Abstract
The short palate lung and nasal epithelial clone 1 (SPLUNC1) protein may be differentially expressed in oral infections, oral inflammatory disorders, or oral malignancies and may be involved in innate immune responses in the oral cavity. However, the actual concentration of SPLUNC1 in saliva has not previously been determined. In this study, we determined the concentrations of SPLUNC1 in saliva using a particle-based antibody capture and detection immunoassay. A commercial goat anti-rhSPLUNC1 polyclonal antibody (AF1897) was linked to fluorescent polystyrene microspheres and used as the capture antibody. A commercial mouse IgG2b anti-rhSPLUNC1 monoclonal antibody (MAB1897) was biotinylated and used as the detection antibody. Western blot and 2-dimensional fluorescence difference gel electrophoresis (2-D DIGE) analysis of immunoprecipitated rhSPLUNC1 and SPLUNC1 from saliva were used to show that the capture AF1897 and detection MAB1897 antibodies both recognized SPLUNC1. Protein concentrations in saliva from 20 subjects ranged from 0.9 to 23.9 mg/ml; SPLUNC1 concentrations ranged from 34.7 ng/ml to 13.8 μg/ml; and SPLUNC concentrations normalized per mg of total salivary protein ranged from 4.7 ng/ml to 5.3 μg/ml. These results show that SPLUNC1 is detected in saliva in a variety of concentrations. This immunoassay may prove to be useful in determining the concentration of SPLUNC1 in saliva for assessing its role in the pathogenesis of oral infections, oral inflammatory disorders, or oral malignancies.
Keywords: SPLUNC, SPLUNC1, Luminex, xMAP, saliva, innate immunity
1. Introduction
The short palate lung and nasal epithelial clone 1 (SPLUNC1) protein is a member of the lipid transfer/LPS binding protein (LT/LBP) family of proteins produced in tissues of the oronasal cavity, respiratory tract, and digestive tract. The LT/LBP family of proteins includes several proteins with lipid binding (PLTP, CETP, LBP, BPI) and immunomodulatory properties (BPI, LBP) and is part of a cluster of at least 14 genes on chromosome 201, 2. SPLUNC1 and LPLUNC1 are speculated to have innate immune functions1, 3. SPLUNC1 (also known as PLUNC, LUNX, SPURT, or NASG) concentrations are increased in the sputum of patients with chronic obstructive pulmonary disease4, increased 3–20 fold in the nasal lavage fluid following exposure to chemical irritants5, and increased in smokers6. Furthermore, SPLUNC1 mRNA expression is elevated in non-small cell lung carcinomas (NSCLC), adenocarcinomas, mucoepidermoid carcinomas, bronchoalveolar carcinomas, and oral squamous cell carcinomas (OSCC)7–10.
In the oral cavity, SPLUNC1 is expressed in the tonsil, tongue, and parotid salivary glands7, 11 and can be detected in saliva by western blot analysis12. However, the actual concentrations of SPLUNC1 in saliva have not previously been quantified. The objective of this study was to develop an xMAP particle-based antibody capture and detection immunoassay to determine SPLUNC1 concentrations in human saliva.
2. Materials and methods
2.1 Saliva
Approximately 3–6 ml of unstimulated saliva was collected from subjects referred for evaluation to The University of Iowa Department of Periodontics. Informed written consent was obtained from each subject and saliva samples were collected, stored, and used in accordance with guidelines established by the University of Iowa Institutional Review Board in our approved protocol (ID# 199811030). Before periodontal examination, all subjects were asked to submit a naïve saliva sample. For this study, subjects were not asked to rinse prior to saliva collection and no considerations were made for the time of day the sample was collected or time of previous meal, snack, or consumption of beverages. All subjects were systemically healthy and had not taken antibiotics during the 6 months prior to participating in the study. Subjects who were pregnant, immunosuppressed, had gingival hypertrophy, were on anti-inflammatory medication, or required premedication with a systemic antibiotic were excluded. After collection, saliva was centrifuged at 2,750 × g for 30 minutes at 4°C and supernatants were transferred to new vials. Total protein concentration was performed ten times on each salivary supernatant (NanoDrop products, Wilmington, DE) and the supernatants were stored at −80°C.
2.2 Gingival crevicular fluid
Gingival crevicular fluid samples from 78 subjects of a recent study at The University of Iowa Department of Periodontics were used 13, 14. Informed written consent was obtained from each subject and gingival crevicular fluid samples were collected, stored, and used in accordance with guidelines established by the University of Iowa Institutional Review Board in our approved protocol (ID# 200603706). Briefly, the collection site was isolated with cotton rolls and gently air-dried. A saliva ejector was used to avoid salivary contamination of the samples. Gingival crevicular fluid samples were collected with a paper strip (Periopaper, Amityville, NY, USA) by inserting the strip for 30 seconds into the gingival crevice until mild resistance was felt. In cases of visible contamination with blood, the strip was discarded and a new site was sampled. Gingival crevicular fluid volume was immediately determined using a Periotron 8000 (Oraflow Inc., Plainview, NY, USA), which had been calibrated using known volumes of the buffer. Strips from each subject were then placed into 300 ul of 0.01 M sodium phosphate buffer, pH 7.2, containing 140 mM NaCl and protease inhibitor (Complete Mini; protease inhibitor cocktail tablets; Roche Applied Science, Indianapolis, IN, USA). After shaking for 20 minutes, the strips were removed and the eluates were centrifuged for 5 minutes at 5,800 × g to remove plaque and cellular elements. The samples were frozen at −80°C until further analysis.
