet to be identified. We can exclude the possibility of a second Ii locus that eluded the Southern probing as it would have hybridized to the same probe on the northern blot (Supplemental Figure 4).
3.8 Cathepsin degradation and evolution of Ii regulation
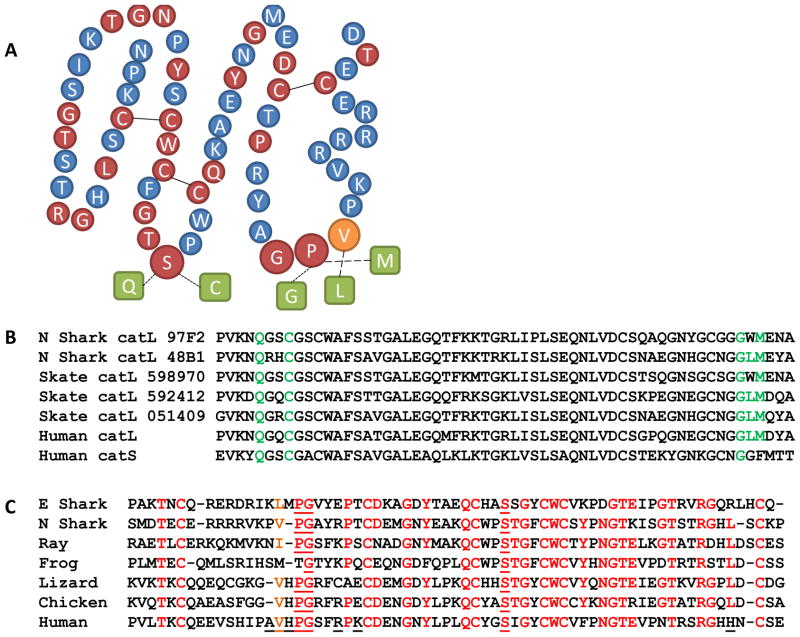
Since the Tg-like domain is a putative component of Ii of all jawed vertebrate groups back to shark, we searched for possible interactions with cathepsins. Several cathepsin ESTs were found from nurse shark, two of which were more similar to the ancient L cathepsin lineage, implicated in Ii proteolysis, than to the primeval B lineage that are not (Figure 7, Supplemental Figure 5) (Uinuk-Ool et al., 2003). Additional putative cathepsin L/S EST sequences were mined from the little skate (Leucoraja erinacea). Based on the crystal structure of the mammalian Ii Tg domain with cathepsin L we modeled similar interactions between the Ii and cathepsin orthologs from cartilaginous fish (Guncar et al., 1999a).
Figure 7. Predicted cathepsin L inhibition by Tg domain in cartilaginous fish.
A. Bead amino acid schematic of Tg-like domain of nurse shark. Amino acids of Ii predicted from mammalian crystal structures to be important contacts with cathepsin L and conserved in shark are shown as larger circles. Conserved cathepsin amino acids that are predicted to interact with Ii are shown as green boxes, dotted lines show hydrogen bonds and electrostatic interactions, dashed lines show hydrophobic interactions. Disulfide bonded cysteines are shown joined by a line. B. Five cathepsins from elasmobranchs having similarities to cathepsin L aligned with cathepsin L and S from human. Residues predicted to form key conserved interactions are in green. C. Amino acid alignment of Tg – like domain from elephant shark, nurse shark, Pacific electric ray, frog (X. laevis), anole lizard, chicken and human. Residues conserved in at least 5 of the 7 aligned sequences are shown in red in panel C and red beads in panel A. Nurse shark Val255 and orthologous residues are highlighted in orange as this position was usually maintained as an aliphatic, hydrophobic residue. Underlined amino acids make key contacts between Ii Tg-like domain and cathepsin L in human. Structure interactions adapted from Guncar and Turk (Guncar et al., 1999b).
As described above, the Tg-like domain of mammalian Ii forms a wedge-shaped conformation of three loops stabilized by three disulfide bonds between conserved cysteines (Turk et al., 1999). The inhibited papain-like cysteine proteases including cathepsin L share a common fold of two domains, which separate on the top in a “V” shaped active site cleft (Coulombe et al., 1996). Several interactions identified in the solved mammalian cathepsin L – Tg domain structure could be maintained by residues found in the Tg domains and putative cathepsin L in cartilaginous fish.
3.9 Ii evolution
Besides the structural data described above, two additional lines of evidence from phylogenetic and syntenic analyses suggested that the canonical functions of Ii arose at the origins of RAG/AID/MHC-based adaptive immunity.
