Abstract
Sex chromosome dosage compensation in both eutherian and marsupial mammals is achieved by X chromosome inactivation (XCI)—transcriptional repression that silences one of the two X chromosomes in the somatic cells of females. We recently used RNA fluorescent in situ hybridization (FISH) to show, in individual nuclei, that marsupial X inactivation (in the absence of XIST) occurs on a gene-by-gene basis, and that escape from inactivation is stochastic and independent of gene location. In the absence of similar data from fibroblast cell lines of eutherian representatives, a meaningful comparison is lacking. We therefore used RNA-FISH to examine XCI in fibroblast cell lines obtained from three distantly related eutherian model species: African savannah elephant (Loxodonta africana), mouse (Mus musculus) and human (Homo sapiens). We show that, unlike the orthologous marsupial X, inactivation of the X conserved region (XCR) in eutherians generally is complete. Two-colour RNA-FISH on female human, mouse and elephant interphase nuclei showed that XCR loci have monoallelic expression in almost all nuclei. However, we found that many loci located in the evolutionarily distinct recently added region (XAR) displayed reproducible locus-specific frequencies of nuclei with either one or two active X alleles. We propose that marsupial XCI retains features of an ancient incomplete silencing mechanism that was augmented by the evolution of the XIST gene that progressively stabilized the eutherian XCR. In contrast, the recently added region of the eutherian X displays an incomplete inactivation profile similar to that observed on the evolutionarily distinct marsupial X and the independently evolved monotreme X chromosomes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00412-011-0343-8) contains supplementary material, which is available to authorized users.
Introduction
The mammalian X and Y chromosomes are morphologically and genetically differentiated. The X chromosome is large and gene rich, whereas the Y chromosome is highly degenerated and bears few functional genes. This chromosomal sexual dimorphism introduces disequilibrium of X-borne gene dose between females (with two Xs) and males (with just one X). In therian mammals, subclasses Eutheria (‘placental’ mammals) and Metatheria (marsupials), the imbalance is corrected by X chromosome inactivation (XCI), a paradigmatic epigenetic phenomenon in which two homologous X chromosomes are expressed differently within the same cell. The result is transcriptional silencing of one of the two X chromosomes in female somatic cells (Graves and Gartler 1986).
Most data on XCI have been gathered for humans and mice (which belong to the same superordinal eutherian clade). It was originally proposed that inactivation of one X occurs randomly early in embryogenesis, and is stable and somatically heritable (Lyon 1961). Expression of the non-coding regulatory RNA, Xist (X-inactive specific transcript), initiates XCI during early embryogenesis, although its expression is regulated differently between eutherian mammals (Okamoto et al. 2011). The accumulation and spread of Xist from an X chromosome inactivation centre (XIC) rapidly ensures chromosome-wide transcriptional silencing by triggering recruitment of inactive histone modifications and variants to the inactive X (Xi), and to DNA methylation sites on Xi (reviewed in Heard 2005). The transcriptional silencing is almost complete along the mouse X chromosome (Yang et al. 2010), and RNA-fluorescent in situ hybridization (FISH) for five loci showed monoallelic expression of these genes in the majority of cells (Chaumeil et al. 2006). In contrast, approximately 15% of genes are expressed from the inactive X in humans (Carrel and Willard 2005). Most of these genes are located on the short arm of the X, reflecting the different evolutionary history of the eutherian Xp (see below).
Comparative genomics (Veyrunes et al. 2008) has dated the origin of the therian sex chromosomes after the divergence of Prototheria (monotremes) from the therian ancestor (~166 million years ago), but before the therian radiation (~148 million years ago; dates from Bininda-Emonds et al. 2007). Comparative mapping between marsupials and eutherians reveals that the eutherian X consists of two evolutionary blocks (Graves 1995)—(1) an ancient conserved region (X conserved region (XCR)) that encompasses the entire long arm (Xq) and proximal short arm (Xp) of human X that is homologous to the marsupial X, and (2) a region recently added (XAR) to the sex chromosomes in the eutherian ancestor that is autosomal in marsupials. This latter region constitutes most of human Xp, where most human ‘escapee’ genes are located. Almost complete conservation of the gene content of the eutherian X (excluding the atypical mouse (Amar et al. 1988)), and even of gene order (Rodriguez Delgado et al. 2009; Raudsepp et al. 2004), suggests that there is strong selection against rearrangements on the X, consistent with the hypothesis that these would interfere with XCI (Ohno 1967).