2.3 Culture of human airway epithelia (HAE)
Secretions from primary cultures of human airway epithelia (HAE) were prepared as previously described15 and used as a source of native SPLUNC1 protein in accordance with guidelines established by the University of Iowa Institutional Review Board. For this, epithelial cells were dissociated and seeded onto collagen-coated, semipermeable membranes with a 0.4-μm pore size (Millicell-HA; surface area, 0.6 cm2, Millipore Corp., Bedford, MA, USA). Cells were maintained in 2% Ultroser G medium at 37°C with 5% CO2. Twenty-four hours after seeding, the mucosal medium was removed, and the cells were allowed to grow at the air-liquid interface. Only well-differentiated cultures (>2 weeks old) were used. To collect airway epithelial secretions, the apical surfaces of polarized HAE were rinsed sequentially with PBS containing Mg2+ and Ca2+, as described in Bartlett et al15. Samples were collected in a total volume of 200 μl per 24-well plate. Samples were then centrifuged at 10,000 × g for 10 minutes, and clarified supernatants were transferred to fresh tubes. Samples were stored at 4°C until use.
2.4 rhSPLUNC
Recombinant human SPLUNC1 (rhSPLUNC1) was prepared as previously described15. Briefly, the cDNA for human SPLUNC1 (National Center for Biotechnology Information, NCBI accession number NM 016583) was cloned into the plasmid vector pMAL-c2x (New England Biolabs, Ipswich, MA, USA) for the expression of a fusion protein containing an N-terminal maltose-binding protein tag and a C-terminal 6xHis tag. Full-length fusion protein was expressed in the Escherichia coli strain BL21 Star (DE3, Gibco, Invitrogen Corp., Carlsbad, CA, USA). rhSPLUNC1 was purified first, by passing the fusion protein preparation through a column containing amylose resin (New England Biolabs) and then, by passing the fraction containing rhSPLUNC1 through a nickel resin column (Ni Sepharose 6 Fast Flow, GE Healthcare Biosciences Corp., Piscataway, NJ, USA). The maltose-binding protein tag was cleaved from the purified rhSPLUNC1 using Factor Xa protease (New England Biolabs).
2.5 Antibodies
Goat anti-rhSPLUNC1 antibody (AF1897, R&D Systems, Minneapolis, MN) and mouse IgG2b anti-rhSPLUNC1 monoclonal antibody (MAB1897, R&D Systems, Minneapolis, MN) were used. Horseradish peroxidase conjugated anti-murine IgG secondary antibody (Pierce, Rockford, IL) was used.
2.6 Determination of antibody specificity for rhSPLUNC
To confirm that the antibodies were specific, rhSPLUNC was precipitated by both the capture AF1897 and the detection MAB1897 antibodies and compared. SPLUNC1 was also precipitated from saliva by both the capture AF1897 and the detection MAB1897 antibodies and compared. For the latter, 6 ml of saliva was centrifuged at 2,750 × g for 30 minutes at 4°C to remove particulates and filtered (100 KDa MWCO Centricon, Millipore, Billerica, MA). The filtrate was diluted 1:2 with Cytokine Assay Buffer (CA buffer) containing PBS, pH 7.4, 1% BSA, 0.05% Tween 20, and 0.05% sodium azide (Millipore, Billerica, MA). The diluted filtrate was split into two tubes of 6 ml each. AF1897 was added to one tube and MAB1897 was added to the other tube. Both solutions were mixed well and refrigerated. After 16 hours incubation at 4°C, the mixtures were again filtered (100 KDa MWCO Centricon, Millipore, Billerica, MA). The retentate (e.g., precipitate containing the antibody + SPLUNC1) was washed with 1 ml CA buffer. 1.0 ml 0.1 M glycine HCl, (pH 2.5) was added and the SPLUNC1 was separated from the antibody by filtration (100 KDa MWCO Centricon, Millipore, Billerica, MA). The pH of the filtrate containing SPLUNC1 was adjusted to pH 7.0 by adding 0.05 ml of 1.0 M Tris, pH 9.0. Eluted SPLUNC1was dialyzed against distilled water (3.5 KDa MWCO, Slide-a-Lyzer Dialysis Cassette, Thermo Fisher Scientific, Rockford, IL) and lyophilized. As a control, rhSPLUNC was precipitated similarly by both the capture AF1897 and the detection MAB1897 antibodies.
2.7 Western blot
Western blot analysis of immunoprecipitated rhSPLUNC1 was used to show that the capture and detection antibodies both recognized SPLUNC1. For this, immunoprecipitated rhSPLUNC1 samples were diluted in sample buffer to 1.25 μg/ul and denatured by boiling for ten minutes. 12% denaturing SDS polyacrylamide gels were loaded with 20 μl of each preparation (twice on the gel), resolved at 200 volts for 40 minutes, transferred to polyvinylidene difluoride membranes electrophoretically, and incubated overnight in blocking buffer containing 5% dry milk in 1X TBS with 0.05% Tween 20. The membrane was cut into two equal portions, washed, incubated with AF1897 or MAB1897, washed, and incubated with horseradish peroxidase conjugated anti-goat or murine IgG antibody for 1.5 hours at room temperature. SPLUNC1 positive bands were visualized using enhanced chemiluminescent substrate (Thermo Fisher Scientific Inc., Rockford, IL USA) and images were captured on a digital imaging system (Fotodyne, Inc., Hartland, WI).