3.9.1 Phylogenetic analysis
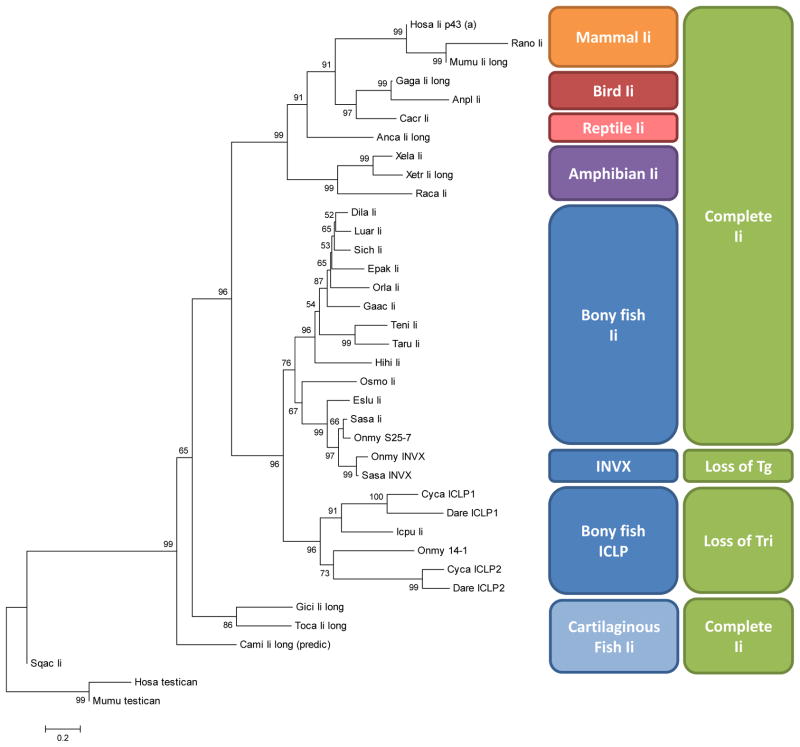
We performed many phylogenetic analyses with different alignments, excluding various domains, and with several matrix- and tree-building algorithms. The tree with the most support at significant nodes includes the entire longest form (Tg domain encoding exon spliced in) of the proteins (Figure 8). Ii from chondrichthyes clustered basal to Ii and Ii paralogs from bony vertebrates. The incomplete sequence from the dogfish shark behaved erratically with different tree building methods, whereas the incomplete elephant shark repeatedly fell basal to all the other vertebrate Ii, as expected for the primitive Holocephalian. As suggested by sequence analysis, domain structure, and intron splice sites; cardinal Ii emerged in the cartilaginous fish.
Figure 8. Cartilaginous fish Ii groups with other Ii in phylogenetic analysis.
Neighbor joining tree of Ii amino acid sequences aligned and analyzed in MEGA. Alignment (with sequences not included in this tree) is found in Supplemental Figure 1, sequence accession numbers and names given in Supplemental Table 2. Bootstrap values at nodes inferred from 1000 replicates. Evolutionary distance is shown with the scale bar in the units of amino acid substitutions per site. Boxes to the center-right highlight different vertebrate phylogenetic clusters of Ii and far right mark loss of Tg and trimerization domains in some teleost Ii forms, compared to the complete Ii.
On the other hand, teleost sequences fell into two well supported clades: those ICLP-like sequences lacking the trimerization domains and those more typical of other vertebrate Ii. A duplication leading to the ICLP genes likely occurred in the ancestors of protacanthopterygii (including samonids) and ostariophysi (including catfish and cyprinids). The INVX duplication generating Tg-less bony fish Ii may have occurred more recently in the salmonid lineage, as they have thus far only been found in trout and salmon.
3.9.2 Genomic syntenies
We used available genome projects to study the Ii locus. The amniota include the (paraphyletic) reptiles, birds and mammals, showed conservation of syntenic genes up- and downstream of the Ii locus (Supplemental Figure 6). However, we identified only one neighboring gene (Tcof1, encoding the treacle protein associated with Treacher Collins Syndrome) that was conserved between the amphibian Xenopus and the amniotes. The gene distal to Ii on the other side of Tcof1 in frog was csf1r (colony stimulating factor receptor), that linked to the flanking regions of zebrafish ICLPs. The additional genome-wide duplication in bony fish likely allowed for much divergence, but clear conservation of synteny was found among frog, lizard, bird, mammals, and one of the bony fish forms. Surprisingly, the cod has lost Ii as well as class II and functional CD4 (Star et al., 2011).
4. Discussion
In characterizing the expressed Ii gene in nurse shark and other cartilaginous fish we found general conservation of sequence, splice variants, expression, genomic organization, predicted structure, and function with Ii from tetrapods. Evidence was also found of an ancient relationship between the specialized cathepsin L and the Ii as well as the regulation of the former’s action by the Ii’s own Tg domain. Lockstep expression of Ii and class II was observed and a putative CIITA was identified from cartilaginous fish. Genomic syntenies, exon-intron structure and Southern blotting suggest that one Ii locus emerged early in the Ig/TCR/MHC based adaptive immune system of jawed vertebrates and was maintained in all major groups. Additional genome wide duplication(s) afforded bony fish multiple Ii loci encoding different domain structures, presumably with distinct functions, one of which is found in Ii splice forms of all other vertebrates.
Class II groove occupancy by CLIP is a hallmark property of the invariant chain, yet we found little strict conservation of this sequence between vertebrate classes. However methionine residues were common in shark as other vertebrate CLIP regions. Two human Ii CLIP methionine residues (in position127 and 138 of Figure 2) are the most important occupiers of conserved class II pockets (Ghosh et al., 1995), and meta comparative analyses may reveal a broader “super-motif” among vertebrate class II of deep pockets that recognize these CLIP methionine residues (Malcherek et al., 1995). Other residues such as alanine, arginine, proline and serine also are common in CLIP. We suspect that the high positive selection exerted on the class II peptide binding groove over the half-billion years of Ii evolution has forced CLIP to change accordingly. Additionally, temperature may pose very different requirements on the CLIP-class II interaction in endothermic birds and mammals versus poikilothermic vertebrates. CLIP and the region linking CLIP with the trimerization domain are where the teleost Ii homologs show significant differences from other vertebrate Ii, suggestive of distinct physiology.
