SUMMARY
The hematopoietic Colony Stimulating Factor-1 receptor (CSF-1R or FMS) is essential for the development of diverse cell types central to the immune system. Here we report a structural and mechanistic consensus for the assembly of hematopoietic human and mouse CSF-1:CSF-1R complexes. The EM structure of the complete extracellular assembly of the human CSF-1:CSF-1R complex reveals how receptor dimerization by CSF-1 invokes a ternary complex featuring extensive homotypic receptor contacts that contribute 15-fold to the affinity of the complex, and striking structural plasticity at the extremities of the complex. Small-angle X-ray scattering analysis of unliganded hCSF-1R points to large domain rearrangements upon CSF-1 binding, and provides structural evidence for the relevance of receptor predimerization at the cell-surface. Comparative structural and binding studies of human and mouse CSF-1R complexes, including a quantification of the CSF-1/CSF-1R species cross-reactivity, show that bivalent cytokine binding to receptor is a common denominator in complex formation independent of receptor homotypic interactions.
INTRODUCTION
Receptor tyrosine kinases (RTKs) are a large family of metazoan-specific cell-surface receptors that play essential roles in diverse cellular processes (Lemmon and Schlessinger, 2010). The hallmark of signaling via RTKs lies in cytokine-induced activation of the receptor extracellular segments, which initiates a cascade of intracellular signaling following activation of the intrinsic tyrosine kinase activity of RTKs. Class III RTK (RTKIII) groups four pleiotropic hematopoietic receptors: the prototypic Platelet-Derived Growth factor Receptor (PDGFR), CSF-1R, KIT, and fms-like tyrosine kinase III receptor (Flt3). Collectively, intracellular signaling via RTKIII has a major impact in the development and homeostasis of the cellular repertoire throughout the hematopoietic system. RTKIII are characterized by a modular structure featuring five extracellular Ig-like domains followed by a single transmembrane helix (TM) and intracellular split kinase domains (Lemmon and Schlessinger, 2010). A remarkable aspect of RTKIII activation is that the cognate protein ligands are all dimeric with similar dimensions despite their grouping into two fundamentally different folds (4-helix bundles versus all-β cystine-knot scaffolds) (Jiang et al., 2000; Oefner et al., 1992; Pandit et al., 1992; Savvides et al., 2000; Wiesmann et al., 1997; Zhang et al., 2000). Recently, interleukin-34 (IL-34) was identified as a second ligand to CSF-1R (Lin et al., 2008), thus adding a perplexing dimension to RTKIII signaling as IL-34 bears no sequence similarity to the currently known cytokine ligands for RTKIII/V or other proteins.
Activation of the extracellular segment of hCSF-1R by its two cytokine ligands, hCSF-1 and interleukin 34 (IL-34), is the cornerstone of signaling cascades central to immunity, as CSF-1R:cytokine signaling complexes are essential for the proliferation, differentiation and functionality of cells derived from the mononuclear phagocytic lineage, such as monocytes, tissue macrophages, microglia, osteoclasts, and antigen-presenting dendritic cells (Chihara et al., 2010; Chitu and Stanley, 2006; Lin et al., 2008; Wei et al., 2010). Furthermore, signaling via wild-type hCSF-1R and mutants thereof has been implicated in a wide range of pathologies in humans such as, arthritis, atherosclerosis, tumor growth, and metastasis (Chitu and Stanley, 2006).
CSF-1R is arguably the most intriguing member of the RTKIII family for two main reasons: (i) CSF-1R is the only known RTK that is activated by two unrelated protein ligands, and (ii) CSF-1R activation demonstrates restrictive species specificity. For instance, mouse CSF-1 (mCSF-1) does not signal through hCSF-1R and other primate CSF-1R, yet hCSF-1 can activate CSF-1R from all primate and non-primate species tested thus far (Garceau et al., 2010). IL-34, the recently identified second ligand for CSF-1R appears to follow suit, in that human IL-34 does not activate mCSF-1R while murine IL-34 does signal through hCSF-1R (Wei et al., 2010).
Despite the prominence of hCSF-1R and hCSF-1 in the biomedical literature over the last three decades, structural characterization of the extracellular complex has remained elusive while structures of the intracellular kinase domain have only recently become available (Schubert et al., 2007; Walter et al., 2007). Such insights are the missing link to the structural and functional diversity of RTKIII/V extracellular complexes, and would help provide a nearly complete picture of the entire CSF-1 ligand-receptor signaling complex given the available structure of the CSF-1R intracellular kinase domains (Griffith et al., 2004). A recent flurry of studies of RTKIII/V extracellular complexes led to a structural paradigm for RTKIII/V activation, whereby the receptors bind via their N-terminal Ig-like domains to the activating dimeric cytokine and concomitantly make homotypic contacts between their membrane-proximal domains (Chen et al., 2008; Leppanen et al., 2010; Liu et al., 2007; Ruch et al., 2007; Shim et al., 2010; Verstraete et al., 2011; Yang et al., 2010; Yang et al., 2008; Yuzawa et al., 2007).
A recent structural study of mCSF-1 in complex with the first three extracellular domains of mCSF-1R (mCSF-1RD1-D3) revealed unexpected monovalent binding of mCSF-1 to one mCSF-1RD1-D3 molecule leading to a binary complex (Chen et al., 2008), in contrast to predictions based on earlier studies of the homologous murine and human c-kit receptors in complex with Stem Cell Factor (SCF). While this first structural snapshot of a partial mCSF-1R complex is informative in its own right, it cannot be readily extrapolated to represent CSF-1R activation in general, given the complexity of species cross-reactivity in CSF-1R signaling. Furthermore, the reported binary mCSF-1RD1-D3:mCSF-1 complex does not offer realistic insights into possible homotypic receptor interactions, a likely critical element of receptor activation.
Here, we dissect the structural modularity and thermodynamic binding fingerprints of the extracellular human and mouse CSF-1:CSF-1R assemblies. Together, our comparative studies provide a comprehensive set of structural and mechanistic insights that now help establish a consensus for the assembly of hematopoietic CSF-1 ligand-receptor complexes.
RESULTS AND DISCUSSION
Biochemical and thermodynamic characterization of full-length CSF-1R ectodomain complexes (CSF-1:CSF-1RD1-D5)
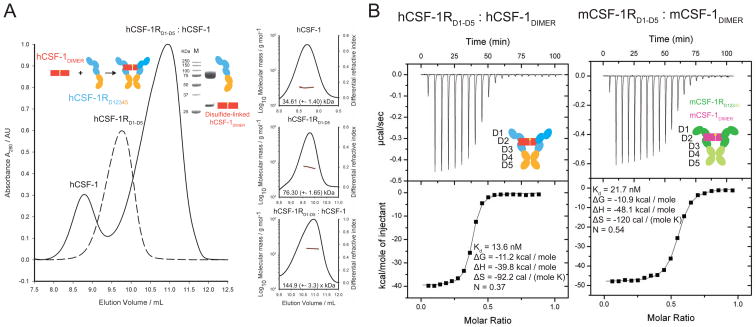
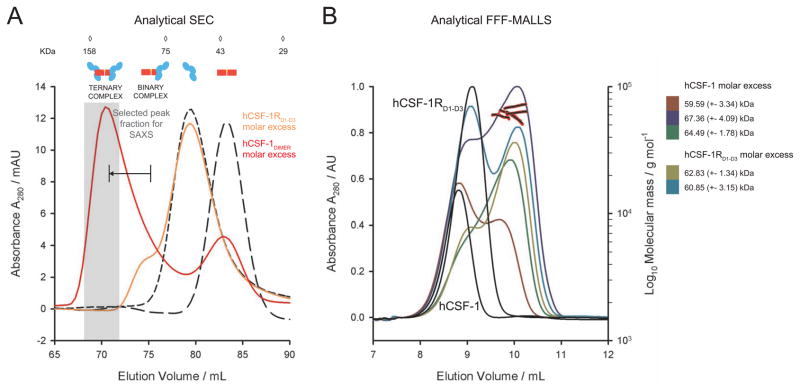
To enable structural and biophysical studies of human and mouse CSF-1:CSF-1RD1-D5 complexes, we produced recombinant glycosylated human and mouse CSF-1RD1-D5 in transiently transfected HEK293T cells in the presence of kifunensine, which limits N-linked glycosylation to Man5–9GlcNAc2 glycan structures (Chang et al., 2007). Recombinant human and mouse CSF-1 were produced by in vitro refolding of inclusion bodies after protein expression in E. coli. Preparations of purified recombinant hCSF-1 and glycosylated hCSF-1RD1-D5 were analytically fractionated by Field-Flow Fractionation (FFF), followed by quantification of their molecular weights (MW) via online Multi-Angle Laser Light Scattering (MALLS). This led to MW determinations of 35 kDa and 76 kDa, for hCSF-1 and hCSF-1RD1-D5, respectively. These values are in excellent agreement with the electrophoretic mobility of dimeric hCSF-1 and monomeric glycosylated hCSF-1RD1-D5 on SDS-PAGE (Figure 1A). Titration of human CSF-1RD1-D5 with excess molar amounts of cognate CSF-1 resulted in a monodisperse molecular species that exhibited a marked shift in elution profile to a much larger particle (145 kDa as determined by MALLS) when compared to the unbound CSF-1R ectodomain (Figure 1A). Considering the experimental accuracy of MW determination by MALLS, we could infer that the apparent CSF-1:CSF-1RD1-D5 complex could be rationalized in terms of 1 hCSF-1 dimer and 2 copies of hCSF-1RD1-D5.