Much less is known about the nature and molecular mechanisms of XCI outside of eutherian mammals. The marsupial XCI is paternally imprinted (Cooper et al. 1971; Sharman 1971) and appears to be incomplete, unstable and tissue specific (reviewed in Deakin et al. 2009). Like the eutherian inactive X, the marsupial X also becomes heterochromatic and exhibits marks of transcriptionally silent chromatin; there are, however, some fundamental epigenetic differences between the Xi in the two lineages (Chaumeil et al. 2011; Rens et al. 2010). Most interestingly, marsupial XCI occurs in the absence of an XIST homologue (Duret et al. 2006; Hore et al. 2007). Moreover, RNA-FISH recently demonstrated that the penetrance of XCI in marsupials is far from complete (Al Nadaf et al. 2010). Whereas X-borne genes are thought to be transcribed from one allele in human and mouse cells (with the exception of escapee genes), marsupial cells show reproducible locus-specific frequencies of nuclei that display either mono- or biallelic expression. This is similar to RNA-FISH results for X loci in monotremes (platypus; Deakin et al. 2008a), even though their sex chromosomes share no homology to those of therians (Veyrunes et al. 2008).
Despite the enormous amount of work done on XCI in mouse and human cells, it is not possible to make a direct comparison with the cell-specific data obtained from marsupials (Al Nadaf et al. 2010) and monotremes (Deakin et al. 2008a). Moreover, equivalent data are lacking from eutherians more distantly related to humans and mice. Using two-colour RNA-FISH, we examine XCI at the cellular level in mouse (Mus musculus) and human (Homo sapiens), and a representative basal eutherian mammal, the African savannah elephant (Loxodonta africana). We show that inactivation of genes on the ancient region (XCR) of the eutherian X is complete in almost all nuclei. However, many genes located in the evolutionarily distinct added region (XAR) showed reproducible frequencies of nuclei with either one or two active X alleles, a pattern similar to that of XCR genes in marsupials.
Materials and methods
Cell culture and coverslip preparation
Human cell lines were supplied courtesy of Dr. Jeff Craig, Murdoch Children's Research Institute, Australia. Male and female mouse and elephant fibroblasts cell lines were established from ear clips. All cell lines were cultured (in DMEM/10% foetal bovine serum) to a density of 70–80% on gelatine-coated coverslips at 37°C in 5% CO2. Coverslips were rinsed with phosphate-buffered saline (PBS) and then permeabilized for 10 min on ice in CSK buffer (100 mM NaCl, 300 mM sucrose, 10 mM PIPES pH 6.8)/0.5% Triton X 100 (Sigma)/2 mM vanadyl ribonucleoside complex (Sigma). Cells were subsequently fixed in 3% paraformaldehyde/1× PBS for 10 min at room temperature, and then washed twice for 5 min each in 70% ethanol, and stored at −20°C. Before hybridization, coverslips were dehydrated in 80%, 95% and 100% ethanol washes (3 min each) and air-dried.