Western blot analysis was also used to detect the presence of SPLUNC1 in saliva collected from 20 subjects. For this, immunoprecipitated rhSPLUNC1 and saliva supernatants were diluted in sample buffer to 1.25 μg/ul and denatured by boiling for ten minutes. 12% denaturing SDS polyacrylamide gels were loaded with 20 μl containing 25 μg salivary protein, resolved at 200 volts for 40 minutes, transferred to polyvinylidene difluoride membranes electrophoretically, and incubated overnight in blocking buffer containing 5% dry milk in 1X TBS with 0.05% Tween 20. Membranes were washed, incubated with MAB1897, washed, and incubated with horseradish peroxidase conjugated anti-murine IgG antibody for 1.5 hours at room temperature. SPLUNC1 positive bands were visualized using enhanced chemiluminescent substrate (Thermo Fisher Scientific Inc., Rockford, IL USA) and images were captured on a digital imaging system (Fotodyne, Inc., Hartland, WI).
2.8 Two-Dimensional Fluorescence Difference Gel Electrophoresis (2-D DIGE)
2-DIGE of rhSPLUNC1, SPLUNC1 immunoprecipitated for saliva with AF1897, and SPLUNC1 immunoprecipitated from saliva with MAB1897 was used to show that the capture and detections antibodies identified the same protein profile (Applied Biomics, Hayward, CA). For this rhSPLUNC1 and the immunoprecipitates were labeled with CyDyes and analyzed together by 2-D DIGE (Applied Biomics, Inc., Hayward, CA).
2.9 Determination of SPLUNC1 in human saliva
Fluorescent polystyrene microspheres (5.6 μm, bead #24) were coated with AF1897 and adjusted to contain 4.0 × 106 microspheres/ml (50x stock solution of coated microspheres)(Millipore, Billerica, MA). MAB1897 was biotinylated and adjusted to 0.37 mg/ml (Millipore, Billerica, MA).
Stock solutions of rhSPLUNC1 were prepared and diluted three-fold in CA buffer from 50 to 0.0008 μg/ml and from 2.500 to 0.001 μg/ml. Standards (50 μl/well) were added to a filtration plate in duplicate and saliva supernatants (50 μl/well) were added to the filtration plate in triplicate. Anti-SPLUNC1 coated polystyrene microspheres were diluted in CA buffer to contain 80,000 microspheres/ml (1x solution of coated microspheres) and 25 μl were added per well and mixed on a plate shaker overnight at 4°C in the dark. CA buffer was removed from the plate by vacuum and the microspheres were washed by passing 50 μl CA buffer through each well. 75 μl CA buffer and 25 μl of biotinylated MAB1897 (2.0 μg/ml CA buffer) were added per well. The plate was incubated for 1.5 hrs in the dark at room temperature on a plate shaker. 25 μl of Strepavidin-Phycoerythrin was added per well, and mixed. After incubation for 30 minutes in the dark at room temperature, 25 μl of Stop Solution was added per well. After incubation for 5 minutes in the dark at room temperature, the liquid was removed by vacuum filtration. 150 μl CA buffer was added per well and gently mixed for 1 minute. The median fluorescent intensity (MFI) was determined using the Luminex ®100™ Instrument (Luminex, Austin, TX). Concentrations of SPLUNC1 in each sample were interpolated from the standard curves stated above using Beadview software (Millipore, Billerica, MA) and multiplied by the dilution factor.
3 Results
Western blots of rhSPLUNC1 and immunoprecipitated rhSPLUNC1 had nearly identical profiles suggesting that both the capture and detection antibodies recognized rhSPLUNC1 (Figure 1). A western blot of rhSPLUNC; rhSPLUNC1 immunoprecipitated with MAB1897; and rhSPLUNC1 immunoprecipitated with AF1897 and probed with AF1897 was similar to the western blot of rhSPLUNC; rhSPLUNC1 immunoprecipitated with MAB1897; and rhSPLUNC1 immunoprecipitated with AF1897 and probed with MAB1897. Likewise, rhSPLUNC1, SPLUNC1 immunoprecipitated from saliva with AF1897, and SPLUNC immunoprecipitated from saliva with MAB1897 had nearly identical profiles when examined by 2D-DIGE (Figure 2). These results suggested that the capture and detection antibodies both identified SPLUNC1.
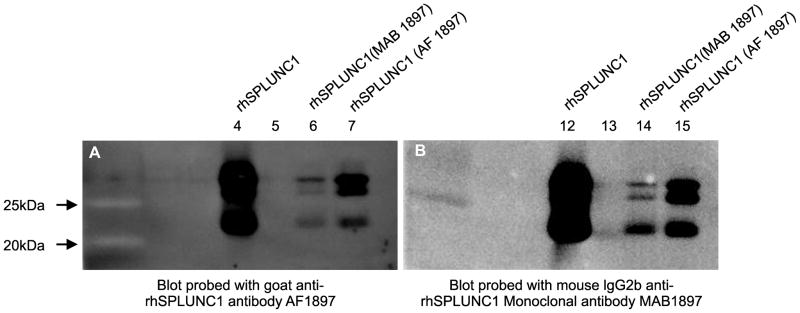
Figure 1.