Figure 1. hCSF-1RD1-D5 forms a ternary assembly with hCSF-1.
(A) Isolation of hCSF-1RD1-D5:hCSF-1 by field-flow fractionation (FFF). Formation of the complex leads to a marked shift in elution profile away from the individual protein components after titration with a molar excess of hCSF-1. The different protein components employed are annotated. The inset shows an SDS-PAGE strip of the isolated complex. The disulfide-linked dimeric nature of hCSF-1 is confirmed since the samples are lacking β-mercaptoethanol (BME). Slight smearing of the hCSF-1RD1-D5 band is due to a certain level of heterogeneous glycosylation (Aricescu et al., 2006). The insets show molecular mass determination by MALLS. The measurements confirm the dimeric nature of hCSF-1 and suggest a hCSF-1RD1-D5 monomer and a hCSF-1:hCSF-1RD1-D5 1:2 stoichiometry of binding. Derived molecular masses and fits to the experimental LS data are shown. (B) Titration of hCSF-1 into hCSF-1RD1-D5 (left panel) and mCSF-1 into mCSF-1RD1-D5 (right panel). Both CSF-1 ligands form a high-affinity ternary complex (N=1:2) with their cognate receptors.
We employed isothermal titration calorimetry (ITC) to establish the affinity, thermodynamic profile and stoichiometry of the CSF-1:CSF-1RD1-D5 complex. Our results show that the complex is characterized by bivalent binding of hCSF-1 to the receptor ectodomain (1 hCSF-1 dimer to 2 molecules of hCSF-1RD1-D5) and that the ensuing high-affinity complex (KD=13.6 nM) is the result of a markedly exothermic binding event coupled to an entropic penalty (Figure 1B, Supplementary Table 2). The nM affinity value we report here for the soluble full-length extracellular complex differs significantly from previously reported Kd values of ~50–100 pM for native hCSF-1R based on cell-assays (Roussel et al., 1988). Similar differences have already been observed for a number of systems, including the homologous KIT and Flt3 (Graddis et al., 1998; Lemmon et al., 1997; Lev et al., 1992; Streeter et al., 2001; Verstraete et al., 2011) and can be attributed to the absence of the TM region and the 2D spatial confinement of the membrane. Upon extending our analysis to the mouse CSF-1:CSF-1R ectodomain complex, we found that mCSF-1 also binds its cognate mCSF-1RD1-D5 in a bivalent fashion to form a high-affinity ternary complex (KD = 21.7 nM) (Figure 1B) with a similar thermodynamic profile, indicating that the assembly of human and mouse ectodomain complexes is likely based on common principles.
Characterization of the CSF-1 ligand-receptor species cross-reactivity
We took advantage of the availability of human and mouse extracellular CSF-1R receptors and ligands to quantify their cross-reactivity and to lend further cross-validation of the binding stoichiometries determined for the human and mouse complexes. To our knowledge, this has never been reported while the biomedical literature is heavily populated by studies of hCSF-1 activity in a murine cellular background and vice-versa. Such information could have important implications in the design and interpretation of cellular assays testing cytokine:receptor activity from a particular species in a heterologous background. Our experiments revealed bivalent binding of CSF-1 ligands to receptors in both cross-reactivity experiments, consistent with the binding behavior of human and mouse CSF-1R to their cognate ligands (Figure 2). We calculated a KD=66.2 nM for the hCSF-1:mCSF-1RD1-D5 interaction, which agrees well with the ability of hCSF-1 to activate all non-primate CSF-1R tested so far. On the other hand, mCSF-1 binds nearly 500-fold less tightly to hCSF-1RD1-D5 (Kd=2.8 μM) than to its cognate receptor thus corroborating the observation that mCSF-1 is not able to activate primate CSF-1R in a cellular setting (Figure 2). Together, our binding studies on the assembly of cognate and non-cognate CSF-1 ligand-receptor complexes show that bivalent cytokine binding to receptor is a conserved mechanistic aspect of the extracellular ligand-receptor interaction.
Figure 2. Thermodynamic characterization of non-cognate extracellular human and mouse CSF-1 receptor-ligand complexes.
Thermodynamic measurements of the human and mouse CSF-1RD1-D5:CSF-1 species cross-reactivity. In each case CSF-1 was titrated into non-cognate CSF-1R. hCSF-1 is able to form a high-affinity complex with mCSF-1R D1-D5 (left panel), while the mCSF-1:hCSF-1RD1-D5 interaction is of much lower strength (right panel). Both complexes display a 1:2 CSF-1:CSF-1R stoichiometry of binding.
EM structure of the complete extracellular assembly of the human CSF-1:CSF-1R complex
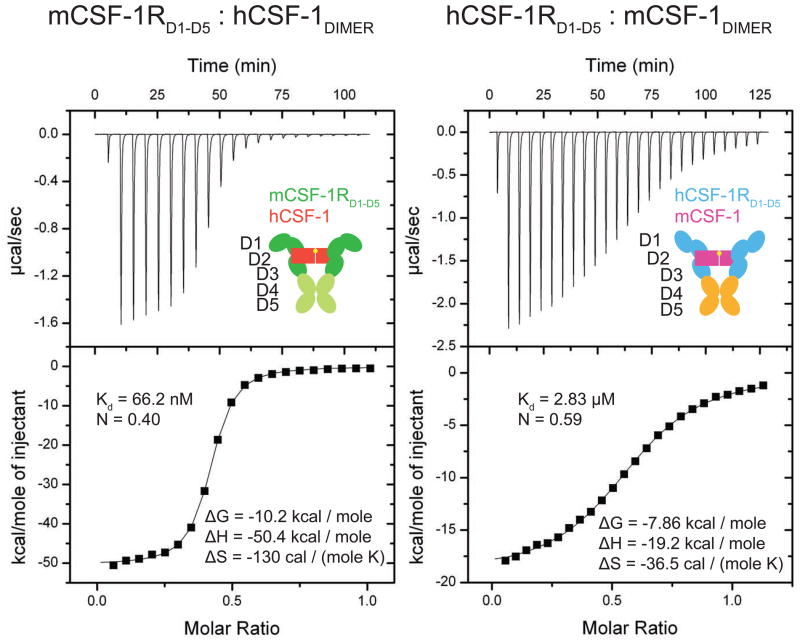
We approached structural characterization of the complete extracellular signaling complex of hCSF-1R with hCSF-1, based on images of negatively-stained hCSF-1RD1-D5:hCSF-1 complex obtained by electron microscopy (EM). The recombinant hCSF-1RD1-D5:hCSF-1 complex used in the EM analysis was obtained by preparative size-exclusion chromatography (SEC) as a highly monodisperse molecular species. Multivariate statistical analysis and classification of circa 18,500 particles indicated the presence of a two-fold symmetry axis. Thus, an ab initio three-dimensional reconstruction was produced by angular reconstitution with imposed C2 symmetry and further improved by iterative projection matching to generate a 3D reconstruction of the hCSF-1RD1-D5:hCSF-1 extracellular complex to ~23 Å resolution (Figure 3A,B).