Probes and probe preparation
Probes used in this study are large BAC clones containing the genes of interest. These genes are distributed along the length of the X chromosome of both the XCR and XAR (Online Resource 1 and 4). BACs bearing 12 human and mouse genes of interest (11 of which were in common) were identified using BAC tracks on the Ensembl or UCSC genome browsers (mouse RPCI-23 and RPCI-24, human RP-11; BAC PAC Resources, CHORI). For elephant, 24 X borne BAC clones (representing 16 X-specific loci; some BACs overlapped; Rodriguez Delgado et al. 2009), isolated from a male-derived BAC library (VMRC-15; BAC PAC Resources, CHORI), were used as probes. However, BACs containing five genes (AMELX, AR, PLP1, RBMX and SOX3) produced no RNA-FISH signals. Results are only presented for the 11 expressed loci. Of these 11 elephant markers, six were in common with both human and mouse (Table 1). For the other five elephant-specific markers, RNA-FISH was not conducted in human and mouse because they were close to loci already included in this study. One to two micrograms of BAC DNA was prepared (as described in Veyrunes et al. 2008) and labelled by nick translation reaction with SpectrumOrange dUTP or SpectrumGreen dUTP (Enzo life Sciences, New York, NY, USA).
Table 1.
Quantitative analysis of female fibroblast RNA-FISH data
| Gene name | Percent nuclei with 1 signal | ||
|---|---|---|---|
| HSA | LAF | MMU | |
| STS | 59 | – | – |
| EIF1AX | 74 | 25 | 96 |
| ZFX | – | 19 | – |
| GK | – | 58 | – |
| USP9X | 69 | 24 | 91 |
| AMELX | NE | NE | 92 |
| POLA1 | NE | – | 68 |
| UBA1 | 93 | 76 | 97 |
| KDM5C | 65 | 55 | 94 |
| HUWE1 | 96 | 97 | 84 |
| ATRX | 98 | 90 | 62 |
| HPRT | – | – | 94 |
| PHF6 | 99 | – | 90 |
| RBMX | 93 | NE | 89 |
| XIAP | – | 92 | – |
| FMR1 | – | 82 | – |
| G6PD | – | 92 | – |
| MECP2 | 90 | – | 91 |
Frequency of nuclei with a single signal for X-borne loci investigated in this study. At least 100 female nuclei were scored (with two signals for the control autosomal gene) for each locus. All data are collected from one cell line in each species. XCR genes are rendered in bold italic
NE no expression in fibroblasts, ‘–’ no data, HSA Homo sapiens, LAF Loxodonta africana, MMU Mus musculus
RNA fluorescence in situ hybridization (FISH)
In all instances, 1 μg of labelled test gene DNA was co-precipitated with 1 μg of the control gene DNA, and RNA-FISH was performed as described in Al Nadaf et al. (2010). Images were captured on a SPOT RT Monochrome charge-coupled device camera (Diagnostic Instruments Inc., Sterling Heights, MI, USA) and analysed in IP lab (Scanalytics Inc., Fairfax, VA, USA).
Hybridization efficiencies for the X-linked test genes were determined by the frequency of male nuclei with one signal. Using this efficiency measure, the binomial p 2 + 2pq + q 2 = 1 was used to determine the expected frequency of female nuclei that have one signal due to inefficient hybridization.
Results
We tested inactivation status of X-borne loci in fibroblasts cultured from three eutherian species (human, mouse and elephant) using RNA-FISH (Fig. 1). The probes used in this study are derived from genes distributed along the length of the X chromosome of both the XCR and XAR (Fig. 2 and Online Resource 1). To control for probe accessibility and cell polyploidy, two-colour RNA-FISH was utilized; two probes, one containing the X-borne gene of interest, and the second containing the autosomal housekeeping gene, GBA, were labelled with different colour fluorochromes, in human and mouse. A pseudoautosomal region gene (XG) was used for elephant cell lines (Fig. 1 and Online Resource 1). Both alleles of the control gene needed to be detected before the cell was included in the analysis. At least 100 diploid nuclei were scored for each test gene (Online Resource 2). We used these data to create an XCI activity map for each species (Fig. 2).
Fig. 1.