Western blot analysis of rhSPLUNC1 and immunoprecipitated rhSPLUNC1 to show that the capture and detection antibodies both recognized rhSPLUNC1. (A) Western blot showing rhSPLUNC (lane 4); rhSPLUNC1 immunoprecipitated with mouse IgG2b anti-rhSPLUNC1 monoclonal antibody (MAB1897 (lane 6); and rhSPLUNC1 immunoprecipitated with goat anti-rhSPLUNC1 antibody (AF1897) (lane 7) and probed with AF1897. (B) Western blot showing rhSPLUNC (lane 12); rhSPLUNC1 immunoprecipitated with mouse IgG2b anti-rhSPLUNC1 monoclonal antibody (MAB1897) (lane 14); and rhSPLUNC1 immunoprecipitated with AF1897 (lane 15) and probed with mouse IgG2b anti-rhSPLUNC1 monoclonal antibody (MAB1897). Both blots had nearly identical profiles suggesting that both antibodies recognized rhSPLUNC1.
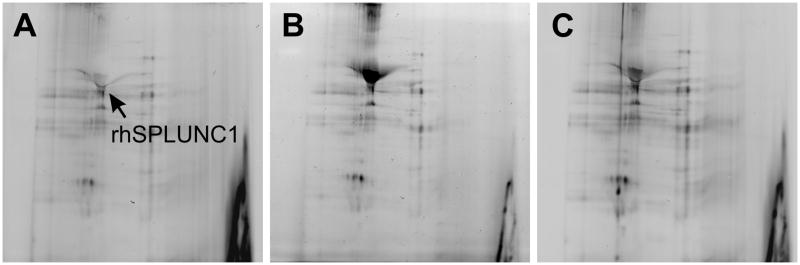
Figure 2.
Two-Dimensional Fluorescence Difference Gel Electrophoresis (2-D DIGE) gels comparing rhSPLUNC1 (A) with SPLUNC1 immunoprecipitated from a saliva sample with commercial goat anti-rhSPLUNC1 antibody (AF1897) (B) and mouse IgG2b anti-rhSPLUNC1 monoclonal antibody (MAB1897) (C). rhSPLUNC1 and immunoprecipitated SPLUNC1 were labeled with dyes, separated in a pH 3–10 linear range in the first dimension, and separated in 10.5% gel in the second dimension. Nearly identical 2-D DIGE profiles were seen among rhSPLUNC1, rhSPLUNC1 immunoprecipitated with AF1897, and rhSPLUNC1 immunoprecipitated with MAB1897 suggesting that both antibodies recognized SPLUNC1 in saliva. The relative location of SPLUNC1 is shown. Other spots represent a carry-over of protein from the immunoprecipitation process. Trace contaminants like albumin, salivary alpha amylase, MMP27, immunoglobulin kappa light chain variable region, etc were all detected.
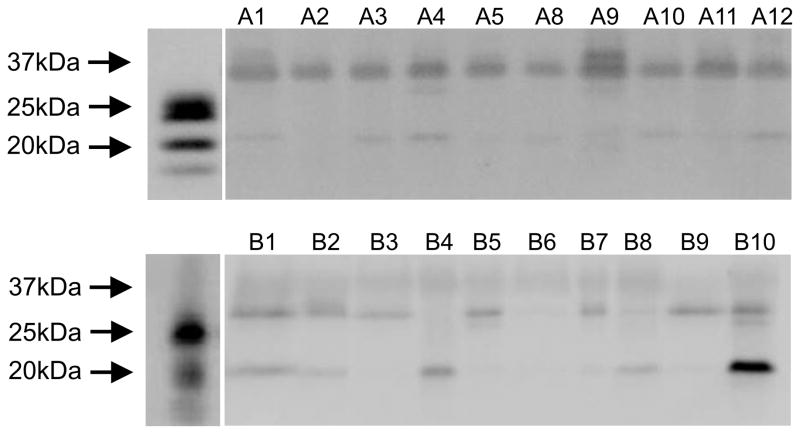
Also using western blot analysis, SPLUNC1 was detected in saliva from 20 systemically healthy subjects (Figure 3). There were bands indicative of SPLUNC1 in all saliva samples. It is worth noting that human saliva SPLUNC1 runs higher on SDS PAGE gels than rhSPLUNC1 possibly because of glycosylation. The 17–20 kDa lower band is likely a breakdown product of full length SPLUNC1 often observed with both rhSPLUNC1 and native SPLUNC1 and recognized by the SPLUNC1 antibodies.
Figure 3.
Western blots comparing SPLUNC1 in human saliva from 20 systemically healthy subjects. There were bands indicative of SPLUNC1 in all saliva supernatants. Note that saliva SPLUNC1 runs higher on the SDS PAGE gel than rhSPLUNC1 possibly because of glycosylation. Also there is a 17–20 kDa lower band that is a breakdown product of full length SPLUNC1 often observed with both rhSPLUNC1 and native SPLUNC1.