All current data suggest that the trimerization domain is a fixed component of all Ii from all groups except teleost fish, as in the cyprinid ICLPs and trout 14-1 sequence. In some of these fish sequences an extended region is substituted for the trimerization domain, most evident as positions 222–253 of zebrafish ICLP-1 (Figure 2). It is noteworthy that nurse shark and electric ray Ii have extended regions following their trimerization domains. Although the teleost sequence replacing the trimerization domain in ICLP-1 may be similar to prokaryotic catalase (Dijkstra et al., 2003), it is possible this was due to untrimmed vector deposited in the databases. These unique portions of the bony fish sequences and the extended region after the trimerization domain of cartilaginous fish are similar neither to each other nor any to any other protein.
Interestingly, cathepsin S, the cathepsin free of Tg domain inhibition, could not be identified from cartilaginous fish, suggesting that it may have evolved after the emergence of the class II pathway. Cathepsin S is pH independent and is upregulated by interferon γ (Chapman, 1998), characteristics of a highly specialized adaptive cathepsin. In mammals the regulation afforded by cathepsin S and L (or S, L and V (Sevenich et al., 2010)) may be used for modulation of T cell selection in the thymus (Lombardi et al., 2005), as cathepsin L is necessary for optimal positive selection by cortical thymic epithelial cells (Nakagawa et al., 1998). Perhaps this was coincident with the neofunctionalization of cathepsin K in osteoclasts. Other components of the class II pathway arose after the emergence of adaptive immunity, DM in amphibians and DO in mammals.
At least twice in teleost Ii evolution new Ii loci emerged that have been specialized by loss of exons: trout and salmon INVX have no Tg domain (like alternative splicing in other vertebrates) and the ICLPs (including trout 14-1 and a catfish cDNA) have no trimerization domain. Loss of the trimerization domain in the ICLPs would be expected to alter the class II processing pathway significantly. Trimerization is necessary for release of the Ii/class II complex from the ER and protection from rapid degradation, yet the Ii TM domain is capable of trimerizing as well (Dixon et al., 2006). The self-affinity of Ii in the membrane is attributed to a glutamine and two threonine residues, of which the glutamine is well conserved from shark to man (Figure 2). The teleost ICLPs that lack the trimerization domain do have one of the less conserved threonines as well as the glutamine. Future work must address how MHCII/Ii trimers (instead of nonamers) articulate with calnexin and are transported to the MIIC. Interestingly, some bony fish appear to make multiple Ii forms with diverse domain configurations (trout having full length S25-7, INVX lacking the Tg and 14-1 lacking the trimerization), while other (perhaps most) fish seem restricted to just one or two variants. The two genes in one fish species are often very similar, apparently without subfunctionalization. But maintenance of two ICLP forms in the cyprinids suggests the possibility of distinct physiology for ICLP-1 and ICLP2.
A wealth of circumstantial evidence suggests the Tg domain is resistant to proteolysis which shifts the control to the regulation of cathepsin S which is exempt from the Tg domain’s inhibition. But certainly the smaller forms must be favored in some circumstances? Evidence for this is not easy to find in the literature, but Ii p31 (without Tg) predominates in B cells and the longer form enhances antigen processing in macrophages and dendritic cells (Ye et al., 2003a). At a gross tissue level, nurse shark certainly seems to express the two isoforms defined here at similar levels (Figure 5) as neither the middle nor the lower “bands” representing differential polyadenylation sites is tight, signifying that both contain the isoforms with and without the Tg domain.
This Tg domain near the carboxy-terminus of Ii (Katunuma et al., 1994) is a member of the cystatin superfamily (Brown and Dziegielewska, 1997) and this suggests that Ii could have evolved from a stefin or cystatin for chaperone function. As CLIP became more specialized for the class II peptide binding groove, the Tg domain could have coevolved with a specialized cathepsin. Work in Ii/H2-M−/− mice with restoration of p43 suggested that, like DM, the Ii Tg domain may augment peptide-CLIP exchange (Bikoff et al., 1998), which might be the ancestral method of facilitating peptide/CLIP exchange prior to the emergence of DM in amphibians.
Several search strategies failed to identify an Ii ortholog in jawless chordates (Supplemental Table 3). To obtain the remnant genes containing functional domains in Ii, the Tg and trimerization domains of nurse shark and electric ray, as well as the ‘transmembrane to CLIP’ and carboxy terminus from nurse shark were used as bait in BLAST searches against lamprey genomic scaffolds, lamprey ESTs, hagfish ESTs and genomic scaffolds from the urochordate Ciona intestinalis and cephalochordate Branchiostoma floridae. That no likely candidates were revealed may indicate great sequence divergence between the gnathostome Ii and the fragmented loci that were assembled for the genesis of Ii; such ancestral loci would likely have encoded genes with quite different functions before the emergence of the class II antigen presentation system.
However, syntenic clusters of genes may be maintained in cartilaginous fish and even jawless vertebrates to the cystatin family member or other gene(s) that later gave rise to the Ii. We used these identified syntenic genes (Arsi, Tcof1, Rps14, Ndst1, Synpo and Csf1r) from more recent vertebrate groups to search Version 3.0 of the lamprey (Petromyzon marinus) genome project supercontigs. Despite finding good candidates for human Tcof1 (lamprey contig 40760), mouse Arsi (lamprey contig 7255), zebrafish Csf1r (amphioxus unassigned chromosomal scaffold 617826) and human RPS14 (lamprey contig 47654, and many lesser candidates for these and other syntenics in lamprey) we were unable to identify a neighboring gene on these short contigs that bore domains or characteristics likely to be co-opted into the Ii (trimerization, Tg-like, type II transmembrane with di-leucine motifs). We do not think this effort will remain futile; completion of the lamprey genome as well as those of other vertebrate genomes may yet yield an Ii antecedent. The location of the CD74 gene in humans (5q11-23) is intriguing, while not on an MHC paralogon it is near a region that seems to have broken off from the MHC paralogous region on chromosome 9 (Lundin et al., 2003).