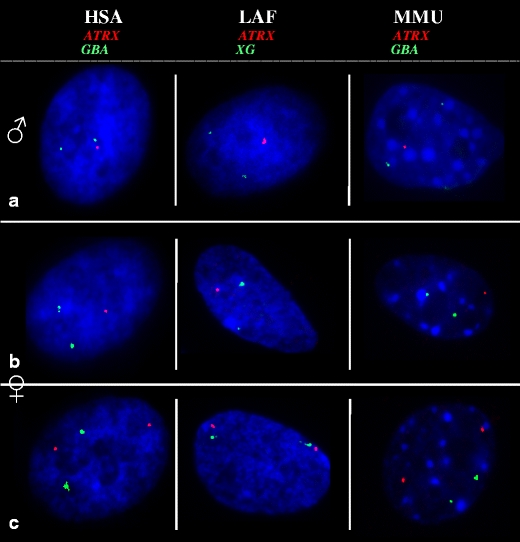
RNA-FISH of an X-borne gene (red) and an autosomal control (green) in male and female fibroblasts of three eutherian species. Transcription from the two autosomal alleles (green) is detected in all nuclei. a Male fibroblast nuclei show transcription from the single X-borne allele, whereas female fibroblast nuclei show transcription from either one (b) or two (c) X-borne ATRX alleles. Nuclei are counterstained with DAPI (blue). HSA Homo sapiens, LAF Loxodonta africana, MMU Mus musculus
Fig. 2.
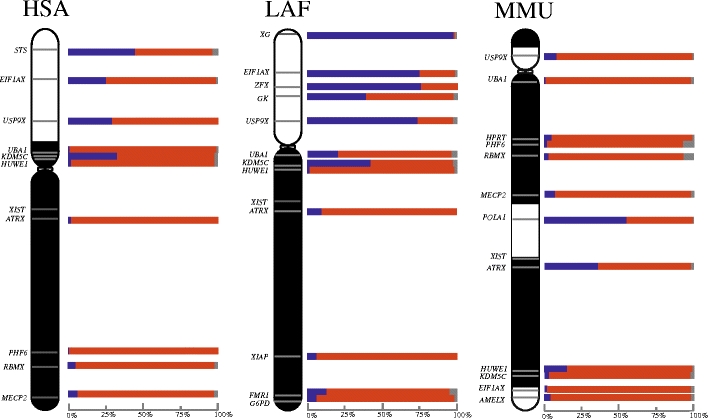
Activity maps of the human (HSA), elephant (LAF) and mouse (MMU) X chromosomes. Bars represent percentage of nuclei in which the X-borne test loci are transcribing from two (blue), one (red) or zero (grey) alleles. X conserved region is indicate in black. X added region is indicated in white
Expression from X chromosomes
Male interphase cells (XY) were used to test the efficiency and specificity of probe hybridization (a single signal is expected for an X-borne gene; Fig. 1a and Online Resource 2). Loci with no X-gene expression in male nuclei were probably not expressed in fibroblasts and were eliminated from the analysis (Online Resource 2).
In female cells (XX), nuclei expressing a gene from only one of the two X chromosomes (1X-active) were observed as a single signal, whereas cells expressing a gene from both X chromosomes (2X-active) were observed as two signals (Fig. 1b, c). Our RNA-FISH results identified a number of genes in all three species that displayed complete inactivation (one signal in >90% female nuclei, comparable to the frequency in male cells; Table 1 and Online Resource 2). Loci with two signals in more than 10% of cells in the population were considered to escape inactivation to varying extents (13–81% for different loci). Importantly, none of the loci showed two signals in all nuclei (complete escape from inactivation; Table 1 and Online Resource 2).
