Abstract
The first enzyme in the shikimic acid biosynthetic pathway, 3-deoxy-D-arabinoheptulosonate 7-phosphate synthase (DAH7PS), varies significantly in size and complexity in the bacteria and plants that express it. The DAH7PS from the archaebacterium Aeropyrum pernix (DAH7PSAp) is among the smallest and least complex of the DAH7PS enzymes, leading to the hypothesis that DAH7PSAp would not be subject to feedback regulation by shikimic acid pathway products. We overexpressed DAH7PSAp in Escherichia coli, purified it, and characterized its enzymatic activity. We then solved its X-ray crystal structure with a divalent manganese ion and phosphoenolpyruvate bound (PDBID: 1VS1). DAH7PSAp is a homodimeric metalloenzyme in solution. Its enzymatic activity increases dramatically above 60°C, with optimum activity at 95°C. Its pH optimum at 60°C is 5.7. DAH7PSAp follows Michaelis-Menten kinetics at 60° C, with a KM for erythrose 4-phosphate of 280 μM, a KM for phosphoenolpyruvate of 891 μM, and a kcat of 1.0 s-1. None of the downstream products of the shikimate biosynthetic pathway we tested inhibited the activity of DAH7PSAp. The structure of DAH7PSAp is similar to the structures of DAH7PS from Thermatoga maritima (PDB ID: 3PG8) and Pyrococcus furiosus (PDB ID: 1ZCO), and is consistent with its designation as an unregulated DAH7PS.
Keywords: Aeropyrum pernix, 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase, shikimate pathway, crystal structure, feedback regulation
1. INTRODUCTION
The shikimate pathway produces chorismate in seven steps, starting with D-erythrose 4-phosphate (E4P) and phosphoenolpyruvate (PEP) (1, 2). Chorismate is the common precursor for the biosynthesis of the aromatic amino acids phenylalanine, tyrosine and tryptophan, and it is the precursor to the important molecules ubiquinone, and para-amino benzoic acid. The shikimate pathway is essential in plants and microorganisms but absent in mammals, making it a potential target for the development of antibiotics and herbicides. Glyphosate, for example, is a commercially important herbicide that inhibits 5-enolpyruvlyshikimate-3-phosphate synthase (also known as EPSP synthase), which catalyzes the sixth step in the pathway (3).
The first enzyme in the shikimate pathway, DAH7PS (EC 4.1.2.15), catalyzes the condensation of E4P and PEP to form 3-deoxy-D-arabino-heptulosonate 7-phosphate and inorganic phosphate. The DAH7PS enzymes are diverse, with some organisms, including Escherichia coli, containing more than one DAH7PS. The structures of all DAH7P enzymes share a core (α/β)8 catalytic core, but there are many additions to this core that subject the enzymes to a variety of different regulatory schemes. The DAH7PS from B. subtilis, for example, has an active chorismate mutase domain fused to the N-terminus of the (α/β)8 catalytic core. This domain confers feedback inhibition by chorismate and prephenate (4). The DAH7PS from Thermatoga maritima (DAH7PSTm, (5)) also has a domain fused to the N-terminus of the (α/β)8 catalytic core that is involved in its regulation by tyrosine and phenylalanine (6). This domain is similar to the ACT domain (7) that regulates the activity of many amino acid metabolizing enzymes (8). The DAH7PS from Mycobacterium tuberculosis presents a much more complex regulatory mechanism. This enzyme has both an N-terminal extension and an added internal loop that provide independent binding sites for regulatory amino acids. Enzyme activity is modulated by combinations of phenylalnine and tryptophan, or tyrosine and tryptophan, but not by the individual amino acids alone. We originally chose to study the DAH7PSAp because it is among the shortest of the bacterial DAH7Ps enzymes, and A. pernyx is a very primitive microbial species. The enzyme's small size (276 amino acids) and lack of N-terminal, C-terminal or internal additions to the basic (α/β)8 catalytic core suggested to us that it would be an unregulated enzyme. Its primitive origin suggested to us that its structure would help unravel the mechanisms by which other enzymes are regulated. We therefore cloned DAH7PSAp, expressed it in E. coli, and characterized the purified enzyme. We then solved its X-ray crystal structure with divalent Mn ion and PEP bound. The structure was deposited (PDB ID: 1VS1) in 2006. This paper describes the cloning, expression, and characterization of the enzyme, and discusses its structure in relation to those of other DAH7PS enzymes.
2. Experimental Procedures
2.1. Materials
A. pernix genomic DNA (ATCC 700893D) was purchased from ATCC (Manassas, VA). The pCR®T7 TOPO®TA expression kit, isopropyl-β-D-thiogalactoside, and the PCR/sequencing primers were purchased from Invitrogen (Carlsbad, CA). FailSafe® PCR premix selection kit was purchased from Epicentre Technologies (Madison, WI). Plasmid preparations were performed utilizing a Promega (Madison, WI) Wizard DNA purification kit. XL1-Blue and BL21-CodonPlus®(DE3)-RIL E. coli competent cells were purchased from Stratagene (La Jolla, CA). Restriction enzymes, Vent DNA polymerase, and T4 ligase were purchased from New England Biolabs (Beverly, MA). Enzymatic reactions were performed in an MJ Research PTC-200 Peltier Thermal Cycler. The University of Michigan Biomedical Resources Core Facility performed the DNA sequencing. PEP mono (cyclohexylammonium) salt, E4P sodium salt, chorismate, shikimate, prephenate, and 1,3-bis[tris(hydroxymethyl)methylamino]propane (BTP) were obtained from Sigma (St. Louis, MO). KCl and (NH4)2SO4 were purchased from Fisher Scientific (Fairlawn, NJ). EDTA disodium salt was obtained from Mallinckrodt (Phillipburg, NJ). Tris base was purchased from Research Organics (Cleveland, OH). High grade Spectra/Por® 7 dialysis tubing (10,000 molecular weight cut-off and metal-free) was obtained from VWR Scientific (Chicago, IL). Centriprep YM-10 concentrators were purchased from Millipore (Billerica, MA). Phenyl Superose (HR10/10) and UNO® Q6 chromatography column were purchased from Amersham Pharmacia Biotech (New York, NY) and Bio-Rad (Richmond, CA), respectively. Bio-Rad Protein Assay Reagent was used to determine the concentration of the protein, with bovine serum albumin (from Sigma) as the standard. The Mini-PROTEAN II electrophoresis unit from Bio-Rad (Richmond, CA) was used to perform SDS-PAGE. The samples were resolved on a 12% polyacrylamide gel and dyed with 0.25% Coomassie brilliant blue R from Aldrich Chemical Company (St. Louis, MO). Optical spectroscopy was performed using an HP 8453 UV-visible spectrometer. All preparations and dialyses were performed at room temperature unless otherwise indicated. All reaction buffers were adjusted to pH 7.0 at the specified reaction temperature, unless otherwise noted.