In considering the evolution of the class II system, we should note that many functions have been described for Ii. As mentioned, macrophage migration inhibitory factor (MIF) binds the extracellular portion of Ii and then signals by the complex associating with CD44 (Leng et al., 2003). Both mouse Ii isoforms in the lung can mediate allergen-induced lung inflammation and eosinophilia, but the p41 is necessary for IgE response and airway hyper responsiveness (Ye et al., 2003b). The free cytoplasmic domain of Ii has been shown to induce B cell differentiation via NF-κB (Matza et al., 2002a; Matza et al., 2002b). These or other functions of Ii suggest an alternative physiology that may have preceded its more famed roles as chaperone and peptide cleft occupier, and may also expose the ancestral loci. Cathepsins are ancient and were clearly co-opted for class II antigen generation. Sea urchin cathepsins (L and B) are up-regulated in LPS activated coelomocytes (Nair et al., 2005). Cathepsin S cleavage of the Ii cytoplasmic tail even can regulate the motility of dendritic cells via the tail’s interactions with myosin II. There are many leads to follow in many model organisms.
4.1 Conclusion
Now that the third chain of MHC class II has been identified in sharks, focus can turn to how this antigen presentation system arose to play a major role in adaptive immunity. As comparative genetic data continues to grow, so should our ability test evolutionary hypotheses. Exciting advances in the jawless agnathans suggest their variable lymphocyte receptor (VLR) system uses VLR-B for free receptors in a humoral response and VLR-A (and perhaps VLR-C) in cellular responses (Pancer et al., 2005; Rogozin et al., 2007). These T cell-like VLR-A bearing lymphocytes develop in a “thymoid” region of the lamprey pharynx (Bajoghli et al., 2011). Apparently lacking MHC and Ii, are these T cell analogs of the jawless fish lacking a pathway of processing antigen from the endosomal pathway that only evolved in the jawed gnathostomes? A similar question can now be asked of the adaptive immune system of the Atlantic cod, whose genome lacks MHC class II, Ii and functional CD4 (Star et al., 2011). Much remains to be learned of the adaptive immune system of this fish, but it is clear that this is a derived loss of the class II/Ii/CD4 arm of adaptive immunity in a subset of teleosts.
What genomic loci in animals with only innate or innate and a VLR based adaptive system (reviewed in (Saha et al., 2010)) were exploited by the fledgling adaptive system that uses MHC restricted T cells and RAG mediated rearrangement of Ig superfamily genes? The peptide binding regions of MHC and the Ii had definitive ancestors, as did immunoglobulins and T cell receptors. Many cathepsin genes are encoded in MHC paralogous regions which are thought to have originated from two rounds of full genome duplication early in vertebrate evolution (Flajnik and Kasahara, 2010). Several other recent studies point to earlier genomic concentrations of genes that are not linked in mammals, such as Ig-like variable domains in T cell receptors (Criscitiello et al., 2010; Parra et al., 2010) and B2-microglobulin in the MHC (Ohta et al., 2011). With the tightly coordinated expression of class II and Ii and the finding of a partial CIITA locus in shark reported here, there are new reasons to investigate the origins of the antigen presenting cell. Large scale genomics and RNAi screens are beginning to elucidate how CIITA is regulated, casting spotlights on the RMND5B and MAPK1 (Paul et al., 2011). Lower vertebrates may tell us if and how CIITA was pirated from an innate NLR (NOD, LRR receptor) locus early in the gnathostomes to serve as a master regulator in early antigen presenting cells to allow their presentation to, and activation of, helper T cells.
Supplementary Material
Highlights.
The MHC class II associated invariant chain is well conserved from shark to man
CIITA appears to coordinately regulate MHC II and Ii in early vertebrates
Regulation of cathepsin degradation was an ancestral feature of Ii
Bony fish have unique forms of Ii and sometimes even do without it
Alternative splicing of the thyroglobulin domain encoding exon is a fundamental feature of the Ii
Ii evolved in jawed cartilaginous fish
Acknowledgments
This work was supported by the NIH through grants to MFC (AI56963) and MFF (AI027877), and is dedicated to the memory of Matt Graham.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Michael F. Criscitiello, Email: mcriscitiello@cvm.tamu.edu.
Yuko Ohta, Email: yota@som.umaryland.edu.
Jeannine O. Eubanks, Email: jeubanks@cvm.tamu.edu.
Patricia L. Chen, Email: pchen@cvm.tamu.edu.
Martin F. Flajnik, Email: mflajnik@som.umaryland.edu.