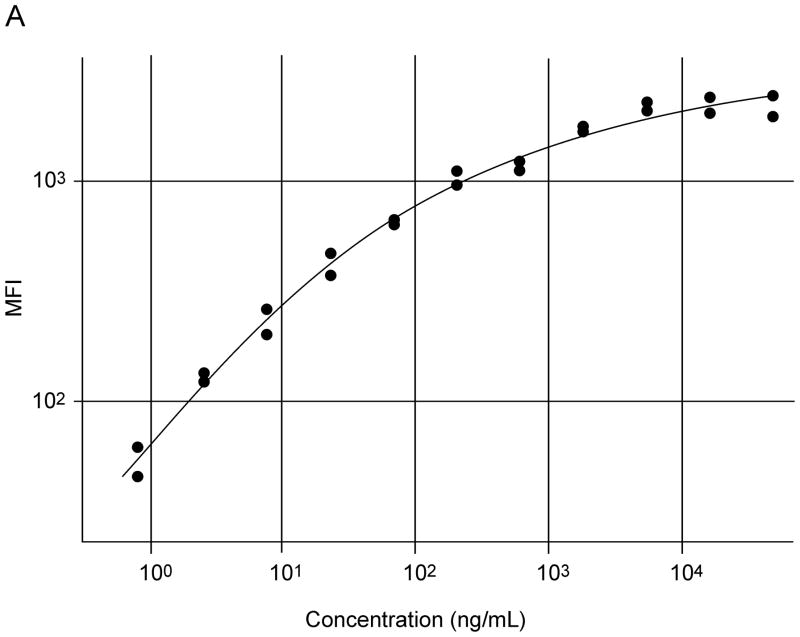
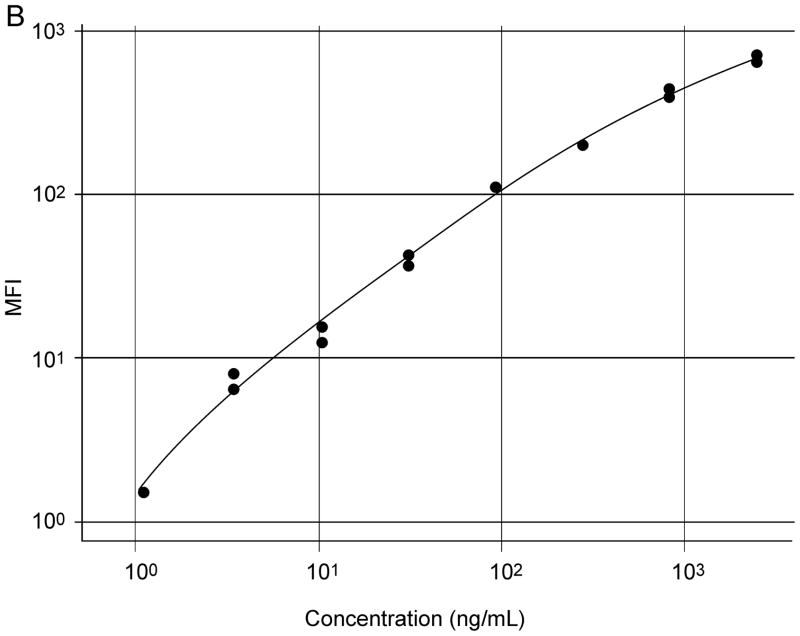
AF1897 and MAB1897 worked well in a particle-based antibody capture and detection immunoassay (Figure 5). Initially, a checkerboard configuration was used to establish the optimum concentrations of reagents for the capture and detection of SPLUNC1. Dilutions of the 200 μg/ml rhSPLUNC1 standard protein stock solution (diluted 1:3 from 50 μg/ml to 0.85 ng/ml); dilutions of the 50X stock solution containing microspheres coated with AF1897; and dilutions of the stock solution containing biotinylated MAB1897 (4, 2, or 1 μg/ml) were all assessed. An optimized assay contained a standard curve of rhSPLUNC1 diluted 1:3 from 50 to 0.0008 μg/ml and from 2.5 to 0.001 μg/ml; a 1:50 dilution of the AF1897 antibody coated microspheres; and 2 μg/ml of biotinylated antibody MAB1897 (Figure 4). Increasing biotinylated antibody MAB1897 above 2 μg/ml did not increase sensitivity. Saliva was diluted 1:2, 1:10, or 1:100 with CA buffer.
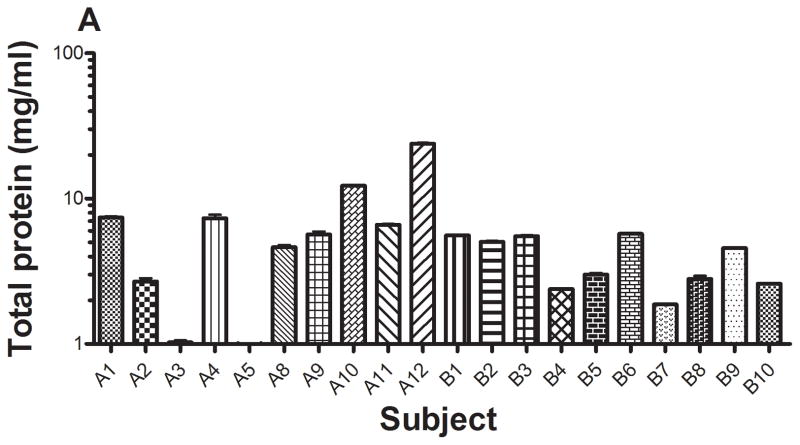
Figure 5.