2.2. Plasmid construction
The gene encoding DAH7PS (gi:14600379) (9) was amplified from A. pernix genomic DNA using standard PCR methodologies. The forward primer was 5’-CATATGTGGAGGTGGCTCCCA GTGGCCG-3’, and the reverse primer was 5’-GAATTCAAGCTTGGATCCTACAGCAGCCTGTGCCACC-3’. The PCR product was isolated and ligated into the linear pCR®T7/CT-TOPO® vector according to the manufacturer's specifications. This ligation mixture was used to transform E. coli TOP10F’ competent cells. Plasmids isolated from these cells were sequenced and one plasmid with the correct sequence was restricted with endonucleases NdeI and BamHI (restriction sites underlined above). The restricted product was ligated into a similarly restricted pT7-7 vector that had also been treated with calf alkaline phosphatase. The ligation mixture was used to transform E. coli XL1-Blue competent cells. A plasmid isolated from one transformant, containing the correct sequence was used to transform E. coli BL21-CodonPlus®(DE3)-RIL competent cells.
2.3. Protein expression and purification
E. coli cells harboring the DAH7PSAp plasmid were grown in 2xYT media (100 mg/L ampicillin) or M9 minimal media (100 mg/L ampicillin) with shaking (300 rpm) at 37°C. The cultures were grown until A600 = 1.4 and expression was induced by addition of isopropyl-β-D-thiogalactoside to a final concentration of 0.4 mM. The E. coli in 2xYT media were allowed to grow for an additional 4 hours at 37°C while the E. coli in M9 minimal media were allowed to grow for an additional 36 hours at room temperature. E. coli cells were pelleted by centrifugation (6000 rpm, 6 min, 4° C). The cell pellet was resuspended in buffer A (20 mM Tris-HCl, pH 8.0 for DAH7PSAp) and sonicated at 4°C in an ice bath (four 30 s pulses with a 2 min rest in between pulses). Cell debris was removed by centrifugation (18000 rpm, 30 min, 4°C). Solid NaCl was added to the supernatant to a final concentration of 0.1 M, and the supernatant was rapidly heated to 80°C in a boiling water bath. The supernatant was heated for an additional 10 minutes in an 80°C water bath with gentle swirling. The denatured proteins were removed by centrifugation (18000 rpm, 30 min, 4°C), and solid (NH4)2SO4 was added to the supernatant to a final concentration of 20% (w/v). The sample was passed through a 0.22 μm filter and loaded onto a Phenyl Superose column (HR 10/10) pre-equilibrated with 20% (NH4)2SO4 in buffer A. The protein was eluted at a flow rate of 1 ml/min; first utilizing a reverse gradient from 20% to 0% (NH4)2SO4 in buffer A over 60 min followed by buffer A for 15 min. Fractions with DAH7PS activity were pooled and dialyzed overnight against 2 L of buffer A at 4°C. The dialyzed preparation was filtered using a 0.22 μm filter and applied onto a UNO® Q6 column pre-equilibrated with buffer A. The protein was eluted at a flow rate of 2 ml/min using a linear gradient from 0 to 0.5 M potassium chloride in buffer A over 60 min. SDS-PAGE was used to verify the purity of each fraction, and homogeneous fractions containing DAH7PS activity were pooled and dialyzed in 2 L of buffer A overnight at 4°C. The purified enzymes were aliquoted, quickly frozen in an ethanol and dry ice bath, and stored at -80°C.
2.4. Molecular weight determination
The native molecular weight of the enzyme was determined by gel filtration on a Superose 12 column (HR10/30) as previously described (10). Cytochrome c (12.4 kDa), carbonic anhydrase (29 kDa), albumin (66 kDa), alcohol dehydrogenase (141 kDa), and β-amylase (200 kDa) were used as molecular weight standards. Blue dextran (2000 kDa) was used to determine the column void volume. Standards and samples of 5-10 mg/ml were injected into a 50 μl sample loop for holding before being loaded onto the Superose 12 column (HR10/30).
2.5. Activity assay
Enzyme (various concentrations), 3 mM PEP, and 100 mM Tris-acetate (pH 7.0 at the desired reaction temperature) were incubated for 2 min in a thin-walled PCR tube at the desired temperature. The reaction was initiated by the addition of 3 mM E4P, with the final volume being 50 μl. The reaction was quenched after 2 min by the addition of 50 μl of 10% (v/v) cold trichloroacetic acid. The amount of DAH7P produced was quantified using a modified Aminoff periodate-thiobarbituric acid assay (11). The production of 1 μmol of DAH7P per min was defined as one unit of activity.
2.6. Temperature Optimum, pH dependence, and Thermostability
To determine the optimum reaction temperature of the desired enzyme, the enzyme was allowed to react with PEP and E4P in 100 mM Tris-acetate at pH 7.0 at temperatures between 30 and 100°C (10°C increments) utilizing the discontinuous colorimetric assay as described above. The buffer pH was adjusted to 7.0 at the desired temperatures to account for temperature dependent pH changes. For pH dependence studies, the enzymatic activity was measured at 60°C utilizing a series of 100 mM bistris propane buffers in the pH range of 5.0 to 9.0. The pH of the buffers was adjusted at 60°C. Assays were performed as described above. Thermostability was determined by incubating the enzyme in the presence or absence of 1 mM PEP at 90°C for DAH7PSAp. At different time intervals, aliquots were taken. The residual activity of the enzyme was measured using the discontinuous colorimetric assay at the same temperature as its incubation temperature.
2.7. Metal analysis
The identification and quantification of the metal(s) in the enzyme “as isolated” was determined by high-resolution inductively coupled plasma-mass spectrometry on a Finnigan MAT ELEMENT instrument. To determine the effect of metal chelating agents on the enzyme, a mixture of 10 mM EDTA and 1 mM dipicolinic acid (DPA) was incubated with enzyme, 100 mM Tris-acetate (pH 7.0 at 60° C), and 3 mM PEP for 1.0 hour at room temperature. The residual activity was assayed as described above.