References
- Ashman JB, Miller J. A role for the transmembrane domain in the trimerization of the MHC class II-associated invariant chain. J Immunol. 1999;163:2704–2712. [PubMed] [Google Scholar]
- Bajoghli B, Guo P, Aghaallaei N, Hirano M, Strohmeier C, McCurley N, Bockman DE, Schorpp M, Cooper MD, Boehm T. A thymus candidate in lampreys. Nature. 2011;470:90–94. doi: 10.1038/nature09655. [DOI] [PubMed] [Google Scholar]
- Bakke O, Dobberstein B. MHC class II-associated invariant chain contains a sorting signal for endosomal compartments. Cell. 1990;63:707–716. doi: 10.1016/0092-8674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- Bartl S, Baish MA, Flajnik MF, Ohta Y. Identification of class I genes in cartilaginous fish, the most ancient group of vertebrates displaying an adaptive immune response. J Immunol. 1997;159:6097–6104. [PubMed] [Google Scholar]
- Bernstein RM, Schluter SF, Lake DF, Marchalonis JJ. Evolutionary Conservation and Molecular-Cloning of the Recombinase Activating Gene-1. Biochem Bioph Res Co. 1994;205:687–692. doi: 10.1006/bbrc.1994.2720. [DOI] [PubMed] [Google Scholar]
- Bevec T, Stoka V, Pungercic G, Dolenc I, Turk V. Major histocompatibility complex class II-associated p41 invariant chain fragment is a strong inhibitor of lysosomal cathepsin L. J Exp Med. 1996;183:1331–1338. doi: 10.1084/jem.183.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlmakers MJ, Benaroch P, Ploegh HL. Mapping functional regions in the lumenal domain of the class II-associated invariant chain. J Exp Med. 1994;180:623–629. doi: 10.1084/jem.180.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikoff EK, Huang LY, Episkopou V, van Meerwijk J, Germain RN, Robertson EJ. Defective major histocompatibility complex class II assembly, transport, peptide acquisition, and CD4+ T cell selection in mice lacking invariant chain expression. J Exp Med. 1993;177:1699–1712. doi: 10.1084/jem.177.6.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikoff EK, Kenty G, Van Kaer L. Distinct peptide loading pathways for MHC class II molecules associated with alternative Ii chain isoforms. J Immunol. 1998;160:3101–3110. [PubMed] [Google Scholar]
- Blander JM, Medzhitov R. On regulation of phagosome maturation and antigen presentation. Nat Immunol. 2006;7:1029–1035. doi: 10.1038/ni1006-1029. [DOI] [PubMed] [Google Scholar]
- Blum JS, Cresswell P. Role for intracellular proteases in the processing and transport of class II HLA antigens. Proc Natl Acad Sci U S A. 1988;85:3975–3979. doi: 10.1073/pnas.85.11.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AM, Wright KL, Ting JP. Human major histocompatibility complex class II-associated invariant chain gene promoter. Functional analysis and in vivo protein/DNA interactions of constitutive and IFN-gamma-induced expression. J Biol Chem. 1993;268:26328–26333. [PubMed] [Google Scholar]
- Brown WM, Dziegielewska KM. Friends and relations of the cystatin superfamily--new members and their evolution. Protein Sci. 1997;6:5–12. doi: 10.1002/pro.5560060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman HA. Endosomal proteolysis and MHC class II function. Curr Opin Immunol. 1998;10:93–102. doi: 10.1016/s0952-7915(98)80038-1. [DOI] [PubMed] [Google Scholar]
- Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- Coulombe R, Grochulski P, Sivaraman J, Menard R, Mort JS, Cygler M. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 1996;15:5492–5503. [PMC free article] [PubMed] [Google Scholar]
- Criscitiello MF, Flajnik MF. Four primordial immunoglobulin light chain isotypes, including lambda and kappa, identified in the most primitive living jawed vertebrates. Eur J Immunol. 2007;37:2683–2694. doi: 10.1002/eji.200737263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscitiello MF, Kamper SM, McKinney EC. Allelic polymorphism of TCRalpha chain constant domain genes in the bicolor damselfish. Dev Comp Immunol. 2004;28:781–792. doi: 10.1016/j.dci.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Criscitiello MF, Ohta Y, Saltis M, McKinney EC, Flajnik MF. Evolutionarily conserved TCR binding sites, identification of T cells in primary lymphoid tissues, and surprising trans-rearrangements in nurse shark. J Immunol. 2010;184:6950–6960. doi: 10.4049/jimmunol.0902774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscitiello MF, Saltis M, Flajnik MF. An evolutionarily mobile antigen receptor variable region gene: doubly rearranging NAR-TcR genes in sharks. Proc Natl Acad Sci U S A. 2006;103:5036–5041. doi: 10.1073/pnas.0507074103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra JM, Kiryu I, Kollner B, Yoshiura Y, Ototake M. MHC class II invariant chain homologues in rainbow trout (Oncorhynchus mykiss) Fish Shellfish Immunol. 2003;15:91–105. doi: 10.1016/s1050-4648(02)00141-9. [DOI] [PubMed] [Google Scholar]
- Dixon AM, Stanley BJ, Matthews EE, Dawson JP, Engelman DM. Invariant chain transmembrane domain trimerization: a step in MHC class II assembly. Biochemistry. 2006;45:5228–5234. doi: 10.1021/bi052112e. [DOI] [PubMed] [Google Scholar]
- Flajnik MF. Comparative analyses of immunoglobulin genes: surprises and portents. Nat Rev Immunol. 2002;2:688–698. doi: 10.1038/nri889. [DOI] [PubMed] [Google Scholar]
- Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freisewinkel IM, Schenck K, Koch N. The segment of invariant chain that is critical for association with major histocompatibility complex class II molecules contains the sequence of a peptide eluted from class II polypeptides. Proc Natl Acad Sci U S A. 1993;90:9703–9706. doi: 10.1073/pnas.90.20.9703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki K, Smith CM, Liu L, Sundick RS, Dixon B. Alternate forms of MHC class II-associated invariant chain are not produced by alternative splicing in rainbow trout (Oncorhynchus mykiss) but are encoded by separate genes. Dev Comp Immunol. 2003;27:377–391. doi: 10.1016/s0145-305x(02)00119-2. [DOI] [PubMed] [Google Scholar]
- Ghosh P, Amaya M, Mellins E, Wiley DC. The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3. Nature. 1995;378:457–462. doi: 10.1038/378457a0. [DOI] [PubMed] [Google Scholar]
- Guncar G, Pungercic G, Klemencic I, Turk V, Turk D. Crystal structure of MHC class II-associated p41 Ii fragment bound to cathepsin L reveals the structural basis for differentiation between cathepsins L and S. EMBO J. 1999a;18:793–803. doi: 10.1093/emboj/18.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guncar G, Pungercic G, Klemencic I, Turk V, Turk D. Crystal structure of MHC class II-associated p41 Ii fragment bound to cathepsin L reveals the structural basis for differentiation between cathepsins L and S. EMBO J. 1999b;18:793–803. doi: 10.1093/emboj/18.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- Jasanoff A, Wagner G, Wiley DC. Structure of a trimeric domain of the MHC class II-associated chaperonin and targeting protein Ii. EMBO J. 1998;17:6812–6818. doi: 10.1093/emboj/17.23.6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PP, Murphy DB, Hewgill D, McDevitt HO. Detection of a common polypeptide chain in I--A and I--E sub-region immunoprecipitates. Mol Immunol. 1979;16:51–60. doi: 10.1016/0161-5890(79)90027-0. [DOI] [PubMed] [Google Scholar]
- Kasahara M, Vazquez M, Sato K, McKinney EC, Flajnik MF. Evolution of the major histocompatibility complex: isolation of class II A cDNA clones from the cartilaginous fish. Proc Natl Acad Sci U S A. 1992;89:6688–6692. doi: 10.1073/pnas.89.15.6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katunuma N, Kakegawa H, Matsunaga Y, Saibara T. Immunological significances of invariant chain from the aspect of its structural homology with the cystatin family. FEBS Lett. 1994;349:265–269. doi: 10.1016/0014-5793(94)00657-1. [DOI] [PubMed] [Google Scholar]
- Koch N, Moldenhauer G, Hofmann WJ, Moller P. Rapid intracellular pathway gives rise to cell surface expression of the MHC class II-associated invariant chain (CD74) J Immunol. 1991;147:2643–2651. [PubMed] [Google Scholar]
- Kongsvik TL, Honing S, Bakke O, Rodionov DG. Mechanism of interaction between leucine-based sorting signals from the invariant chain and clathrin-associated adaptor protein complexes AP1 and AP2. J Biol Chem. 2002;277:16484–16488. doi: 10.1074/jbc.M201583200. [DOI] [PubMed] [Google Scholar]
- Lamb CA, Cresswell P. Assembly and transport properties of invariant chain trimers and HLA-DR-invariant chain complexes. J Immunol. 1992;148:3478–3482. [PubMed] [Google Scholar]
- Lenarcic B, Bevec T. Thyropins--new structurally related proteinase inhibitors. Biol Chem. 1998;379:105–111. [PubMed] [Google Scholar]
- Leng L, Metz CN, Fang Y, Xu J, Donnelly S, Baugh J, Delohery T, Chen Y, Mitchell RA, Bucala R. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197:1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong JS, Jantzen SG, von Schalburg KR, Cooper GA, Messmer AM, Liao NY, Munro S, Moore R, Holt RA, Jones SJ, Davidson WS, Koop BF. Salmo salar and Esox lucius full-length cDNA sequences reveal changes in evolutionary pressures on a post-tetraploidization genome. BMC Genomics. 2010;11:279. doi: 10.1186/1471-2164-11-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi G, Burzyn D, Mundinano J, Berguer P, Bekinschtein P, Costa H, Castillo LF, Goldman A, Meiss R, Piazzon I, Nepomnaschy I. Cathepsin-L influences the expression of extracellular matrix in lymphoid organs and plays a role in the regulation of thymic output and of peripheral T cell number. J Immunol. 2005;174:7022–7032. doi: 10.4049/jimmunol.174.11.7022. [DOI] [PubMed] [Google Scholar]
- Long EO, Strubin M, Wake CT, Gross N, Carrel S, Goodfellow P, Accolla RS, Mach B. Isolation of cDNA clones for the p33 invariant chain associated with HLA-DR antigens. Proc Natl Acad Sci U S A. 1983;80:5714–5718. doi: 10.1073/pnas.80.18.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin LG, Larhammar D, Hallbook F. Numerous groups of chromosomal regional paralogies strongly indicate two genome doublings at the root of the vertebrates. Journal of structural and functional genomics. 2003;3:53–63. [PubMed] [Google Scholar]
- Malcherek G, Gnau V, Jung G, Rammensee HG, Melms A. Supermotifs enable natural invariant chain-derived peptides to interact with many major histocompatibility complex-class II molecules. J Exp Med. 1995;181:527–536. doi: 10.1084/jem.181.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matza D, Kerem A, Medvedovsky H, Lantner F, Shachar I. Invariant chain-induced B cell differentiation requires intramembrane proteolytic release of the cytosolic domain. Immunity. 2002a;17:549–560. doi: 10.1016/s1074-7613(02)00455-7. [DOI] [PubMed] [Google Scholar]