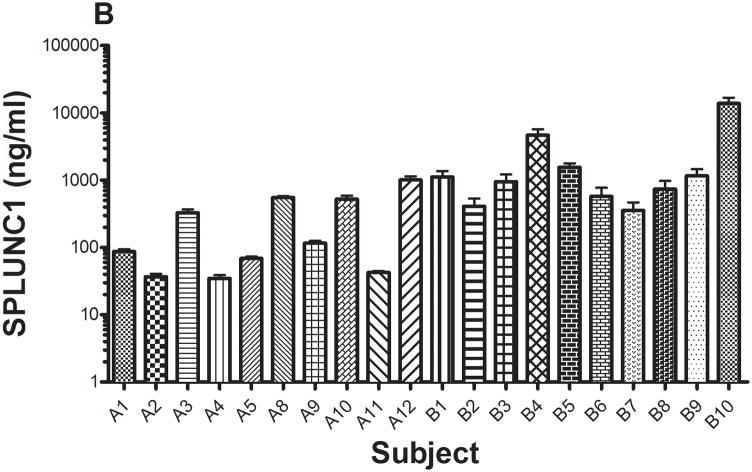
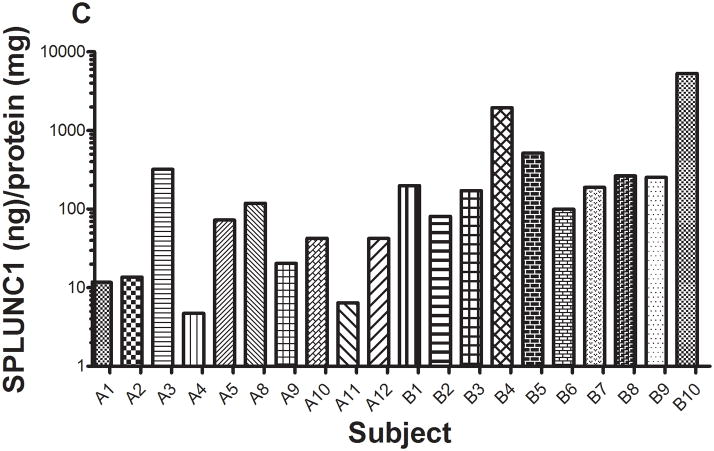
The concentrations of total protein (A) and SPLUNC1 (B) in saliva from 20 subjects were determined using xMAP technology with recombinant rhSPLUNC1 as the standard (Model 100 IS, Luminex Corp., Austin, TX). Total protein concentrations in saliva ranged from 0.9 to 23.9 mg/ml. SPLUNC1 concentrations in saliva ranged from 34.7 ng/ml to 13.8 μg/ml. When normalized to total protein concentration (C), SPLUNC1 concentrations ranged from 4.7 ng/ml to 5.3 μg/ml.
Figure 4.
Typical standard curves of rhSPLUNC1 in an xMAP particle-based assay using immobilized commercial goat anti-rhSPLUNC1 antibody (AF1897) used as the capture antibody and mouse IgG2b anti-rhSPLUNC1 monoclonal antibody (MAB1897) was used as the detection antibody for rhSPLUNC1. Initial standard curves used rhSPLUNC1 diluted 1:3 from 50 to 0.0008 μg/ml (A) and refined standard curves used rhSPLUNC1 diluted 1:3 from 2.5 to 0.001 μg/ml (B).
Total protein concentrations in the saliva from the 20 subjects ranged 0.9 to 23.9 mg/ml (Figure 5a). Total salivary protein varies among individuals and these values were within the normal reported ranges 16, 17. SPLUNC1 also varied in concentration in saliva and ranged from 34.7 ng/ml to 13.8 μg/ml (Figure 5b); and SPLUNC concentrations normalized per mg saliva protein ranged from 4.7 ng/ml to 5.3 μg/ml (Figure 5c)
SPLUNC1 concentrations in 78 human gingival crevicular fluids were less than 0.001 μg/ml.
Protein concentrations in human airway epithelial washes from 3 subjects ranged 0.8 to 2.5 mg/ml; SPLUNC1 concentrations in human airway epithelial washes ranged from 5.6 to 281.0 μg/ml; and SPLUNC1 concentrations normalized per mg protein ranged from 7.0 to 305.4 μg/ml.
4 Discussion
SPLUNC1 is produced in tissues of the oronasal cavity, respiratory tract, and digestive tract. Many studies describe SPLUNC1 expression patterns in the tonsil, tongue7, 11 and in secretions of salivary and parotid glands18; nasal secretions4–6, 19; conducting airways and respiratory secretions7; and in the digestive tract. However, most studies do not quantitate SPLUNC1 concentrations in tissue, secretions, or wash fluids. It has previously been reported that SPLUNC1 can be detected in saliva and airway secretions by western blot and ELISA. However, these assays are not useful in performing mass screening of salivary samples and may only yield semi-quantitative results (e.g., western blots).
The purpose of this study was to develop an xMAP particle-based antibody capture and detection assay to detect SPLUNC1 in saliva. For this, we used saliva samples collected from periodontal patients just entering the periodontal clinic for examination. For this study, we did not collect information about patient disease status nor information pertaining to what the patient had to eat or drink prior to visiting the clinic. First, we wanted to know if the commercially available polyclonal (AF1897) and monoclonal (MAB1897) antibodies could be used as a pair to detect to SPLUNC1. To determine this, we first immunoprecipitated rhSPLUNC with AF1897 and MAB1897 and then separated the immunoprecipitates and rhSPLUNC1 on 12% denaturing SDS polyacrylamide gel. We found that the western blot of rhSPLUNC; rhSPLUNC1 immunoprecipitated with MAB1897; and rhSPLUNC1 immunoprecipitated with AF1897 and probed with AF1897 was similar to the western blot of rhSPLUNC; rhSPLUNC1 immunoprecipitated with MAB1897; and rhSPLUNC1 immunoprecipitated with AF1897 and probed with MAB1897 had nearly identical profiles suggesting that both the capture and detection antibodies recognized rhSPLUNC1 (Figure 1). Second, we wanted to know if AF1897 and MAB1897 could recognize SPLUNC in saliva. To determine this, we first immunoprecipitated SPLUNC from saliva with AF1897 and MAB1897 and then separated the immunoprecipitates and rhSPLUNC1 by 2D-DIGE. We found that rhSPLUNC1, SPLUNC1 immunoprecipitated from saliva with AF1897, and SPLUNC immunoprecipitated from saliva with MAB1897 also had nearly identical separation profiles (Figure 2). Third, we wanted to know if AF1897 on fluorescent polystyrene microspheres and MAB1897, biotinylated, could be used as a capture and detection set to identify SPLUNC in saliva. To determine this, we first established a standard curve using varying concentrations rhSPLUNC1, different dilutions of AF1897 antibody coated microspheres, varying concentrations of biotinylated antibody MAB1897. We found that a standard curve of rhSPLUNC1 diluted 1:3 from 50 to 0.0008 μg/ml; a 1:50 dilution of the AF1897 antibody coated microspheres; and 2 μg/ml of biotinylated antibody MAB1897 were optimum (Figure 4). Fourth, using this assay, we wanted to demonstrate that SPLUNC1 was present in oral samples and cell culture wash fluids. We found that SPLUNC1 was readily detected in saliva and human airway epithelial wash fluids but not so readily detected in gingival crevicular fluid samples.