2.8. Enzyme kinetics
Initial rates were determined at 60° C by varying the concentration of E4P (10-3000 μM) while maintaining the concentration of PEP at 3 mM. For the second substrate, the concentration of PEP was varied over a range from 50 to 6000 μM while the concentration of E4P was kept constant at 3 mM. The amount of DAH7P produced was quantified by the discontinuous colorimetric assay (11). The kinetic constants were determined by fitting the Michaelis-Menten equation using the KaleidaGraph program.
2.9. Feedback inhibition
To identify the allosteric effectors, enzyme, 3 mM PEP, 100 mM Tris-acetate (pH 7.0 at 70° C), and 1 mM inhibitor were incubated on ice for 30 minutes. The mixture was warmed to 70°C for 2 minutes before initiating the reaction with 3 mM E4P. A control reaction was performed because tryptophan interferes with the Aminoff assay. The control reaction was identical to the standard effector-inhibition reaction except 1 mM tryptophan was added after TCA quenching but immediately before the Aminoff assay as opposed to pre-incubating the enzyme with the tryptophan.
2.10. Crystallization, data collection and refinement
All solutions used for crystallization, except for the solutions of protein and MnCl2, were treated with Chelex-100 to remove metal cations. DAH7PSAp-Mn-PEP crystals were grown at room temperature in 5 L hanging drops using the vapor diffusion method. The drops contained 8 mg/ml DAH7PSAp, 4% (w/v) polyethylene glycol (PEG) 8000, 6.3 mM PEP, 0.8 mM MnCl2, and 100 mM LiCl in 50 mM BTP (pH 8.5). The reservoir solution contained 30% PEG 8000 in 50 mM BTP (pH 8.5). The resulting crystals were in space group P21.
X-ray diffraction measurements were performed at the Southeast Regional Collaborative Access Team (SERCAT) beam line 22-BM at the Advanced Proton Source from a single frozen crystal in an evaporating nitrogen stream. The crystal belonged to space group P21 with cell dimensions a = 54.8 Å, b = 130.4 Å, c = 78.4 Å, β = 110.6°, and diffracted to 2.3 Å. The diffraction data were processed with HKL2000 (12) and programs of the CCP4 suite, 1994, (Table 1). Four subunits of DAH7PSAp present in the asymmetric unit were found by molecular replacement search with MOLREP (CCP4 suite, 1994), using the (β/α)8-barrel core of DAH7PSTm (PDB ID 1RZM) as a model. CNS (13) and REFMAC5 (CCP4 suite, 1994) were used for refinement and O (14) was employed for model building and adjustment. The search for bound water molecules was performed with ARP/wARP (15).
Table 1.
Data collection and refinement statistics for DAH7PSAp.
|
A. Diffraction Data
|
||
| Resolution (Å) | 20.0-2.3 | |
| Unique reflections | 45666 | |
| Completeness (%) | 96.5 (89.1) | |
| Multiplicity | 7.5 (7.2) | |
| <I>/<σ(I)> | 19.2 (6.2) | |
| Rmerge (%) | 9.3 (30.5) | |
| Outermost shell (Å) | 2.38-2.30 | |
|
B. Refinement |
||
| rmsd bond length (Å) | 0.019 | |
| rmsd bond angles (deg.) | 1.67 | |
| Average B-factor, protein (Å2) | 36.1 | |
| Rwork/Rfree (%) | 17.3/21.8 | |
| Test set (%) | 5.1 | |
The final model of DAH7PSAp has R/Rfree values of 17.3/21.8% and includes 8187 protein atoms, four Mn2+ cations and PEP molecules, and 191 bound water molecules. The N-terminal segments (from five to nine residues) are disordered in all subunits. The model has good stereochemistry as analyzed by PROCHECK (16) with 93.0% of the residues in the most favored regions and no residues in the disallowed regions of the Ramachandran plot. The coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID 1VS1). LSQMAN was used to superimpose molecules (17). Buried interface surface areas were calculated utilizing Naccess (18).
3. RESULTS
3.1. Overexpression and purification
The gene encoding DAH7PSAp (gi:14600379) was cloned by PCR from genomic DNA and inserted into E. coli expression plasmid pT7-7. Protein expression in E. coli BL21(DE3) was too low to be useful. We attributed this result to the rare (for E. coli) codons that occur at the beginning of the insert's sequence. Protein production increased when we switched to E. coli BL21-CodonPlus®(DE3)-RIL cells. The purification began by heating the cell lysate to denature and precipitate the E. coli proteins. Subsequent chromatography using a Phenyl Superose column and then an UNO® Q6 column, produced homogeneous DAH7PSAp (Fig. 1). The typical yield of purified was approximately 20 mg per liter of culture. The enzyme “as isolated” precipitates when it is concentrated to more than 2 mg/ml, but with PEP present, the enzyme remains soluble in solution to a concentration of 10 mg/ml.
Figure 1.
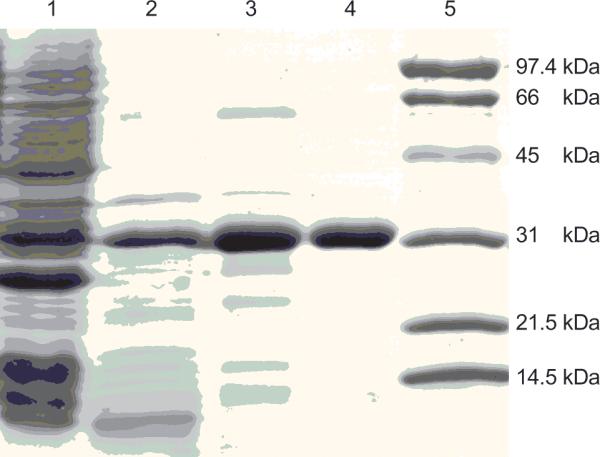
SDS-PAGE of DAHPSAp. Electrophoresis was performed under reducing conditions on a 12% polyacrylamide gel and stained with Coomassie Brilliant Blue R. Lane 1, supernatant after cell disruption; lane 2, heat supernatant; lane 3, phenyl superose column; lane 4, UNOTM Q6 fraction; lane 5, molecular weight standards. DAHPS has an apparent molecular weight of 30 kDa.
3.2. Quaternary structure in solution
The DAH7PSAp monomer migrated as a 30 kDa protein on an SDS-PAGE gel, consistent with the value (29,159 Da) predicted from its sequence. We determined the native molecular weight to be 55.4 kDa, or 1.85 × the monomeric weight, using analytical gel filtration chromatography. These data indicate that DAH7PSAp exists in solution as a homodimer.