Figure 3. Architecture of liganded hCSF-1RD1-D5.
(A) Three-dimensional reconstruction of the hCSF-1RD1-D5:hCSF-1 complex from EM data. A gallery of representative class averages (above) and re-projections of the final 3D reconstruction (below) under similar orientations is shown. (B) Angle, front, top and side orientational views of the reconstructed particle superimposed with computational models of the complex. (C) Conservation of the D4-D4′ dimerization motif across members of the RTKIII and RTKV families. Residues 374–393 present on the D4 βE strand and EF loop of hKIT are aligned with corresponding sequences of h/mCSF-1R, hFlt3, hPDGFR and hVEGFR. Conserved residues are highlighted. hFlt3 lacks the complete motif and has been shown to be devoid of homotypic receptor contacts (Verstraete et al., 2011). See also Figure S1.
The reconstructed 3D molecular envelope of the hCSF-1RD1-D5:hCSF-1 complex reveals a central triangular toroidal structure featuring a pair of appendages extending away from each other at the top of the ring in a plane perpendicular to the toroid, and two in-plane legs of electron density emanating from the bottom of the ring (Figure 3B). Clear features in the electron density strongly suggested that dimeric hCSF-1 binds bivalently to two hCSF-1RD1-D5 receptor molecules at the head of the particle, and that the two receptor molecules engage in homotypic interactions away from the ligand-binding epitope. Manual placement of homology models of hCSF-1RD1-D5 derived from the structure of the extracellular segment of human KIT (Yuzawa et al., 2007), and of the crystal structure of hCSF-1 (Pandit et al., 1992) into the EM map confirmed this initial interpretation, and showed that the volume of the EM map could readily account for all components of the hCSF-1R extracellular complex. To improve our preliminary model against the experimental EM envelope we employed a computational approach based on molecular dynamics protocols, which produced 20 different models that were subsequently averaged to yield the final model (Figure 3B and Supplementary Figure 1A).
The hCSF-1RD1-D5:hCSF-1 complex now joins the human KITD1-D5-SCF (Yuzawa et al., 2007) and the human Flt3 ligand-receptor (Verstraete et al., 2011) complexes as the third complete extracellular RTKIII complex structurally characterized to date, and offers important architectural and functional insights. First, it reveals that the cytokine-binding epitope on hCSF-1R is defined by domains 2 and 3 (Figure 3B). With the exception of the Flt3 ligand-receptor interaction, this feature of receptor-ligand engagement has emerged as a consensus blueprint of RTKIII activation in all other structurally characterized RTKIII complexes thus far (binary mCSF-1RD1-D3:mCSF-1 complex (Chen et al., 2008), KITD1-D3(5):SCF (Liu et al., 2007; Yuzawa et al., 2007) and PDGFRD1-D3:PDGF-B (Shim et al.)). Second, it shows that receptor homotypic interactions can be attributed to a broad interaction interface between the tandem D4 domains of hCSF-1R, while the membrane-proximal D5 domains diverge away to a separation of ~65 Å (Figure 3B). Homotypic receptor interactions have long been considered as the driving force for the cooperative character of extracellular complex formation and activation in RTK. Recent studies on RTKIII receptors KIT and PDGFR showed that receptor contacts mediated by a conserved dimerization sequence fingerprint mapped to the EF loop of D4 are important for receptor activation (Yang et al., 2008; Yuzawa et al., 2007) (Figure 3C). Consistent with the proposed key role of the consensus dimerization motif, structural studies on Flt3, the only RTKIII/V receptor lacking this sequence fingerprint, showed that the Flt3 ligand-receptor assembly is devoid of homotypic receptor interactions (Verstraete et al., 2011).
Whereas our structural studies show that hCSF-1RD4 plays a direct role in the CSF-1 extracellular ternary complex, the possible contribution of D5 still remains unclear. The membrane-proximal D5 in KITD1-D5-SCF does not make interactions with its tandem D5 and the corresponding C-termini come to 15 Å from each other (Yuzawa et al., 2007). Furthermore, the crystal structure of the complete extracellular Flt3 ligand-receptor complex has recently shown that the two Flt3D5 approach each other to about 25 Å (Verstraete et al., 2011). In hCSF-1R this separation is much larger, thus highlighting the possible conformational diversity of the membrane-proximal domains. Reconciling such interdomain distances in terms of growing evidence on the importance of TM domains in RTK activation (Finger et al., 2009; Li and Hristova, 2006), is not obvious. Yet it would appear that the linker regions between D5 and the TM domains of RTKIII (typically 10–15 amino acids) would offer the necessary spatial freedom to allow such intramembrane interactions to take place, while the D4-D5 interface could help orient such associations. Finally, our studies show that the N-terminal D1 extends well away from the core of the complex without making any interactions with the ligand or other receptor domains. Our computational models show considerable rigid-body flexibility around the D1-D2 linker (Supplementary Figure 1A). Indeed, the corresponding negative-stain electron density for D1 only became clear in later rounds of image classification. Interestingly, Flt3D1 in the Flt3 ligand-receptor complex also emanates away from the core of the complex (Verstraete et al., 2011). It is currently not clear what the possible role of such flexible D1 modules might be, but it has been suggested that D1 might participate in intermolecular interactions at the cell surface (Verstraete et al., 2011). However, the apparent conformational independence of D1 in human Flt3 and CSF-1R is not a conserved structural feature within the RTKIII family as structures of the binary mCSF-1RD1-D3:mCSF-1 complex, as well as the ternary KITD1-D3(5):SCF and PDGFRD1-D3:PDGF-B complexes show that D1 bends downwards to interact with D2. We carried out additional measurements on hCSF-1RD1-D5:hCSF-1 by SAXS which consistently corroborate our EM findings, in that the scattering data indicate a P2-symmetrical ternary complex with flexible D1 and large divergence of the membrane-proximal D5 (Supplementary Figure 1B and Supplementary Table 1).
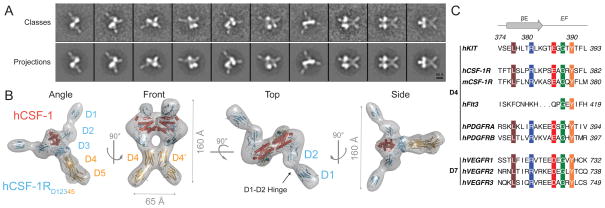
Structural plasticity of human CSF-1RD1-D5 revealed by SAXS analysis of the unbound receptor
We carried out measurements on hCSF-1RD1-D5 by SAXS to generate structural insights into unbound hCSF-1R and any possible domain rearrangements that might occur upon ligand binding. The X-ray scattering by hCSF-1RD1-D5 within a broad concentration range was only consistent with a dimeric species (Figure 4 and Supplementary Table 1). Interestingly, the MW for hCSF-1RD1-D5 as determined based on our SAXS data is exactly twice the MW determined via analytical FFF-MALLS measurements conducted at lower concentrations (Figure 1A). This suggests that monomeric and dimeric species for hCSF-1RD1-D5 can exist in equilibrium, albeit with a rather poor equilibrium dissociation constant. Molecular envelopes derived from ab initio reconstructions and rigid-body modeling agree remarkably well with each other, and point to a well-defined dimeric assembly that lacks internal symmetry (Figure 4). Despite the dramatic deviation from the two-fold symmetry observed in the receptor:ligand complex (Figure 3B), we note that the extended conformation of the unliganded receptor resembles the bound conformation observed in the EM structure, hinting that preferential structural sampling might facilitate productive ligand binding. The observed hCSF-1R dimerization in vitro is consistent with previously reported cellular studies which showed the propensity of CSF-1R to form dimers at the cell surface of CSF-1 dependent BAC1.2F5 cells (Li and Stanley, 1991). Thus, the structural view of unbound hCSF-1R analysis of the SAXS data may represent dimeric forms of hCSF-1R at high levels of receptor expression or when the receptors are constitutively activated in disease scenarios. In this respect, extracellular receptor pre-dimerization could also play an important role in generating the ultra high affinities observed in a physiological setting. Interestingly, a number of other RTKs, such as the IGF1 (Lawrence et al., 2007), EGFR (Chung et al., 2010; Mi et al., 2011) and Eph (Himanen et al., 2007) receptors do form oligomers in the absence of cytokine ligand. Nonetheless, hCSF-1RD1-D5 would have to undergo dramatic domain rearrangements to bind hCSF-1. Such conformational switching has already been observed in the related human KIT (Yuzawa et al., 2007) and human VEGFR (Ruch et al., 2007). Together, our data reinforce the notion that extracellular complex formation is cooperative and relies on an intricate interplay of receptor-ligand interactions, and intramolecular and homotypic receptor contacts.