- Matza D, Lantner F, Bogoch Y, Flaishon L, Hershkoviz R, Shachar I. Invariant chain induces B cell maturation in a process that is independent of its chaperonic activity. Proc Natl Acad Sci U S A. 2002b;99:3018–3023. doi: 10.1073/pnas.052703299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihelic M, Turk D. Two decades of thyroglobulin type-1 domain research. Biol Chem. 2007;388:1123–1130. doi: 10.1515/BC.2007.155. [DOI] [PubMed] [Google Scholar]
- Nair SV, Del Valle H, Gross PS, Terwilliger DP, Smith LC. Macroarray analysis of coelomocyte gene expression in response to LPS in the sea urchin. Identification of unexpected immune diversity in an invertebrate. Physiol Genomics. 2005;22:33–47. doi: 10.1152/physiolgenomics.00052.2005. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Roth W, Wong P, Nelson A, Farr A, Deussing J, Villadangos JA, Ploegh H, Peters C, Rudensky AY. Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science. 1998;280:450–453. doi: 10.1126/science.280.5362.450. [DOI] [PubMed] [Google Scholar]
- Neefjes JJ, Ploegh HL. Inhibition of endosomal proteolytic activity by leupeptin blocks surface expression of MHC class II molecules and their conversion to SDS resistance alpha beta heterodimers in endosomes. EMBO J. 1992;11:411–416. doi: 10.1002/j.1460-2075.1992.tb05069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan DM, Noonan D, Quaranta V. Four Ia invariant chain forms derive from a single gene by alternate splicing and alternate initiation of transcription/translation. J Exp Med. 1987;166:444–460. doi: 10.1084/jem.166.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Landis E, Boulay T, Phillips RB, Collet B, Secombes CJ, Flajnik MF, Hansen JD. Homologs of CD83 from elasmobranch and teleost fish. J Immunol. 2004;173:4553–4560. doi: 10.4049/jimmunol.173.7.4553. [DOI] [PubMed] [Google Scholar]
- Ohta Y, McKinney EC, Criscitiello MF, Flajnik MF. Proteasome, transporter associated with antigen processing, and class I genes in the nurse shark Ginglymostoma cirratum: evidence for a stable class I region and MHC haplotype lineages. J Immunol. 2002;168:771–781. doi: 10.4049/jimmunol.168.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta Y, Shiina T, Lohr RL, Hosomichi K, Pollin TI, Heist EJ, Suzuki S, Inoko H, Flajnik MF. Primordial linkage of beta2-microglobulin to the MHC. J Immunol. 2011;186:3563–3571. doi: 10.4049/jimmunol.1003933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancer Z, Saha NR, Kasamatsu J, Suzuki T, Amemiya CT, Kasahara M, Cooper MD. Variable lymphocyte receptors in hagfish. Proc Natl Acad Sci USA. 2005 doi: 10.1073/pnas.0503792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra ZE, Ohta Y, Criscitiello MF, Flajnik MF, Miller RD. The dynamic TCRdelta: TCRdelta chains in the amphibian Xenopus tropicalis utilize antibody-like V genes. Eur J Immunol. 2010;40:2319–2329. doi: 10.1002/eji.201040515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P, van den Hoorn T, Jongsma ML, Bakker MJ, Hengeveld R, Janssen L, Cresswell P, Egan DA, van Ham M, Ten Brinke A, Ovaa H, Beijersbergen RL, Kuijl C, Neefjes J. A Genome-wide multidimensional RNAi screen reveals pathways controlling MHC class II antigen presentation. Cell. 2011;145:268–283. doi: 10.1016/j.cell.2011.03.023. [DOI] [PubMed] [Google Scholar]
- Pieters J, Bakke O, Dobberstein B. The MHC class II-associated invariant chain contains two endosomal targeting signals within its cytoplasmic tail. J Cell Sci. 1993;106 (Pt 3):831–846. doi: 10.1242/jcs.106.3.831. [DOI] [PubMed] [Google Scholar]
- Pond L, Kuhn LA, Teyton L, Schutze MP, Tainer JA, Jackson MR, Peterson PA. A role for acidic residues in di-leucine motif-based targeting to the endocytic pathway. J Biol Chem. 1995;270:19989–19997. doi: 10.1074/jbc.270.34.19989. [DOI] [PubMed] [Google Scholar]
- Rast JP, Anderson MK, Strong SJ, Luer C, Litman RT, Litman GW. alpha, beta, gamma, and delta T cell antigen receptor genes arose early in vertebrate phylogeny. Immunity. 1997;6:1–11. doi: 10.1016/s1074-7613(00)80237-x. [DOI] [PubMed] [Google Scholar]
- Riese RJ, Chapman HA. Cathepsins and compartmentalization in antigen presentation. Curr Opin Immunol. 2000;12:107–113. doi: 10.1016/s0952-7915(99)00058-8. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Iyer LM, Liang L, Glazko GV, Liston VG, Pavlov YI, Aravind L, Pancer Z. Evolution and diversification of lamprey antigen receptors: evidence for involvement of an AID-APOBEC family cytosine deaminase. Nat Immunol. 2007;8:647–656. doi: 10.1038/ni1463. [DOI] [PubMed] [Google Scholar]
- Romagnoli P, Germain RN. The CLIP region of invariant chain plays a critical role in regulating major histocompatibility complex class II folding, transport, and peptide occupancy. J Exp Med. 1994;180:1107–1113. doi: 10.1084/jem.180.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha NR, Smith J, Amemiya CT. Evolution of adaptive immune recognition in jawless vertebrates. Semin Immunol. 2010;22:25–33. doi: 10.1016/j.smim.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer PH, Green JM, Malapati S, Gu L, Pierce SK. HLA-DM is present in one-fifth the amount of HLA-DR in the class II peptide-loading compartment where it associates with leupeptin-induced peptide (LIP)-HLA-DR complexes. J Immunol. 1996;157:5487–5495. [PubMed] [Google Scholar]