3.3. Metal content
High-resolution inductively coupled plasma mass spectrometry showed that DAH7PSAp, as isolated, contains 0.8 molar equivalents of Zn2+ per subunit and trace amounts of other metals (Table 2). To demonstrate that DAH7PSAp requires a metal ion for activity, we attempted to remove the metal ion with chelating agents and demonstrate that the apoenzyme is inactive. Treatment of DAH7PSAp with 10 mM EDTA and 1 mM DPA for 1 hour reduces the enzymatic activity to 21% of its original activity, but does not fully inactivate the enzyme. Attempts to fully inactivate the enzyme by treating it with higher concentrations of EDTA, or EDTA with DPA, for one hour also failed to remove all the activity (data not shown). Incubation in a buffer containing 10 mM EDTA and 1 mM DPA for 24 hours resulted in enzyme precipitation. From these results, we conclude that active DAH7PSAp is a metalloenzyme that is resistant to treatment with standard chelating agents, and that the inactive apoenzyme is unstable and precipitates.
Table 2.
Metal analysis.
| Metal | Molar equivalent metal per DAH7PSAp subunit |
|---|---|
| Zinc | 0.8 |
| Magnesium | 0.05 |
| Iron | 0.03 |
| Cobalt | NDa |
| Nickel | ND |
| Copper | ND |
| Manganese | ND |
| Cadmium | ND |
None detected (value < 0.01)
3.4 DAH7PS activity
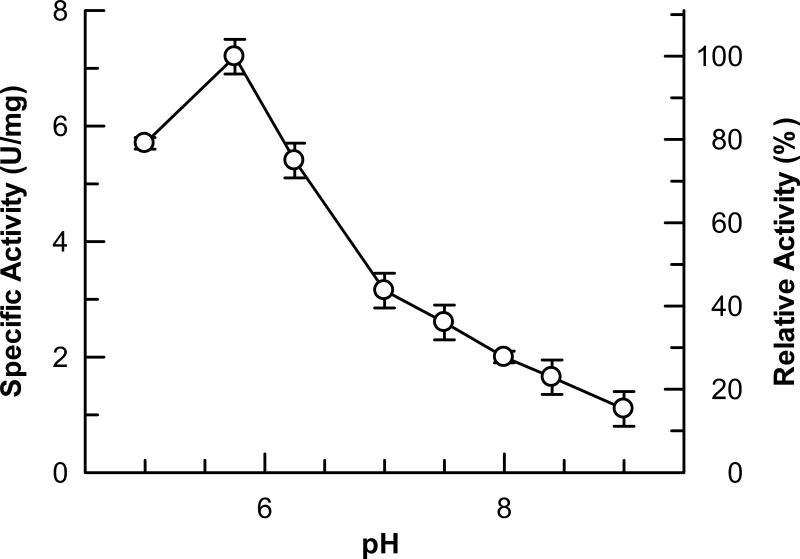
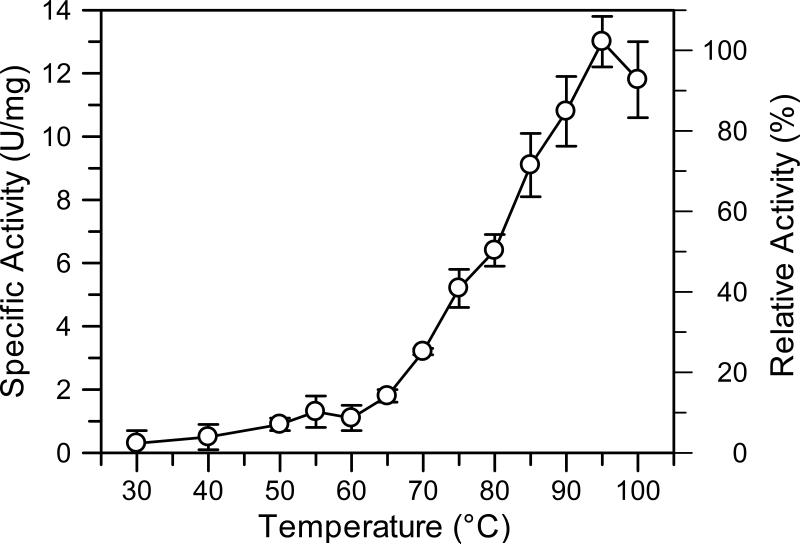
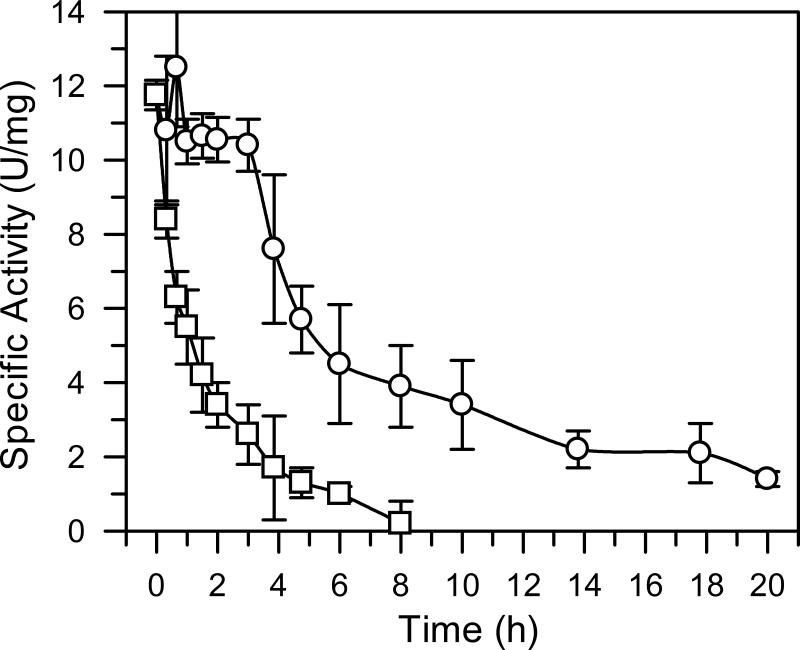
Purified DAH7PSAp exhibits maximal activity at pH 5.7 (Fig. 2) when assayed at 60°C. However, since the enzyme precipitates in an acidic buffer over a period of several hours, we performed subsequent work at pH 7.0, a pH at which the enzyme retains approximately 40% of its maximal activity at 60°C. Enzymatic activity at pH 7.0 increases sharply between 65°C and 95°C. The optimum reaction temperature, 95°C (Fig. 3), corresponds to the growth temperature of A. pernix, which is 90°C to 95°C (19). Incubation of the isolated enzyme at 90°C for 1 hour reduces the activity of DAH7PSAp is reduced by 50% (Fig. 4). Addition of the substrate PEP stabilizes the enzyme significantly, increasing its half-life at 90°C to 5.8 hours. Titration of each substrate demonstrated that the activity of DAH7PAp follows Michaelis-Menten kinetics. Apparent kinetic constants were determined for each substrate, in the presence of a saturating concentration of the other substrate, at pH 7.0 and 60°C. The apparent Km for E4P was 280 ± 20 μM and the apparent Km for PEP was 890 ±30 μM. The apparent kcat value is 1.0 ± 0.2 s-1.