Figure 4. Plasticity of unliganded hCSF-1RD1-D5.
(A) Structural analysis of ligand-free hCSF-1RD1-D5 by SAXS. Experimental scattering curves are shown in black to a maximal momentum transfer of s=0.25 Å−1 (nominal resolution 25 Å) and the individual data:fit pairs are put on an arbitrary y-axis to allow for better visualization. Curve (i): Rigid-body optimized fit of dimeric hCSF-1RD1-D5. Modeling was constrained by specifying ambiguous contact distances for the D45-D45′ module (circled). Curve (ii): Rigid-body optimized fit of receptor domains for monomeric hCSF-1RD1-D5. The upper inset shows the calculated distance distribution function for modeled dimeric and monomeric receptors, and their fits with the experimental function. The rigid body SASREF model and ab initio GASBOR bead model are displayed side-to-side to highlight agreement in overall shape reconstruction. See also Table S1.
Human and mouse CSF-1RD1-D3 can form stable ternary complexes with cognate CSF-1 ligands
A previous structural study of mCSF-1 in complex with the first three extracellular domains of mCSF-1R (mCSF-1RD1-D3) revealed an unexpected binary complex, whereby a mCSF-1 dimer binds monovalently to a single mCSF-1RD1-D3 molecule (Chen et al., 2008). This is in striking contrast to full-length ectodomain that forms a ternary complex with cognate or non-cognate ligand (Figure 1, 2 and 3). To address this apparent discrepancy in behavior between full-length and truncated receptors and to explore the contribution of the D4-D5 module to the mechanism of ternary complex formation, we produced recombinant glycosylated human and mouse CSF-1RD1-D3 to enable structural and biophysical studies.
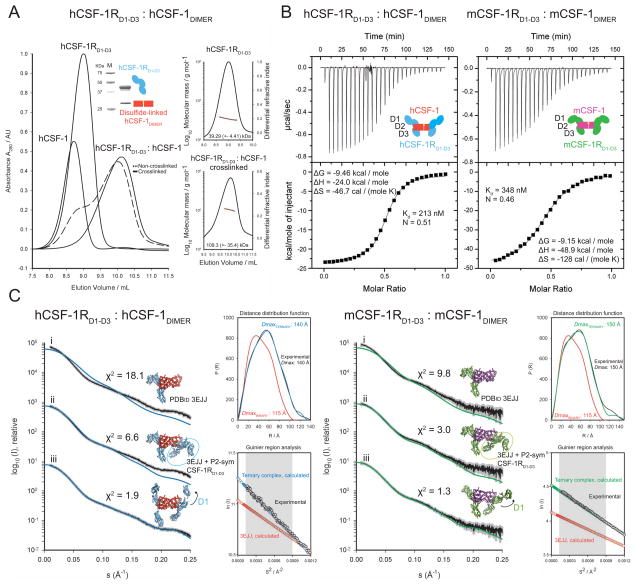
While the full-length ectodomains could readily reach their end-point assembly even with substoichiometric molar amounts of hCSF-1 using either SEC or FFF fractionation methods (Figure 1A), the CSF-1RD1-D3 constructs behaved differently (Figure 5). Titrating hCSF-1 with a molar excess of hCSF-1RD1-D3 only leads to minor shift on SEC as a shoulder peak of the unbound CSF-1RD1-D3 peak (Figure 5A). This behavior is consistent with previous findings (Chen et al., 2008). However, upon titrating hCSF-1RD1-D3 with a stoichiometric excess of cytokine ligand a clear shift can be obtained in the elution profile of hCSF-1RD1-D3 on SEC corresponding to a well-defined and markedly larger molecular species (Figure 5A). We sought to obtain more direct evidence into the molecular composition of the two species observed in SEC by attempting to determine their MW via analytical FFF-MALLS. Preparation of the hCSF-1:hCSF-1RD1-D3 complex by either a molar excess of hCSF-1 or hCSF-1RD1-D3 consistently revealed a ~65 kDa assembly, consistent with binary complex formation (Figure 5B). This clearly contradicted the chromatographic observation of two different kinds of complexes via SEC (Figure 5A). In an effort to resolve this apparent discrepancy, we applied the peak fraction obtained via SEC by titrating a molar excess of hCSF-1 to hCSF-1RD1-D3 to FFF fractionation followed by MALLS measurements. This fraction falls apart into two peaks and the largest molecular species represented a 65 kDa particle as determined by MALLS (Figure 6A). We therefore wondered whether the kinetics of molecular diffusion underlying the FFF method combined with a possible instability of the hCSF-1:hCSF-1RD1-D3 at such low concentrations might affect the integrity of the complex. To address this, we first subjected the distinct peak of the hCSF-1:hCSF-1RD1-D3 complex isolated by SEC (Figure 5A) to cross-linking with formaldehyde followed by fractionation via FFF and MALLS measurements. Indeed, this approach led to a dramatically different elution profile on FFF characterized by a single peak corresponding to a molecular species of 109 kDa (Figure 6A). This indicates that both binary and ternary hCSF-1:hCSF-1RD1-D3 complexes are possible depending on experimental conditions, and that an apparent prerequisite for the formation and stability of the ternary complex is the presence of a stoichiometric excess of ligand.
Figure 5. hCSF-1 can make both a monovalent and bivalent complex with hCSF-1RD1-D3.
(A) Isolation of the hCSF-1RD1-D3:hCSF-1 complex by size-exclusion chromatography (SEC). Titration with either a molar excess of hCSF-1RD1-D3 or hCSF-1 leads to different complexes. A marked shift in elution profile away from the individual protein components can only be observed after titration with a molar excess of hCSF-1. The resulting peak fraction has as such been analyzed by SAXS (Figure 6C). The different protein components employed are annotated. (B) Only the binary complex can be observed by FFF, regardless of stoichiometric excess of any component.
Figure 6. Human and mouse CSF-1RD1-D3 can form ternary complexes with cognate CSF-1 ligands.