Activity maps of eutherian inactive X chromosomes
We constructed an activity map (Fig. 2) for the inactive X of each species in order to determine whether escape from inactivation differed between the evolutionary blocks of the X chromosome, and whether there was any polarity to the frequency of escape (Fig. 2). Genes escaping X inactivation showed an obvious clustering within the XAR of human and elephant (but not mouse), with 29% to 84% nuclei being 2X-active. Conversely, most genes located in the XCR region in all species (except ATRX, FMR1, KDM5C and UBA1 in at least one of the three species) appear to be subject to X inactivation, with 2X-active expression in less than 10% of nuclei. Three of the four XCR escapee genes have Y homologues in humans, mice or marsupials (FMR1 only escapes in elephant, for which there is no Y data). ATRX (with a Y homologue in marsupials) was expressed in mouse fibroblast nuclei from both X chromosomes in 36% of cells. KDM5C, with a Y homologue in most therian species and which escapes inactivation in human and elephant, but not mouse, was expressed from both X chromosomes at frequencies of 33%, 43% and 3%, respectively. UBA1, with a Y homologue in mouse and marsupial but not human, was expressed from both X chromosomes in 21% of elephant, but only in 5% and 1% of human and mouse fibroblast nuclei.
In contrast, all genes within the XAR showed incomplete inactivation to a various extent in human and elephant, but not in mouse. The frequency of nuclei showing inactivation ranged from 48% to 71% (52–29% escape) for human, and 25–60% (75–40% escape) for elephant (Table 1 and Online Resource 2). These ‘escapee’ genes have a reproducible (experimental and biological replicates) locus-specific frequency of nuclei with either one or two active alleles (Online Resource 3). In mouse, however, most XAR genes are fully inactivated (Fig. 2). For instance, E1FAX and USP9X, which escape inactivation in human and elephant, are subject to XCI in mouse.
Discussion
Dosage compensation by means of XCI occurs in all therian mammals, and many of the molecular mechanisms involved in XCI are conserved between human and mouse. Considerable differences exist, however, in the initiation of XCI during early embryogenesis among eutherian mammals (Okamoto et al. 2011), with a different percentage of genes escaping from XCI in mouse (3%) and human (15%) (Carrel et al. 1999; Carrel and Willard 2005; Sudbrak et al. 2001; Yang et al. 2010).
Given that data on escapee genes outside of the rodent and primate lineages are scarce, we studied the expression of loci distributed on the X chromosomes in three eutherian species (human, mouse and elephant). We conducted RNA-FISH experiments on fibroblasts of both sexes, similar to those performed on representative marsupial (the tammar wallaby) and monotreme (the platypus) species (Al Nadaf et al. 2010; Deakin et al. 2008a), to address whether escape from inactivation is due to expression from the inactive X in a proportion of cells or by expression of alleles on Xi in all cells.
Our data show that most genes on the ancient part of the X are completely inactivated (1X-active), but loci on the recently added region showed stochastic escape (2X-active) in a proportion of cells.
Monoallelic expression on XCR and biallelic expression on XAR
Genes subject to XCI were mostly located on the conserved region of the X chromosome (XCR) in each of the three species (Table 1), consistent with previous findings in humans and mice (Carrel and Willard 2005; Chaumeil et al. 2006; Yang et al. 2010). This confirms that X inactivation is complete for most loci on this ancient region of the eutherian X, and is different from the pattern of expression observed for orthologous loci on the marsupial X (Al Nadaf et al. 2010). The three XCR genes that were the exception were ATRX, KDM5C and UBA1. They showed considerable expression from both loci (>20%) in one or more of the eutherian species tested. These loci all have Y homologues in humans, mice or marsupials: KDM5C has an active Y homologue in eutherians (Agulnik et al. 1994) and also a copy on the marsupial Y (Waters et al. 2001). There is a UBA1 Y homologue in mouse and marsupials (Mitchell et al. 1998, 1992), and ATRX has a Y homologue in marsupials (Pask et al. 2000). However, having a Y copy does not necessitate escape. For instance, XCR genes, which also have a Y copy in either eutherians or marsupials (such as PHF6, HUWE1, MECP2 and RBMX), do not escape with high frequencies. The XCR findings contrasted sharply with those for genes on the XAR, which are autosomal in marsupials, and were 2X-active in many nuclei—a pattern indicative of escape from inactivation (Fig. 2).