Figure 2.
Optimum pH. The optimal pH of the DAHPs were determined by measuring enzyme activities in 3 mM PEP, 3 mM E4P, and 100 mM BTP buffer with an adjusted pH of 5 - 9 at 60°C; (•- DAHPSAp).
Figure 3.
Optimum reaction temperature. The enzyme activities were measured in 3 mM PEP, 3 mM E4P, and 100 mM Tris-acetate (pH 7.0 at indicated temperatures) under various reaction temperatures (•- DAHPSAp). Error bars illustrate standard deviation of three sets of data.
Figure 4.
Thermostability. The DAHPSAp in 20 mM Tris-acetate (pH 8.0 adjusted at room temperature) was incubated at 90°C in the presence (□) or absence (○) of 1 mM PEP. At various times, an aliquot was taken out to determine its residual activity at 90°C. Standard deviation of three sets of data is shown by the error bars.
3.5. Feedback inhibition
We tested several downstream products of the shikimante pathway and found that none of them inhibited the activity of DAH7PSAp at physiologically relevant concentrations (Table 3). Tryptophan initially appeared to inhibit the enzymatic activity. However, tryptophan added after the enzymatic reaction but before the Aminoff assay also showed a reduction in pink chromophore, indicating that tryptophan interferes with the Aminoff assay. Since the same apparent inhibition occurred whether the tryptophan was added before or after the enzymatic reaction took place, we conclude that tryptophan does not inhibit the enzymatic process.
Table 3.
Feedback regulation.
| Compounds | DAH7PSAp activity remaining after treatment (%)a |
|---|---|
| None | 100 |
| L-Phenylalanine | 93.9 ± 6.8 |
| L-Tyrosine | 88.9 ± 6.8 |
| Chorismate | 89.0 ± 6.7 |
| Shikimate | 99.9 ± 5.0 |
| Prephenate | 84.0 ± 2.5 |
| L-Tryptophan | 49.4 ± 3.9 |
| L-Tryptophanb | 41.8 ± 8.6 |
Results show the standard deviation of triplicate assays.
added after quenching with 10% TCA, but before the periodate-Aminoff thiobarbiturate assay.
3.6. Crystal structure of DAH7PSAp
Crystals of DAH7PSAp grown in the presence of Mn2+ and PEP contained a tetramer in the asymmetric unit. The four subunits have almost identical conformations. Superposition of 267 Cα atoms of monomers A and B resulted in a root mean squared deviation (rmsd) of 0.08 Å. Superposition of 268 Cα atoms of monomers A and C, or A and D led to rmsds of 0.13 Å and 0.14 Å, respectively. Between five and nine N-terminal residues were disordered in each subunit, including the tryptophan residues at positions two and four. The structure refined well, with an R / Rfree = 17.3 / 21.8%. All ϕ and ψ dihedral angles lie in either the most favored region or the additional allowed regions of the Ramachandran plot. The modes of PEP and metal binding in DAH7PSAp were very similar to those in T. maritima DAH7PS (20). The plane of the carboxylate of PEP is twisted ~20° with respect to the plane of its C-C double bond, as seen in the X-ray crystal structures of T. maritima DAH7PS (20) and the phenylalanine-sensitive DAH7PS from E. coli (21). A water molecule occupies the presumed binding site for the phosphate group of E4P in the DAH7PSAp structure.
4. DISCUSSION
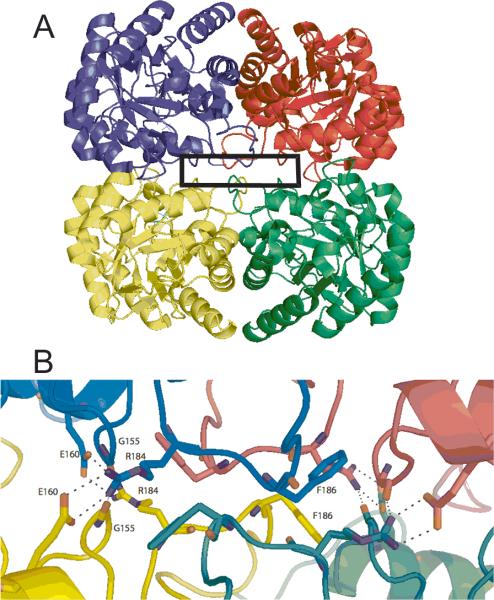
The DAH7PS enzymes are a diverse group of proteins that have incorporated a nuanced set of regulatory mechanisms. The smaller enzymes, classified as Type I (22) and further divided into Types Iα and Iβ (23) using sequence-based phylogenetic algorithms, are either unregulated or regulated by small sequences appended to the N- or C-terminus. DAH7PSAp is categorized as a Type Iβ enzyme, as are DAH7PSTm and the previously described DAH7PS from Pyrococcus furiosus (DAH7PSPf, (24)). DAH7PSAp, like DAH7PSPf and the catalytic domain of DAH7PSTm, has a core (α/β)8 barrel. The DAHPSAp has an extension of several residues at the N-terminus relative to DAH7PSPf, but these residues are disordered in the electron density of A. pernix and do not provide a binding site for a typical feedback inhibitor. DAH7PSAp is a dimer in solution, but the asymmetric unit in the crystal is a tightly associated tetramer (Fig. 5A). DAH7PSPf is also a dimer in solution (25) but packs as an apparent tetramer in the crystal (24). In contrast, DAH7PSTm is a tetramer in solution (5), but packs in the crystal as a dimer (see PDB ID: 3PG8). The buried interface surface areas between the adjacent monomers were calculated for the DAHPSAp, DAHPSPf, and DAHPSTm tetramers (Table 4). The buried interface area between monomers A and D is about 122 Å2 more than the buried interface area between monomers A and C; however, this difference is not significant enough to determine which pair of dimers is the functional dimer for DAHPSAp. A closer examination of the DAHPSAp crystal structure shows highly symmetrical residue interactions (G155, E160, R184, and F186) between monomers A and C and similar residue interactions between monomers B and D (Fig. 5B). The E160 from monomer B is in close proximity to R184 of monomer D and forms hydrogen bonds, thus, monomer A and C are more likely to be the biological dimer for DAHPSAp than monomers A and D.