(A) The ternary hCSF-1RD1-D3:hCSF-1 complex is transient on FFF. Injection of the isolated SEC peak fraction (Figure 5A) on FFF reveals a disassembly of the complex. A 110 kDa species indicative of a ternary complex can only be observed after incubation with a crosslinking agent, suggesting that a ternary complex is inherently less stable. The insets show a SDS-PAGE strip of the isolated non-crosslinked complex and molecular mass determination by MALLS. (B) Thermodynamic profile of hCSF-1RD1-D3:hCSF-1 and mCSF-1RD1-D3:mCSF-1. Both thermograms can be accurately fitted by a “one set of binding sites” model and display a 1:2 CSF-1:CSF-1RD1-D3 stoichiometry of binding. (C) Structural analysis of hCSF-1RD1-D3:hCSF-1 (left panel) and mCSF-1RD1-D3:mCSF-1 (right panel) by SAXS after isolation by SEC (Figure 5A). Experimental scattering curves are shown in black to a maximal momentum transfer of s=0.25 Å−1 (nominal resolution 25 Å) and the individual data:fit pairs are put on an arbitrary y-axis to allow for better visualization. Curves (i): Comparison of the experimental scattering with calculated scattering from the monovalent mCSF-1RD1-D3:mCSF-1 structure (PDB code 3EJJ). This binary model lacks significant scattering mass as judged by the gross incompatibility with the lowest angle experimental data. Curves (ii): Comparison of the experimental scattering with calculated scattering from a bivalent model derived from the mCSF-1RD1-D3:mCSF-1 structure (PDB code 3EJJ) in which an additional CSF-1RD1-D3 arm was generated by applying a pure 2-fold symmetry operation about the ligand dimer interface (circled). Curves (iii): Rigid-body optimized fit of the bivalent CSF-1RD1-D3:CSF-1 complex with specified CSF-1:CSF-1RD2 core contacts and moving domains D1 and D3. The upper insets show the calculated distance distribution function for the modeled ternary complexes (blue or green) and for PDBid 3EJJ (red), and their fits with the experimental function (black). The lower insets display the experimental Guinier region (black) and the calculated Guinier region of the rigid body refined ternary models (blue or green) and the binary PDBid 3EJJ model (red). The shaded area indicates the range of fitting for RG analysis (RG·S ≤ 1.3). See also Figure S2 and Table S2.
We employed isothermal titration calorimetry (ITC) to further characterize the interaction between hCSF-1RD1-D3 with cognate hCSF-1 and to obtain insights into the contribution of the membrane-proximal module D4-D5 to the extracellular assembly. Firstly, the binding isotherm could be accurately fitted using a “one set of binding sites” model, and there was no evidence for two sequential or independent binding sites with different affinities. Importantly, the complex displayed a 1:2 stoichiometry of binding revealing bivalent binding of hCSF-1 to hCSF-1RD1-D3, in complete agreement with the association mode of the full-length ectodomain complex (Figure 1B). Nonetheless, the strength of the interaction and the corresponding thermodynamic profile differs drastically from that of the hCSF-1RD1-D5:hCSF-1 interaction (Figure 6B, Supplementary Table 2). Notably, hCSF-1 binds 15-fold less tightly to hCSF-1RD1-D3 than to full-length extracellular hCSF-1R (Kd=213 nM vs. Kd=13.6 nM). Thus, the absence of the membrane-proximal module D4-D5 provides a significant enthalpic loss of ~15 kcal mol−1 coupled to an entropic gain.
The observation of the bivalent hCSF-1RD1-D3:hCSF-1 complex via ITC (N=0.5) is in stark contrast to the monovalent binding mode reported for the mouse CSF-1RD1-D3:CSF-1 interaction (Chen et al., 2008), thus creating a puzzling paradox with respect to mechanistic aspects of receptor binding and activation. It would indeed seem unlikely that complex formation would bear such fundamental differences in the two homologous systems given the preponderance of conserved sequences on human and mouse CSF-1 and CSF-1R involved at the interaction epitope (Supplementary Figure 2). To resolve the apparent disagreement between the two sets of findings we characterized the assembly of the mouse CSF-1:CSF-1RD1-D3 complex by ITC. Our results based on several experimental replicas show unequivocally that the stoichiometry, corresponding affinities, and thermodynamic profile for mCSF-1RD1-D3:mCSF-1 are equivalent to those of the human counterpart (Figure 6B). Furthermore, we conclude that the relative contribution of the membrane-proximal domains to complex formation is similar in the two systems indicating a conserved role for the D4-D5 in the assembly of the extracellular complex. Thus, both the human and mouse CSF-1 ligand-receptor assemblies appear to share a common interaction mode, based on the inherent capacity of CSF-1 to bind bivalently to its cognate receptor. It is currently unclear why the ITC measurements by Chen et al. on the mouse CSF-1RD1-D3:CSF-1 interaction deviate so fundamentally from the data we present here. Nonetheless, our combined SEC/FFF/MALLS analysis of the CSF-1:CSF-1RD1-D3 complex provides a rationale for the crystallographic observation of the intriguing mouse CSF-1:CSF-1RD1-D3 binary complex (Chen et al., 2008), in the sense that we have shown that both ternary and binary assemblies can be formed for the CSF-1:CSF-1RD1-D3 complex depending on experimental conditions.
To provide further structural insights into extracellular complex formation and to investigate further the bivalent mode of CSF-1 binding to CSF-1R revealed by our ITC analysis (Figure 6B), we measured Small-angle X-ray Scattering (SAXS) data for the hCSF-1RD1-D3:hCSF-1 and mCSF-1RD1-D3:mCSF-1 complexes (Figure 6C and Supplementary Table 1). Both complexes were prepared via SEC by saturating CSF-1RD1-D3 with a molar excess of cognate CSF-1, and were conservatively pooled (Figure 5A). Importantly, our data analysis reveals that the crystal structure of the binary mCSF-1:mCSF-1RD1-D3 complex (Chen et al., 2008) is grossly incompatible with the SAXS data (Figure 6C, curves i), thereby directly challenging the claim that mCSF-1 cannot dimerize mCSF-1R in the absence of the membrane proximal module D4-D5 (Chen et al., 2008). Both the molecular parameters obtained directly via SAXS and structural modeling of the data showed unambiguously that hCSF-1RD1-D3:hCSF-1 and mCSF-1RD1-D3:mCSF-1 can form stable ternary complexes with P2 symmetry in solution (Figure 6C, curves ii-iii), thus providing a structural basis for the observed binding stoichiometries via ITC (Figure 6B). Furthermore, we note that the overall features of hCSF-1RD1-D3:hCSF-1 in solution are consistent with the corresponding segment in the hCSF-1RD1-D5:hCSF-1 EM model, in that D1 points upwards, albeit at a slightly different angle (Figure 6C, curve iii).
A common assembly mechanism for human and mouse CSF-1 ligand-receptor complexes
The integration of our findings on both the human and mouse CSF-1 ligand-receptor complexes puts our study in position to help resolve a puzzling mechanistic paradox for the assembly of extracellular CSF-1 ligand-receptor complexes that arose from a recent study on the mouse system (Chen et al., 2008). The premise of this study was that mouse CSF-1 is unable to dimerize its cognate receptor in the absence of the membrane proximal domains D4 and D5. The authors proposed that formation of a binary complex lowers the affinity of the second binding site on the dimeric cytokine, calling upon a negative cooperativity scenario, and extrapolated their reasoning to a distinct mechanistic proposal for CSF-1R activation entailing two steps. In a first step the ligand and receptor form an initial binary complex with low affinity that can only proceed to the ternary complex upon the simultaneous formation of cytokine-receptor interactions at the opposite binding epitope coupled to homotypic receptor interactions.
The diverse biochemical and structural evidence we reported here illustrate that the assembly of human and mouse extracellular CSF-1 complexes is driven by two common overriding principles. In the first instance, the cytokine ligands have the inherent capacity to offer two receptor binding sites leading to ternary complex formation. Bivalent binding of CSF-1 can thus take place to the pool of monomeric and dimeric CSF-1R at the cell surface. Secondly, assembly of the high-affinity complex is dramatically enhanced as a result of well-defined homotypic interactions between extracellular domain 4 modules. This is an example of positive cooperativity, and in the case of CSF-1 this is reflected in a pronounced enthalpy gain upon formation of the ternary complex. This also implies that binding of cytokine ligand to already predimerized CSF-1R would invoke a reorientation of the ectodomains to prime their role in the signaling complex. Together, these two sequential steps constitute a clear consensus for the binding events that lead to the assembly of high-affinity human and mouse CSF-1 ligand-receptor complexes. It remains to be seen whether IL-34, the newly discovered cytokine ligand for CSF-1R, will follow suit.