Our results suggest that SPLUNC may be differentially expressed among compartments in the oronasal cavity and respiratory tract. SPLUNC1 is expressed in tissues of the oral cavity, respiratory tract, and digestive tract and present in salivary and parotid gland, nasal, and respiratory secretions4–6, 19, 20. In our study, we observed similar results. We detected SPLUNC in saliva and human airway epithelial wash fluids, but not to any extent in gingival crevicular fluid samples. As pointed out by Gorr20, the PLUNC proteins are primarily expressed in oral and airway epithelia and it is thought that they play antimicrobial roles in these tissues. The difference in expression between different PLUNC proteins suggests that each occupies an epithelial niche. We do not believe that the lack of detection in our study is a failure to detect SPLUNC1 as these fluid samples were used in previous studies and were found to contain a number of chemokines and cytokines13, 14. It is more likely that SPLUNC1 was not present at high concentrations in gingival crevicular fluid.
We also detected a wide range of protein and SPLUNC1 concentrations in saliva from a limited number of subjects. In saliva, SPLUNC1 ranged in concentration from 34.7 ng/ml to 13.8 μg/ml (Figure 5b). When normalized to saliva protein, SPLUNC1 ranged in concentration from 4.7 ng/ml to 5.3 μg/ml (Figure 5c). We also detected a wide range of protein and SPLUNC1 concentrations in saliva from a limited number of subjects. In saliva, SPLUNC1 ranged in concentration from 34.7 ng/ml to 13.8 μg/ml (Figure 5b). When normalized to saliva protein, SPLUNC1 ranged in concentration from 4.7 ng/ml to 5.3 μg/ml (Figure 5c). These subjects were referred for evaluation to The University of Iowa Department of Periodontics. It is possible that this vast range of SPLUNC1 concentration may be related to a subject’s periodontal health. Unfortunately, information on their periodontal health was not known and a follow up study should be properly planned to look for such a correlation.
SPLUNC1 is also expressed in non-small cell lung carcinomas, adenocarcinomas, mucoepidermoid carcinomas, bronchoalveolar carcinomas, and oral squamous cell carcinomas7–10.
In summary, we now can quantitate SPLUNC1 in saliva using an xMAP particle-based antibody capture and detection immunoassay. Such an immunoassay may prove to be useful in determining the concentration of SPLUNC1 in saliva for assessing its role in the pathogenesis of oral infections, oral inflammatory disorders, or oral malignancies. For example, we can now determine the effect of patient disease status (e.g., determine periodontal disease status and match subjects with appropriate healthy control subjects), time of saliva sample collection (e.g., collect saliva samples from subjects at specific times throughout the day), and effect of diet (e.g., collect saliva samples from subjects whose diet was documented) on SPLUNC1 production. It is possible that these situations may influence or correlate with SPLUNC1 production.
Acknowledgments
We are grateful to Patricia J. Conrad for preparation of the figures. This work was supported by funds from the training grant NIH/NIDCR T32 DE014678 and the research grant NIH/NIDCR R01 DE014390. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Karl G. Kohlgraf, Email: karl-kohlgraf@uiowa.edu.
Abbey R. Ackermann, Email: abbey-ackermann@uiowa.edu.
Rupasree N. Srikantha, Email: rupasree-srikantha@uiowa.edu.
Sophie A. Joly, Email: sophie-joly@uiowa.edu.
Jennifer Bartlett, Email: jennifer-bartlett@uiowa.edu.
Lokesh Gakhar, Email: lokesh-gakhar@uiowa.edu.
Georgia K. Johnson, Email: georgia-johnson@uiowa.edu.
Paul B. McCray, Jr, Email: paul-mccray@uiowa.edu.
Janet M. Guthmiller, Email: janet_guthmiller@dentistry.unc.edu.
Kim A. Brogden, Email: kim-brogden@uiowa.edu.