Figure 5.
DAHPSAp tetramer. (A). DAHPSAp tetramer structure (monomer A-green, monomer B-cyan, monomer C-magenta, and monomer D-yellow). (B). Magnified interface shown G155, E160, R184, and F186 from each monomer interacting at the monomer interface. Interacting residues are labeled in monomers B and D, and similar interactions occur between monomer A and C.
Table 4.
Interface contact areas for monomers within DAH7PS crystals.
 | |||
|---|---|---|---|
| Functional 4° structure | Interface contact area between | Interface contact area between | |
| DAHPSAp | Dimer | C and A = 1393 Å2 | D and A = 1515 Å2 |
| DAHPSPf | Dimer | B’ and A = 1383 Å2 | B and A = 1266 Å2 |
| DAHPSTm | Tetramer | B and A = 1469 Å2 | A’ and A = 1426 Å2 |
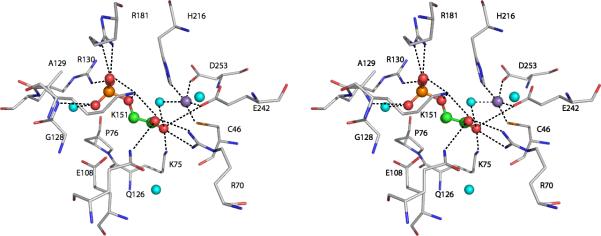
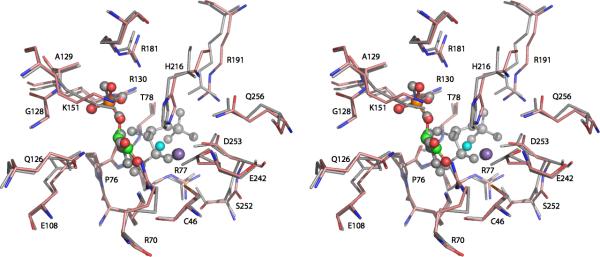
A picture of the active site of DAH7PSAp (Fig. 6) shows the key features involved in catalysis. The four resides that donate metal chelating ligands, H216, D253, E242, and C46, are strictly conserved within DAH7PSAp, DAH7PSTm, and DAH7PSPf, and broadly conserved within the Type I DAH7PS enzymes. All of the characterized DAH7PS enzymes are metalloenzymes, and it is quite likely that every DAH7PS enzyme is a metalloenzyme. The identity of the physiologically relevant metal ion for these enzymes has not been definitively proved. Early studies (26) suggested that the phenylalanineinhibitable DAH7PS from yeast (ARO3) was an iron-containing enzyme. Subsequent experiments (27) demonstrated that apo-enzyme reconstituted with divalent zinc, copper, or iron had similar activity. Divalent cobalt ions gave elevated activity (167%), divalent cadmium or nickel ions returned modest activity (<70%), while divalent magnesium or manganese ions delivered significantly reduced activity (<20%). Structural studies of yeast DAH7PS have been done with divalent cobalt ions present and with divalent manganese ions present (28). The structures of Co2+- and Mn2+-containing enzymes, with bound PEP, are essentially the same. Both exhibit trigonal bipyramidal coordination of the metal ions, with a water molecule in an equatorial position. The metal ion in DAH7PSAp (Fig. 6) also has a water molecule close enough to coordinate the bound Mn2+. Basic residues R181 and R130 form hydrogen bonds to the phosphate group of PEP while K75 and R70 form hydrogen bonds to the carboxyl group of PEP. This type of architecture is typical for Type Iβ DAH7P synthases. Fig.7 presents an overlay of the active sites of DAH7PSAp and DAH7PSTm that demonstrates the near identity of the two active sites.
Figure 6.
Active site of DAHPSAp. A stereoview showing the active site residues interacting with the manganese ion (purple sphere), the PEP substrate (sphere model: orange-phosphorus, red-oxygen, and green-carbon), and water molecules (cyan spheres). Four residues and one water molecule are shown to coordinate to manganese (C46-Mn 2.58Å, H216-Mn 2.44Å, E242-Mn 2.13Å, D253-Mn 2.39Å, H2O-Mn 2.26Å).
Figure 7.
Superposition of the active sites of DAH7PSAp and DAH7PSTm. Stereo view of the DAHPSAp active site residues interacting with bound the manganese ion (purple sphere) and the PEP substrate (sphere model: orange-phosphorus, red-oxygen, and green-carbon). The water molecules are shown as cyan spheres. DAH7PSTm residues with its bound PEP, E4P, and cadmium ion are shown in gray. All active site residues shown here are conserved between the two enzymes.
The biochemical data presented here are also similar to those of other Type I DAH7P synthases. The temperature-activity profile noted for DAH7PSAp is quite similar to that reported for DAH7PSPf, which also has very low activity below 30°C, and escalating activity above 65°C (25). The thermal stabilization of DAH7PS by PEP is not surprising, since thermal stabilization of an enzyme by its substrate has been previously observed with PEP carboxykinase and PEP (29, 30), with aminotransferase and its substrates pyridoxalmonophosphate and 2-ketoglutarate (31), D-glyceraldehyde-3-phosphate dehydrogenase and coenzyme NADP+ (32), and endo-1,4-beta-xylanase and substrate xylan (33). An increased in DAHPS stability provided by the binding of PEP to the active site has also been previously reported for L-Phe sensitive E. coli DAH7PS (34). A more complete examination of the effect of ligands on the thermostability of an enzyme is contained in a paper by Edwards et al. (35) that describes experiments with beta-galactosidase.
The DAH7PS enzymes from P. furiosus and A. pernix, which are members of euryarchaea and crenarchea, respectively, have no allosteric modifier. Neither do the DAH7PS enzymes from several other members of the archaea, based upon the similarity of their primary sequences to those of DAH7PSAp and DAH7PSPf. It seems wasteful for cells to synthesize aromatic compounds and the other metabolites dependent upon the shikimate pathway in an unregulated manner. Perhaps these primitive organisms regulate this pathway at a different step in the pathway, such as the supply of E4P, or perhaps the regulation occurs at the transcriptional level. More research is necessary to resolve this conundrum. Less primitive organisms apparently evolved more complex DAH7PS enzymes that are susceptible to feedback control. This has been accomplished through additions to the primitive (α/β)8 barrel scaffold, like those seen in the structures of DAH7PSTm and E. coli DAH7PS. There is evidence to support the idea that gene duplication and single point mutations within the extensions are responsible for creating DAH7PS with different inhibitor binding abilities (36).