EXPERIMENTAL PROCEDURES
Production of recombinant human and mouse CSF-1, CSF-1R and CSF-1 ligand-receptor complexes
Recombinant human and mouse CSF-1 were produced as inclusion bodies in a prokaryotic expression system based on a previously described approach (Verstraete et al., 2009) and were purified to homogeneity following in vitro refolding. The fragment encoding residues 1–149 corresponding to the alpha splice variant of human and mouse CSF-1, was cloned into the pET-15b vector (Novagen). After expression in the BL21(DE3) CodonPlus-RP (Novagen) E. coli strain, h/mCSF-1 accumulated as inclusion bodies. The inclusion bodies were washed three times and then solubilized in 6.5 M GnHCl, 100 mM NaPO4 pH 8.0, 10 mM Tris and 10 mM β-mercaptoethanol. Next, denatured h/mCSF-1 was refolded by rapid dilution in refolding buffer (100 mM Tris, pH 8.5, 1M L-arginine, 3 mM GSH, 1.5 mM GSSG and 0.2 mM PMSF) at 277K. The clarified refolding mixture was loaded onto a HisTrap FF 5 mL affinity column, eluted and subsequently purified by size exclusion chromatography using a Prep-Grade HiLoad 16/60 Superdex 75 column (GE Healthcare). To remove the N-terminal His-tag, h/mCSF-1 was subsequently incubated overnight at room temperature with one unit of biotinylated thrombin (Novagen) per milligram of h/mCSF-1. Proteolytic cleavage was monitored by SDS–PAGE. Biotinylated thrombin was removed using a streptavidin-agarose column (Novagen). Thrombin-treated h/mCSF-1 were purified using a Source 30Q anion-exchange resin and followed by gel-filtration chromatography on a Superdex-75 column (GE Healthcare). The fractions corresponding to h/mCSF-1 were pooled and used for further experiments. Recombinant glycosylated human and murine CSF-1R ectodomain variants were produced in transiently transfected HEK293T cells in the presence of kifunensine based on established protocols (Aricescu et al., 2006; Chang et al., 2007). The recombinant CSF-1R ectodomains carried a C-terminal 6xHis-tag. h/mCSF-1R were purified by affinity chromatography from the supernatant using a Talon FF column (Clontech). The eluted fractions containing the purified protein were subsequently injected onto a Prep-Grade HiLoad 16/60 Superdex 200 column (GE Healthcare). The fractions corresponding to h/mCSF-1R were pooled and used for further experiments.
Human and murine CSF-1RD1-D5:CSF-1 and CSF-1RD1-D3:CSF-1 complexes were isolated by gel-filtration chromatography on Superdex-200 column (GE Healthcare) after incubation of CSF-1R ectodomains with excess molar amounts of purified cognate CSF-1.
Multi-Angle Laser Light Scattering
The molecular masses of CSF-1, CSF-1R and the CSF-1R:CSF-1 complexes were determined by MALLS. Protein sample was injected into a HPLC-driven (Shimadzu) field-flow fractionation (FFF) module (Wyatt Technology) equilibrated with a 20 mM Hepes pH 7.5, 150 mM NaCl running buffer. The FFF module was coupled to an online UV detector (Shimadzu), an 18-angle light scattering detector (DAWN HELEOS) and a refractive index detector (Optilab T-rEX) (Wyatt Technology). Typical concentrations used were 1 to 10 μM of protein species. A RI increment value (dn/dc value) of 0.185 ml/g was used for the protein concentration and molecular mass determination. FFF cross-flows were varied to optimize the resolution of separation. Data analysis was carried out using the ASTRA V software.
Electron microscopy
For preparation of negatively stained hCSF-1RD1-D5:hCSF-1 complex, purified sample at ~0.2 mg/mL in PBS buffer was applied to the clear side of carbon on a carbon-mica interface and stained with 2 % (w/v) uranyl acetate. Images were recorded under low-dose conditions with a JEOL 1200 EX II microscope at 100 kV and at a nominal 40.000× magnification. Selected negatives were digitized on a Zeiss scanner (Photoscan TD) to a pixel size of 3.5 Å at the specimen level. Image processing was carried out using the boxer routine from the EMAN software package (Ludtke et al., 1999) for particle selection, CTFFIND3 (Mindell and Grigorieff, 2003) for contrast transfer function determination, bctf from the bsoft package (Heymann et al., 2008) for CTF correction, Imagic (van Heel et al., 1996) for multivariate statistical analysis, classification and angular reconstitution, and Spider (Shaikh et al., 2008) for projection matching. UROX (Siebert and Navaza, 2009) was used for structure fitting.
A generous semi-automatic particle selection with the EMAN boxer routine lead to an extraction of a total of 18432 individual particle subframes of 80*80 pixels that were contrast transfer function-corrected and low-path-filtered at 15 Å resolution. The data set was translationally aligned relative to the rotationally averaged total sum of the individual images. The aligned dataset was subjected to multivariate statistical analysis (MSA), which suggested the presence of a two-fold symmetry axis. Characteristic class averages were used as a set of references for multi-reference alignment (MRA) followed by MSA and classification. After several iterations, representative class averages were selected to generate a crude initial model of the hCSF-1RD1-D5:hCSF-1 complex by angular reconstitution in C2 symmetry. Iterative projection matching of the model led to a 3D reconstruction with a well-defined global core corresponding to the ligand and hCSF-1RD2-D5, and a protruding weak density cloud, which we interpreted as D1 linked via a flexible linker to D2 in the complex core. To isolate a population of hCSF-1R1RD2-D5:hCSF-1 particles with a better defined orientation for D1, a set of models with the same core fitting the EM envelope, but different orientations of D1 protruding into the weak density cloud, was created based on the mCSF-1R1RD1-D3-mCSF-1 crystal structure (Chen et al., 2008). These models were converted into EM density and averaged together, which reinforced the density of the core in comparison to D1 thus supporting the notion that D1 is flexible. The average model was used for more rounds of projection matching which allowed a better definition for the position of D1. 9421 particles were included in the final reconstruction, which approached 23 Å resolution as estimated via Fourier shell correlation (FSC) according to the 0.5 criterion.
Modeling of the hCSF-1RD1-D5:hCSF-1 complex into the EM envelope
A homology model for hCSF-1RD1-D5 based on PDB entry 2E9W (Yuzawa et al., 2007) was fit into the 3D envelopes from EM with the EMAP module (Wu et al., 2003) of the CHARMM (Brooks et al., 2009) package to generate initial positions of the complex. A self-guided Langevin dynamics (Wu and Brooks, 2003) simulation of 1000 ps was performed, including an implicit solvation model, to search the conformational space to reach the conformations satisfying the EM map constraints. The final conformation was minimized with constraints to maintain the C2 symmetry.
Small-angle X-ray Scattering
Data were collected at beamlines X33 of the EMBL at DESY (Hamburg) and ID14-3 at ESRF (Grenoble) using a robotic sample changer (Roessle et al., 2008). The measurements were carried out at 298 K, within a momentum transfer range of 0.01 Å−1<s<0.6 Å−1 where s=4πsin(θ)/λ and 2θ is the scattering angle. All samples were measured at solute concentrations ranging from 0.5 to 10.0 mg/ml in 50 mM NaPO4 pH 7.40, 100 mM NaCl, with intermittent buffer solution (50 mM NaPO4, pH 7.40, 100 mM NaCl), and the radiation damage was monitored using standard procedures. The data were processed and extrapolated to infinite dilution and the Guinier region was inspected using the program PRIMUS (Konarev et al., 2003). The radius of gyration Rg and forward scattering I(0), the maximum particle dimension Dmax and the distance distribution function p(r) were evaluated using GNOM (Svergun, 1992). The molecular masses of the different constructs were calculated by comparison with the reference bovine serum albumin (BSA) samples. DAMMIN (Svergun, 1999) and AUTOPOROD were used to obtain the excluded volume and Porod volume of the particles, respectively. GASBOR (Svergun et al., 2001) was used to obtain the higher resolution ab initio bead models for the non-liganded hCSF-1RD1-D5; 15 independent runs with an average NSD value of 2.3 were structurally aligned and averaged with DAMAVER (Volkov and Svergun, 2003). X-ray scattering patterns from structural models were calculated using the program CRYSOL (Svergun et al., 1995). Constrained rigid-body refinement of the h/mCSF-1RD1-D3:h/mCSF-1 complexes was carried out in SASREF7 (Petoukhov and Svergun, 2005) with imposed P2 symmetry and specified CSF-1:CSF-1RD2 contacts. Constrained rigid-body refinement of the hCSF-1RD1-D5:hCSF-1 complex was carried out in SASREF7 with imposed P2 symmetry, specified CSF-1:CSF-1RD2 contacts and ambiguous contact distances for the D4-D4′ interface. Constrained rigid-body refinement of the non-liganded receptor was carried out in P1 symmetry and refinement convergence was optimal upon definition of ambiguous distance contacts at the D4-D4′ interface.