References
- 1.Bingle CD, Gorr SU. Host defense in oral and airway epithelia: chromosome 20 contributes a new protein family. The International Journal of Biochemistry & Cell Biology. 2004;36(11):2144–52. doi: 10.1016/j.biocel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Bingle CD, Craven CJ. Meet the relatives: a family of BPI- and LBP-related proteins. Trends in Immunology. 2004;25(2):53–5. doi: 10.1016/j.it.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Gakhar L, Bartlett JA, Penterman J, Mizrachi D, Singh PK, Mallampalli RK, et al. PLUNC is a novel airway surfactant protein with anti-biofilm activity. PLoS One. 2010;5(2):e9098. doi: 10.1371/journal.pone.0009098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di YP, Harper R, Zhao Y, Pahlavan N, Finkbeiner W, Wu R. Molecular cloning and characterization of spurt, a human novel gene that is retinoic acid-inducible and encodes a secretory protein specific in upper respiratory tracts. The Journal of Biological Chemistry. 2003;278(2):1165–73. doi: 10.1074/jbc.M210523200. [DOI] [PubMed] [Google Scholar]
- 5.Lindahl M, Stahlbom B, Tagesson C. Identification of a new potential airway irritation marker, palate lung nasal epithelial clone protein, in human nasal lavage fluid with two-dimensional electrophoresis and matrix-assisted laser desorption/ionization-time of flight. Electrophoresis. 2001;22(9):1795–800. doi: 10.1002/1522-2683(200105)22:9<1795::AID-ELPS1795>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 6.Ghafouri B, Stahlbom B, Tagesson C, Lindahl M. Newly identified proteins in human nasal lavage fluid from non-smokers and smokers using two-dimensional gel electrophoresis and peptide mass fingerprinting. Proteomics. 2002;2(1):112–20. [PubMed] [Google Scholar]
- 7.Bingle L, Cross SS, High AS, Wallace WA, Devine DA, Havard S, et al. SPLUNC1 (PLUNC) is expressed in glandular tissues of the respiratory tract and in lung tumours with a glandular phenotype. Journal of Pathology. 2005;205(4):491–7. doi: 10.1002/path.1726. [DOI] [PubMed] [Google Scholar]
- 8.Lemaire F, Millon R, Young J, Cromer A, Wasylyk C, Schultz I, et al. Differential expression profiling of head and neck squamous cell carcinoma (HNSCC) British Journal of Cancer. 2003;89(10):1940–9. doi: 10.1038/sj.bjc.6601373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitas M, Hoover L, Silvestri G, Reed C, Green M, Turrisi AT, et al. Lunx is a superior molecular marker for detection of non-small cell lung cancer in peripheral blood. Journal of Molecular Diagnostics. 2003;5(4):237–42. doi: 10.1016/s1525-1578(10)60480-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwao K, Watanabe T, Fujiwara Y, Takami K, Kodama K, Higashiyama M, et al. Isolation of a novel human lung-specific gene, LUNX, a potential molecular marker for detection of micrometastasis in non-small-cell lung cancer. International Journal of Cancer. 2001;91(4):433–7. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1059>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 11.Leclair EE. Four BPI (bactericidal/permeability-increasing protein)-like genes expressed in the mouse nasal, oral, airway and digestive epithelia. Biochemical Society Transactions. 2003;31(Pt 4):801–5. doi: 10.1042/bst0310801. [DOI] [PubMed] [Google Scholar]
- 12.Kohlgraf KG, Joly S, Srikantha R, Guthmiller JM. PLUNC Expression in Human Saliva. IADR/AADR/CADR 85th General Session and Exhibition; 2007; New Orleans, LA. 2007. [Google Scholar]
- 13.Thunell DH, Tymkiw KD, Johnson GK, Joly S, Burnell KK, Cavanaugh JE, et al. A multiplex immunoassay demonstrates reductions in gingival crevicular fluid cytokines following initial periodontal therapy. Journal of Periodontal Research. 2010;45(1):148–52. doi: 10.1111/j.1600-0765.2009.01204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tymkiw KD, Thunell DH, Johnson GK, Joly S, Burnell KK, Cavanaugh JE, et al. Influence of smoking on gingival crevicular fluid cytokines in severe chronic periodontitis. Journal of Clinical Periodontology. 2011;38:219–28. doi: 10.1111/j.1600-051X.2010.01684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartlett JA, Hicks BJ, Schlomann JM, Ramachandran S, Nauseef WM, McCray PB., Jr PLUNC is a secreted product of neutrophil granules. Journal of Leukocyte Biology. 2008;83(5):1201–6. doi: 10.1189/jlb.0507302. [DOI] [PubMed] [Google Scholar]
- 16.Ruhl S, Rayment SA, Schmalz G, Hiller KA, Troxler RF. Proteins in whole saliva during the first year of infancy. Journal of Dental Research. 2005;84(1):29–34. doi: 10.1177/154405910508400104. [DOI] [PubMed] [Google Scholar]
- 17.Bonilla CA. Human mixed saliva protein concentration. J Dent Res. 1972;51(2):664. doi: 10.1177/00220345720510027001. [DOI] [PubMed] [Google Scholar]
- 18.Weston WM, LeClair EE, Trzyna W, McHugh KM, Nugent P, Lafferty CM, et al. Differential display identification of plunc, a novel gene expressed in embryonic palate, nasal epithelium, and adult lung. The Journal of Biological Chemistry. 1999;274(19):13698–703. doi: 10.1074/jbc.274.19.13698. [DOI] [PubMed] [Google Scholar]
- 19.Cole AM, Liao HI, Stuchlik O, Tilan J, Pohl J, Ganz T. Cationic polypeptides are required for antibacterial activity of human airway fluid. Journal of Immunology. 2002;169(12):6985–91. doi: 10.4049/jimmunol.169.12.6985. [DOI] [PubMed] [Google Scholar]
- 20.Gorr SU. Antimicrobial peptides of the oral cavity. Periodontology 2000. 2009;51:152–80. doi: 10.1111/j.1600-0757.2009.00310.x. [DOI] [PubMed] [Google Scholar]