Highlights.
> We investigate the DAH7P synthase from A. pernyx, a primitive archaebacterium. > We present the biochemical characterization and crystal structure of the enzyme. > The enzyme activity is not regulated by metabolites from the shikimate pathway.
ACKNOWLEDGEMENTS
This work was funded by National Institutes of Health grants GM 053069 and AI061531 (to R.W.W.) and National Science Foundation grant MCB-9723633 (to R.H.K.). We thank Dr. Ted Huston (Department of Geology, University of Michigan) for performing the metal analysis studies, and Dr. Tod P. Holler of the Department of Medicinal Chemistry, University of Michigan, for assistance with writing, editing and preparing the manuscript for publication.
Abbreviations
- BTP
1,3-bis[tris(hydroxymethyl)methylamino]propane
- DAH7PS
3-deoxy-D-arabino-heptulosonate 7-phosphate synthase
- DAH7PSAp
the DAH7PS from Aeropyrum pernix
- DAH7PSPf
the DAH7PS from Pyrococcus furiosus
- DAH7PSTm
the DAH7PS from Thermatoga maritima
- DPA
dipicolinic acid
- E4P
D-erythrose 4-phosphate
- EDTA
ethylenediamminetetraacetic acid
- KDO
3-deoxy-D-manno-octulosonate
- PCR
polymerase chain reaction
- PEP
phosphoenolpyruvate
- SDS-PAGE
sodium dodecylsulfate polyacrylamide gel electrophoresis
- rmsd
root mean squared deviation
- TCA
trichloroacetic acid
- Tris
tris(hydroxyethyl)aminomethane
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Present Address: State Key Laboratory of Food Science and Technology and School of Biotechnology, Jiangnan University, 1800 Lihu Ave., Wuxi, Jiangsu 214122, China
Present Address: Dept. of Molecular Physiology and Biological Physics, University of Virginia PO Box 800736, Charlottesville, VA 22908-0736
Contributor Information
Lily Zhou, University of Michigan, Ann Arbor, Michigan 48105-1065 mzhu48105@hotmail.com.
Jing Wu, University of Michigan, Ann Arbor, Michigan 48105-1065 jingwu@jiangnan.edu.cn.
Vijayalakshmi Janakiraman, University of Michigan, Ann Arbor, Michigan 48105-1065.
Igor A. Shumilin, Department of Biology, University of Virginia, Charlottesville, VA 22903 igor@iwonka.med.virginia.edu
Ronald Bauerle, Department of Biology, University of Virginia, Charlottesville, VA 22903 rhb7g@virginia.edu.
Robert H. Kretsinger, Department of Biology, University of Virginia, Charlottesville, VA 22903 rhk5i@virginia.edu
REFERENCES
- 1.Bentley R. The shikimate pathway-a metabolic tree with many branches. Crit Rev Biochem Mol Biol. 1990;25(5):307–84. doi: 10.3109/10409239009090615. [DOI] [PubMed] [Google Scholar]
- 2.Haslam E. Shikimic acid : metabolism and metabolites. John Wiley and Sons, Ltd.; Chichester, UK: 1993. [Google Scholar]
- 3.Steinrucken HC, Amrhein N. The herbicide glyphosate is a potent inhibitor of 5-enolpyruvyl-shikimic acid-3-phosphate synthase. Biochem Biophys Res Commun. 1980;94(4):1207–12. doi: 10.1016/0006-291x(80)90547-1. [DOI] [PubMed] [Google Scholar]
- 4.Wu J, Woodard RW. New insights into the evolutionary links relating to the 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase subfamilies. J Biol Chem. 2006;281(7):4042–8. doi: 10.1074/jbc.M512223200. [DOI] [PubMed] [Google Scholar]
- 5.Wu J, Howe DL, Woodard RW. Thermotoga maritima 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) synthase : the ancestral eubacterial DAHP synthase? J Biol Chem. 2003;278(30):27525–31. doi: 10.1074/jbc.M304631200. [DOI] [PubMed] [Google Scholar]
- 6.Cross PJ, Dobson RC, Patchett ML, Parker EJ. Tyrosine latching of a regulatory gate affords allosteric control of aromatic amino acid biosynthesis. J Biol Chem. 2011;286(12):10216–24. doi: 10.1074/jbc.M110.209924. PMCID : 3060475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chipman DM, Shaanan B. The ACT domain family. Curr Opin Struct Biol. 2001;11(6):694–700. doi: 10.1016/s0959-440x(01)00272-x. [DOI] [PubMed] [Google Scholar]
- 8.Grant GA. The ACT domain : a small molecule binding domain and its role as a common regulatory element. J Biol Chem. 2006;281(45):33825–9. doi: 10.1074/jbc.R600024200. [DOI] [PubMed] [Google Scholar]
- 9.Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, Kosugi H, Hosoyama A, Fukui S, Nagai Y, Nishijima K, Nakazawa H, Takamiya M, Masuda S, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Kikuchi H, et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 1999;6(2):83–101. 45–52. doi: 10.1093/dnares/6.2.83. [DOI] [PubMed] [Google Scholar]
- 10.Duewel HS, Sheflyan GY, Woodard RW. Functional and biochemical characterization of a recombinant 3-Deoxy-D-manno-octulosonic acid 8-phosphate synthase from the hyperthermophilic bacterium Aquifex aeolicus. Biochem Biophys Res Commun. 1999;263(2):346–51. doi: 10.1006/bbrc.1999.1361. [DOI] [PubMed] [Google Scholar]
- 11.Sheflyan GY, Howe DL, Wilson TL, Woodard RW. Enzymatic Synthesis of 3-Deoxy-d-manno-octulosonate 8-Phosphate, 3-Deoxy-d-altro-octulosonate 8-Phosphate, 3,5-Dideoxy-d-gluco(manno)-octulosonate 8-Phosphate by 3-Deoxy-d-arabino-heptulosonate 7-Phosphate Synthase. J Am Chem Soc. 1998;120(43):11027–32. [Google Scholar]
- 12.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystallog, Pt A. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 13.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography F NMR system : A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 14.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–9. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 15.Lamzin VS, Wilson KS. Automated refinement for protein crystallography. Methods Enzymol. 1997;277:269–305. doi: 10.1016/s0076-6879(97)77016-2. [DOI] [PubMed] [Google Scholar]
- 16.Laskowski RA, MacArthur MIW, Moss DS, Thornton JM. PROCHECK : a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–91. [Google Scholar]