Isothermal Titration Calorimetry (ITC)
Calorimetric measurements were carried out using purified h/mCSF-1 and h/mCSF-1R samples dialyzed exhaustively against 20 mM Hepes pH 7.5, 150 mM NaCl. Experiments were carried out using a VP-ITC MicroCalorimeter at 310K, and data was analyzed using the Origin ITC analysis software package. Titrations were always preceded by an initial injection of 3 μL and were carried out using 10 μL injections applied 300 sec. apart, with continuous stirring. The data were fit to the “one binding site model”, and apparent molar reaction enthalpy (ΔH°), apparent entropy (ΔS°), association constant (Ka) and stoichiometry of binding (N) were determined. Several titrations were performed to evaluate reproducibility.
Supplementary Material
HIGHLIGHTS.
Assembly of extracellular human and mouse CSF-1:CSF-1R complexes
Thermodynamics of complex formation and characterization of species cross-reactivity
Bivalent binding of CSF-1 to CSF-1R ectodomains underlies complex formation
A mechanistic consensus for human and mouse CSF-1:CSF-1R complexes
Acknowledgments
JE, KV, BV, and KVC are research fellows of the Research Foundation Flanders, Belgium (FWO). AD is funded by a PhD fellowship of the Rhone-Alpes Region, France. AVS and DIS were supported by HFSP Research Grant Ref. RGP 55/2006. SNS is supported by the FWO (Projects 3G064307 and G059710) and Ghent University (BOF instrument). We thank the EMBL-Hamburg for synchrotron beamtime allocation at DESY (Hamburg, Germany) and Clement Blanchet for technical support at beamline X33. Access to these synchrotron facilities is supported by the European Commission under the 7th Framework Programme: Research Infrastructures, Grant Agreement Number 226716. JE would like to thank Rodrigo Gallardo for help during FFF-MALLS data collection and analysis. JE, NB, BR, AD, IG, AVS, DIS, BV, XW and BRB contributed to experimental design, performed research, analyzed data and contributed to the writing of the manuscript. KV and KVC provided experimental tools and assistance with tissue-culture and mammalian protein expression. SNS contributed to experiment design, analyzed data, directed the study, and contributed to the writing of the manuscript.
Footnotes
ACCESSION NUMBERS
The EM map for the 3D-reconstruction of the hCSF-1RD1-D5:hCSF-1 complex has been deposited in the EMDB under accession code EMD-1977.
Supplemental information includes two figures and two tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- Brooks BR, Brooks CL, 3rd, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang VT, Crispin M, Aricescu AR, Harvey DJ, Nettleship JE, Fennelly JA, Yu C, Boles KS, Evans EJ, Stuart DI, et al. Glycoprotein structural genomics: solving the glycosylation problem. Structure. 2007;15:267–273. doi: 10.1016/j.str.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liu H, Focia PJ, Shim AH, He X. Structure of macrophage colony stimulating factor bound to FMS: diverse signaling assemblies of class III receptor tyrosine kinases. Proc Natl Acad Sci U S A. 2008;105:18267–18272. doi: 10.1073/pnas.0807762105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara T, Suzu S, Hassan R, Chutiwitoonchai N, Hiyoshi M, Motoyoshi K, Kimura F, Okada S. IL-34 and M-CSF share the receptor Fms but are not identical in biological activity and signal activation. Cell Death Differ. 2010 doi: 10.1038/cdd.2010.60. [DOI] [PubMed] [Google Scholar]
- Chitu V, Stanley ER. Colony-stimulating factor-1 in immunity and inflammation. Curr Opin Immunol. 2006;18:39–48. doi: 10.1016/j.coi.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- Finger C, Escher C, Schneider D. The Single Transmembrane Domains of Human Receptor Tyrosine Kinases Encode Self-Interactions. Science Signaling. 2009;2 doi: 10.1126/scisignal.2000547. [DOI] [PubMed] [Google Scholar]
- Garceau V, Smith J, Paton IR, Davey M, Fares MA, Sester DP, Burt DW, Hume DA. Pivotal Advance: Avian colony-stimulating factor 1 (CSF-1), interleukin-34 (IL-34), and CSF-1 receptor genes and gene products. J Leukoc Biol. 2010;87:753–764. doi: 10.1189/jlb.0909624. [DOI] [PubMed] [Google Scholar]
- Graddis TJ, Brasel K, Friend D, Srinivasan S, Wee S, Lyman SD, March CJ, McGrew JT. Structure-function analysis of FLT3 ligand-FLT3 receptor interactions using a rapid functional screen. J Biol Chem. 1998;273:17626–17633. doi: 10.1074/jbc.273.28.17626. [DOI] [PubMed] [Google Scholar]
- Griffith J, Black J, Faerman C, Swenson L, Wynn M, Lu F, Lippke J, Saxena K. The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Mol Cell. 2004;13:169–178. doi: 10.1016/s1097-2765(03)00505-7. [DOI] [PubMed] [Google Scholar]
- Heymann JB, Cardone G, Winkler DC, Steven AC. Computational resources for cryo-electron tomography in Bsoft. J Struct Biol. 2008;161:232–242. doi: 10.1016/j.jsb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen JP, Saha N, Nikolov DB. Cell-cell signaling via Eph receptors and ephrins. Curr Opin Cell Biol. 2007;19:534–542. doi: 10.1016/j.ceb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Gurel O, Mendiaz EA, Stearns GW, Clogston CL, Lu HS, Osslund TD, Syed RS, Langley KE, Hendrickson WA. Structure of the active core of human stem cell factor and analysis of binding to its receptor kit. EMBO J. 2000;19:3192–3203. doi: 10.1093/emboj/19.13.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of Applied Crystallography. 2003;36:1277–1282. [Google Scholar]
- Lawrence MC, McKern NM, Ward CW. Insulin receptor structure and its implications for the IGF-1 receptor. Curr Opin Struct Biol. 2007;17:699–705. doi: 10.1016/j.sbi.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Pinchasi D, Zhou M, Lax I, Schlessinger J. Kit receptor dimerization is driven by bivalent binding of stem cell factor. J Biol Chem. 1997;272:6311–6317. doi: 10.1074/jbc.272.10.6311. [DOI] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppanen VM, Prota AE, Jeltsch M, Anisimov A, Kalkkinen N, Strandin T, Lankinen H, Goldman A, Ballmer-Hofer K, Alitalo K. Structural determinants of growth factor binding and specificity by VEGF receptor 2. Proc Natl Acad Sci U S A. 2010;107:2425–2430. doi: 10.1073/pnas.0914318107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S, Yarden Y, Givol D. A recombinant ectodomain of the receptor for the stem cell factor (SCF) retains ligand-induced receptor dimerization and antagonizes SCF-stimulated cellular responses. J Biol Chem. 1992;267:10866–10873. [PubMed] [Google Scholar]
- Li E, Hristova K. Role of receptor tyrosine kinase transmembrane domains in cell signaling and human pathologies. Biochemistry. 2006;45:6241–6251. doi: 10.1021/bi060609y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Stanley ER. Role of dimerization and modification of the CSF-1 receptor in its activation and internalization during the CSF-1 response. EMBO J. 1991;10:277–288. doi: 10.1002/j.1460-2075.1991.tb07948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Lee E, Hestir K, Leo C, Huang M, Bosch E, Halenbeck R, Wu G, Zhou A, Behrens D, et al. Discovery of a cytokine and its receptor by functional screening of the extracellular proteome. Science. 2008;320:807–811. doi: 10.1126/science.1154370. [DOI] [PubMed] [Google Scholar]