- Schutze MP, Peterson PA, Jackson MR. An N-terminal double-arginine motif maintains type II membrane proteins in the endoplasmic reticulum. EMBO J. 1994;13:1696–1705. doi: 10.1002/j.1460-2075.1994.tb06434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz R, Dayhoff M. Matrices for detecting distant relationships. In: MD, editor. Atlas of protein sequences. National Biomedical Research Foundation; 1979. pp. 353–358. [Google Scholar]
- Sevenich L, Hagemann S, Stoeckle C, Tolosa E, Peters C, Reinheckel T. Expression of human cathepsin L or human cathepsin V in mouse thymus mediates positive selection of T helper cells in cathepsin L knock-out mice. Biochimie. 2010;92:1674–1680. doi: 10.1016/j.biochi.2010.03.014. [DOI] [PubMed] [Google Scholar]
- Shachar I, Elliott EA, Chasnoff B, Grewal IS, Flavell RA. Reconstitution of invariant chain function in transgenic mice in vivo by individual p31 and p41 isoforms. Immunity. 1995;3:373–383. doi: 10.1016/1074-7613(95)90121-3. [DOI] [PubMed] [Google Scholar]
- Shi X, Leng L, Wang T, Wang W, Du X, Li J, McDonald C, Chen Z, Murphy JW, Lolis E, Noble P, Knudson W, Bucala R. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25:595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva DS, Reis MI, Nascimento DS, do Vale A, Pereira PJ, dos Santos NM. Sea bass (Dicentrarchus labrax) invariant chain and class II major histocompatibility complex: sequencing and structural analysis using 3D homology modelling. Mol Immunol. 2007;44:3758–3776. doi: 10.1016/j.molimm.2007.03.025. [DOI] [PubMed] [Google Scholar]
- Star B, Nederbragt AJ, Jentoft S, Grimholt U, Malmstrom M, Gregers TF, Rounge TB, Paulsen J, Solbakken MH, Sharma A, Wetten OF, Lanzen A, Winer R, Knight J, Vogel JH, Aken B, Andersen O, Lagesen K, Tooming-Klunderud A, Edvardsen RB, Tina KG, Espelund M, Nepal C, Previti C, Karlsen BO, Moum T, Skage M, Berg PR, Gjoen T, Kuhl H, Thorsen J, Malde K, Reinhardt R, Du L, Johansen SD, Searle S, Lien S, Nilsen F, Jonassen I, Omholt SW, Stenseth NC, Jakobsen KS. The genome sequence of Atlantic cod reveals a unique immune system. Nature. 2011 doi: 10.1038/nature10342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strubin M, Berte C, Mach B. Alternative splicing and alternative initiation of translation explain the four forms of the Ia antigen-associated invariant chain. EMBO J. 1986;5:3483–3488. doi: 10.1002/j.1460-2075.1986.tb04673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaesu NT, Lower JA, Robertson EJ, Bikoff EK. Major histocompatibility class II peptide occupancy, antigen presentation, and CD4+ T cell function in mice lacking the p41 isoform of invariant chain. Immunity. 1995;3:385–396. doi: 10.1016/1074-7613(95)90122-1. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Ting JP, Trowsdale J. Genetic control of MHC class II expression. Cell. 2002;109(Suppl):S21–33. doi: 10.1016/s0092-8674(02)00696-7. [DOI] [PubMed] [Google Scholar]
- Turk D, Guncar G, Turk V. The p41 fragment story. IUBMB Life. 1999;48:7–12. doi: 10.1080/713803477. [DOI] [PubMed] [Google Scholar]
- Turk V, Turk B, Turk D. Lysosomal cysteine proteases: facts and opportunities. EMBO J. 2001;20:4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uinuk-Ool TS, Takezaki N, Kuroda N, Figueroa F, Sato A, Samonte IE, Mayer WE, Klein J. Phylogeny of antigen-processing enzymes: cathepsins of a cephalochordate, an agnathan and a bony fish. Scand J Immunol. 2003;58:436–448. doi: 10.1046/j.1365-3083.2003.01322.x. [DOI] [PubMed] [Google Scholar]
- Viville S, Neefjes J, Lotteau V, Dierich A, Lemeur M, Ploegh H, Benoist C, Mathis D. Mice lacking the MHC class II-associated invariant chain. Cell. 1993;72:635–648. doi: 10.1016/0092-8674(93)90081-z. [DOI] [PubMed] [Google Scholar]
- Wahle E, Keller W. The biochemistry of 3′-end cleavage and polyadenylation of messenger RNA precursors. Annual review of biochemistry. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- Ye Q, Finn PW, Sweeney R, Bikoff EK, Riese RJ. MHC class II-associated invariant chain isoforms regulate pulmonary immune responses. J Immunol. 2003a;170:1473–1480. doi: 10.4049/jimmunol.170.3.1473. [DOI] [PubMed] [Google Scholar]
- Ye Q, Finn PW, Sweeney R, Bikoff EK, Riese RJ. MHC class II-associated invariant chain isoforms regulate pulmonary immune responses. J Immunol. 2003b;170:1473–1480. doi: 10.4049/jimmunol.170.3.1473. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Haire RN, Litman GW. Cloning of two zebrafish cDNAs that share domains with the MHC class II-associated invariant chain. Immunogenetics. 1999;50:84–88. doi: 10.1007/s002510050691. [DOI] [PubMed] [Google Scholar]
- Zhong G, Castellino F, Romagnoli P, Germain RN. Evidence that binding site occupancy is necessary and sufficient for effective major histocompatibility complex (MHC) class II transport through the secretory pathway redefines the primary function of class II-associated invariant chain peptides (CLIP) J Exp Med. 1996;184:2061–2066. doi: 10.1084/jem.184.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.