- 17.Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr. 1994;50(Pt 2):178–85. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 18.Hubbard KL, Thornton JM. NACCESS. Dept. of Biomol. Sci., UMIST; Manchester, UK: 1993. [Google Scholar]
- 19.Sako Y, Nomura N, Uchida A, Ishida Y, Morii H, Koga Y, Hoaki T, Maruyama T. Aeropyrum pernix gen. nov., sp. nov., a novel aerobic hyperthermophilic archaeon growing at temperatures up to 100 degrees C. Int J Syst Bacteriol. 1996;46(4):1070–7. doi: 10.1099/00207713-46-4-1070. [DOI] [PubMed] [Google Scholar]
- 20.Shumilin IA, Bauerle R, Wu J, Woodard RW, Kretsinger RH. Crystal structure of the reaction complex of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from Thermotoga maritima refines the catalytic mechanism and indicates a new mechanism of allosteric regulation. J Mol Biol. 2004;341(2):455–66. doi: 10.1016/j.jmb.2004.05.077. [DOI] [PubMed] [Google Scholar]
- 21.Shumilin IA, Bauerle R, Kretsinger RH. The high-resolution structure of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase reveals a twist in the plane of bound phosphoenolpyruvate. Biochemistry. 2003;42(13):3766–76. doi: 10.1021/bi027257p. [DOI] [PubMed] [Google Scholar]
- 22.Walker GE, Dunbar B, Hunter IS, Nimmo HG, Coggins JR. Evidence for a novel class of microbial 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase in Streptomyces coelicolor A3(2), Streptomyces rimosus and Neurospora crassa. Microbiology. 1996;142(Pt 8):1973–82. doi: 10.1099/13500872-142-8-1973. [DOI] [PubMed] [Google Scholar]
- 23.Gosset G, Bonner CA, Jensen RA. Microbial origin of plant-type 2-keto-3-deoxy-D-arabino-heptulosonate 7-phosphate synthases, exemplified by the chorismate- and tryptophan-regulated enzyme from Xanthomonas campestris. J Bacteriol. 2001;183(13):4061–70. doi: 10.1128/JB.183.13.4061-4070.2001. PMCID : 95290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schofield LR, Anderson BF, Patchett ML, Norris GE, Jameson GB, Parker EJ. Substrate ambiguity and crystal structure of Pyrococcus furiosus 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase : an ancestral 3-deoxyald-2-ulosonate-phosphate synthase? Biochemistry. 2005;44(36):11950–62. doi: 10.1021/bi050577z. [DOI] [PubMed] [Google Scholar]
- 25.Schofield LR, Patchett ML, Parker EJ. Expression, purification, and characterization of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase from Pyrococcus furiosus. Protein Expres Purif. 2004;34(1):17–27. doi: 10.1016/j.pep.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 26.Paravicini G, Schmidheini T, Braus G. Purification and properties of the 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase (phenylalanine-inhibitable) of Saccharomyces cerevisiae. Eur J Biochem. 1989;186(1-2):361–6. doi: 10.1111/j.1432-1033.1989.tb15217.x. [DOI] [PubMed] [Google Scholar]
- 27.Schnappauf G, Hartmann M, Kunzler M, Braus GH. The two 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase isoenzymes from Saccharomyces cerevisiae show different kinetic modes of inhibition. Arch Microbiol. 1998;169(6):517–24. doi: 10.1007/s002030050605. [DOI] [PubMed] [Google Scholar]
- 28.Konig V, Pfeil A, Braus GH, Schneider TR. Substrate and metal complexes of 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from Saccharomyces cerevisiae provide new insights into the catalytic mechanism. J Mol Biol. 2004;337(3):675–90. doi: 10.1016/j.jmb.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 29.Ravanal MC, Goldie H, Cardemil E. Thermal stability of phosphoenolpyruvate carboxykinases from Escherichia coli, Trypanosoma brucei, and Saccharomyces cerevisiae. J Protein Chem. 2003;22(4):311–5. doi: 10.1023/a:1025306105105. [DOI] [PubMed] [Google Scholar]
- 30.Takai K, Sako Y, Uchida A, Ishida Y. Extremely thermostable phosphoenolpyruvate carboxylase from an extreme thermophile, Rhodothermus obamensis. J Biochem. 1997;122(1):32–40. doi: 10.1093/oxfordjournals.jbchem.a021737. [DOI] [PubMed] [Google Scholar]
- 31.Andreotti G, Cubellis MV, Nitti G, Sannia G, Mai X, Adams M1W, Marino G. An extremely thermostable aromatic aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus. Biochim Biophys Acta. 1995;1247(1):90–6. doi: 10.1016/0167-4838(94)00211-x. [DOI] [PubMed] [Google Scholar]
- 32.Fabry S, Hensel R. Purification and characterization of D-glyceraldehyde-3-phosphate dehydrogenase from the thermophilic archaebacterium Methanothermus fervidus. Eur J Biochem. 1987;165(1):147–55. doi: 10.1111/j.1432-1033.1987.tb11205.x. [DOI] [PubMed] [Google Scholar]
- 33.Simpson HD, Haufler UR, Daniel RM. An extremely thermostable xylanase from the thermophilic eubacterium Thermotoga. Biochem J. 1991;277(Pt 2):413–7. doi: 10.1042/bj2770413. PMCID : 1151249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park OK, Bauerle R. Metal-catalyzed oxidation of phenylalanine-sensitive 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase from Escherichia coli : inactivation and destabilization by oxidation of active-site cysteines. J Bacteriol. 1999;181(5):1636–42. doi: 10.1128/jb.181.5.1636-1642.1999. PMCID : 93556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edwards RA, Jacobson AL, Huber RE. Thermal denaturation of beta-galactosidase and of two site-specific mutants. Biochemistry. 1990;29(49):11001–8. doi: 10.1021/bi00501a019. [DOI] [PubMed] [Google Scholar]
- 36.Hartmann M, Schneider TR, Pfeil A, Heinrich G, Lipscomb WN, Braus GH. Evolution of feedback-inhibited beta /alpha barrel isoenzymes by gene duplication and a single mutation. Proc Natl Acad Sci USA. 2003;100(3):862–7. doi: 10.1073/pnas.0337566100. PMCID : 298692. [DOI] [PMC free article] [PubMed] [Google Scholar]