- Liu H, Chen X, Focia PJ, He X. Structural basis for stem cell factor-KIT signaling and activation of class III receptor tyrosine kinases. EMBO J. 2007;26:891–901. doi: 10.1038/sj.emboj.7601545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- Mi LZ, Lu C, Li Z, Nishida N, Walz T, Springer TA. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat Struct Mol Biol. 2011;18:984–989. doi: 10.1038/nsmb.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of Structural Biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- Oefner C, D’Arcy A, Winkler FK, Eggimann B, Hosang M. Crystal structure of human platelet-derived growth factor BB. EMBO J. 1992;11:3921–3926. doi: 10.1002/j.1460-2075.1992.tb05485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit J, Bohm A, Jancarik J, Halenbeck R, Koths K, Kim SH. Three-dimensional structure of dimeric human recombinant macrophage colony-stimulating factor. Science. 1992;258:1358–1362. doi: 10.1126/science.1455231. [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophysical Journal. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessle M, Round AR, Franke D, Moritz S, Huchler R, Fritsche M, Malthan D, Klaering R, Svergun DI. Automated sample-changing robot for solution scattering experiments at the EMBL Hamburg SAXS station X33. Journal of Applied Crystallography. 2008;41:913–917. doi: 10.1107/S0021889808021018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel MF, Downing JR, Rettenmier CW, Sherr CJ. A point mutation in the extracellular domain of the human CSF-1 receptor (c-fms proto-oncogene product) activates its transforming potential. Cell. 1988;55:979–988. doi: 10.1016/0092-8674(88)90243-7. [DOI] [PubMed] [Google Scholar]
- Ruch C, Skiniotis G, Steinmetz MO, Walz T, Ballmer-Hofer K. Structure of a VEGF-VEGF receptor complex determined by electron microscopy. Nat Struct Mol Biol. 2007;14:249–250. doi: 10.1038/nsmb1202. [DOI] [PubMed] [Google Scholar]
- Savvides SN, Boone T, Andrew Karplus P. Flt3 ligand structure and unexpected commonalities of helical bundles and cystine knots. Nat Struct Biol. 2000;7:486–491. doi: 10.1038/75896. [DOI] [PubMed] [Google Scholar]
- Schubert C, Schalk-Hihi C, Struble GT, Ma HC, Petrounia IP, Brandt B, Deckman IC, Patch RJ, Player MR, Spurlino JC, Springer BA. Crystal structure of the tyrosine kinase domain of colony-stimulating factor-1 receptor (cFMS) in complex with two inhibitors. J Biol Chem. 2007;282:4094–4101. doi: 10.1074/jbc.M608183200. [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Gao HX, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nature Protocols. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim AH, Liu H, Focia PJ, Chen X, Lin PC, He X. Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proc Natl Acad Sci U S A. 107:11307–11312. doi: 10.1073/pnas.1000806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim AH, Liu H, Focia PJ, Chen X, Lin PC, He X. Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex. Proc Natl Acad Sci U S A. 2010;107:11307–11312. doi: 10.1073/pnas.1000806107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebert X, Navaza J. UROX 2.0: an interactive tool for fitting atomic models into electron-microscopy reconstructions. Acta Crystallographica Section D-Biological Crystallography. 2009;65:651–658. doi: 10.1107/S0907444909008671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter PR, Minster NI, Kahn LE, Hood WF, Vickery LE, Thurman TL, Monahan JB, Welply JK, McKearn JP, Woulfe SL. Progenipoietins: biological characterization of a family of dual agonists of fetal liver tyrosine kinase-3 and the granulocyte colony-stimulating factor receptor. Exp Hematol. 2001;29:41–50. doi: 10.1016/s0301-472x(00)00616-0. [DOI] [PubMed] [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. Journal of Applied Crystallography. 1995;28:768–773. [Google Scholar]
- Svergun DI. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. Journal of Applied Crystallography. 1992;25:495–503. [Google Scholar]
- Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing (vol 76, pg 2879, 1999) Biophysical Journal. 1999;77:2896–2896. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MHJ. Determination of domain structure of proteins from X-ray solution scattering. Biophysical Journal. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- Verstraete K, Koch S, Ertugrul S, Vandenberghe I, Aerts M, Vandriessche G, Thiede C, Savvides SN. Efficient production of bioactive recombinant human Flt3 ligand in E. coli. Protein J. 2009;28:57–65. doi: 10.1007/s10930-009-9164-5. [DOI] [PubMed] [Google Scholar]
- Verstraete K, Vandriessche G, Januar M, Elegheert J, Shkumatov AV, Desfosses A, Van Craenenbroeck K, Svergun DI, Gutsche I, Vergauwen B, Savvides SN. Structural insights into the extracellular assembly of the hematopoietic Flt3 signaling complex. Blood. 2011;118:60–68. doi: 10.1182/blood-2011-01-329532. [DOI] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Lucet IS, Patel O, Broughton SE, Bamert R, Williams NK, Fantino E, Wilks AF, Rossjohn J. The 2.7 A crystal structure of the autoinhibited human c-Fms kinase domain. J Mol Biol. 2007;367:839–847. doi: 10.1016/j.jmb.2007.01.036. [DOI] [PubMed] [Google Scholar]
- Wei S, Nandi S, Chitu V, Yeung YG, Yu W, Huang M, Williams LT, Lin H, Stanley ER. Functional overlap but differential expression of CSF-1 and IL-34 in their CSF-1 receptor-mediated regulation of myeloid cells. J Leukoc Biol. 2010;88:495–505. doi: 10.1189/jlb.1209822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesmann C, Fuh G, Christinger HW, Eigenbrot C, Wells JA, de Vos AM. Crystal structure at 1.7 A resolution of VEGF in complex with domain 2 of the Flt-1 receptor. Cell. 1997;91:695–704. doi: 10.1016/s0092-8674(00)80456-0. [DOI] [PubMed] [Google Scholar]
- Wu X, Milne JL, Borgnia MJ, Rostapshov AV, Subramaniam S, Brooks BR. A core-weighted fitting method for docking atomic structures into low-resolution maps: application to cryo-electron microscopy. J Struct Biol. 2003;141:63–76. doi: 10.1016/s1047-8477(02)00570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu XW, Brooks BR. Self-guided Langevin dynamics simulation method. Chemical Physics Letters. 2003;381:512–518. [Google Scholar]
- Yang Y, Xie P, Opatowsky Y, Schlessinger J. Direct contacts between extracellular membrane-proximal domains are required for VEGF receptor activation and cell signaling. Proc Natl Acad Sci U S A. 2010;107:1906–1911. doi: 10.1073/pnas.0914052107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yuzawa S, Schlessinger J. Contacts between membrane proximal regions of the PDGF receptor ectodomain are required for receptor activation but not for receptor dimerization. Proc Natl Acad Sci U S A. 2008;105:7681–7686. doi: 10.1073/pnas.0802896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzawa S, Opatowsky Y, Zhang Z, Mandiyan V, Lax I, Schlessinger J. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell. 2007;130:323–334. doi: 10.1016/j.cell.2007.05.055. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Zhang R, Joachimiak A, Schlessinger J, Kong XP. Crystal structure of human stem cell factor: implication for stem cell factor receptor dimerization and activation. Proc Natl Acad Sci U S A. 2000;97:7732–7737. doi: 10.1073/pnas.97.14.7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.