Our human and mouse RNA-FISH results are in broad agreement with expression data (microarray and RNA-seq) from populations of cells and from cell hybrids (Carrel et al. 1999; Carrel and Willard 2005; Sudbrak et al. 2001; Yang et al. 2010), although there were some inconsistencies. In mouse, ATRX displayed biallelic expression in 40% of nuclei; this contrasts with previous findings that ATRX is subject to XCI in mouse (Yang et al. 2010). This could be due to the tissue-specific escape of ATRX from XCI in mice (Garrick et al. 2006; Patrat et al. 2009). Moreover, we detected only 7% biallelic expression of UBA1 in human fibroblast nuclei which was previously reported to escape inactivation in human (Carrel and Willard 2005), but not mouse (Yang et al. 2010). Finally, in contrast to an earlier study (Yang et al. 2010), we did not detect biallelic expression of Kdm5c in mouse. This is unlikely to be due to a low efficiency of hybridization (measure as 98% in controls) and is probably due to the tissue and strain specificity of Kdm5c escape from XCI in mice (Carrel et al. 1996; Sheardown et al. 1996).
Expression from the inactive X in different eutherian species
It is noteworthy that escape from inactivation appears to be much more common in cells derived from human and elephant than from cells of the mouse (Fig. 2). This is consistent with previous observations (Yang et al. 2010) where few genes escape inactivation on the mouse X. This difference in frequencies of escape from inactivation might explain the less severe phenotype in XO mice (Ashworth et al. 1991) than XO humans (with Turner's syndrome).
Experimental and biological replicates indicate that all but one gene in this study escape X inactivation at a reproducible frequency. The exception to this was the variable expression of USP9X in human, which showed almost complete inactivation in one cell line (91%), and some escape from inactivation in two others (69% and 70%). Variable XCI status in approximately 10% of X-linked genes in females has also been identified (Carrel and Willard 2005). Our study was performed on fibroblast cell lines, and since the extent of escape of individual loci often varies between different marsupial tissues (Cooper 1971), replicating it using cells from other tissue sources is a priority.
Gene content (and even gene order) on the eutherian X chromosome is conserved, representing the largest conserved block in the eutherian genome. This probably reflects selection against rearrangements that could disrupt the regulation of the XCI system. Mouse is a notable exception to this in that the ancient and added regions of the X have been rearranged—perhaps facilitating the spread of tight repressive control into the XAR in this species. The marsupial X, however, has been significantly rearranged between the opossum and wallaby (Deakin et al. 2008b), which may reflect an incomplete dosage compensation system (Al Nadaf et al. 2010; Chaumeil et al. 2011).
Stochastic expression of genes on the X
Our observation of the frequencies of human, elephant and mouse nuclei with 1X-active and 2X-active expression patterns implies that escape from inactivation is a stochastic (probabilistic) process in eutherians (Fig. 2). Different genes show reproducible frequency (between experimental replicates and among individuals) of 2X-active and 1X-active nuclei in a population of female fibroblast cells. Thus, escape from inactivation of a locus on the eutherian XAR is controlled by the probability of its expression, rather than its reduced expression from the inactive X in all nuclei.
A strikingly similar conclusion was drawn from data describing the inactivation of loci on the marsupial X (genes on the marsupial X chromosome were expressed from one or both alleles at a frequency characteristic of each locus; Al Nadaf et al. 2010). However, genes that escape inactivation on the marsupial X lie on the ancient conserved region of the eutherian X which, with the exception of few genes, are subject to inactivation (see above). Even more remarkable, the pattern of inactivation on the eutherian XAR and the marsupial XCR is similar to inactivation on the monotreme X chromosomes (Deakin et al. 2008a)—a chromosome that shares no homology with marsupial or eutherian sex chromosomes. Thus, we have observed almost identical inactivation patterns on the independently evolved (evolutionarily distinct) regions of mammalian sex chromosomes.
Evolution of X chromosome inactivation
We propose, therefore, that the stochastic inactivation that characterizes XCR genes in marsupials, and XAR genes in eutherians, represents the original mechanism of X inactivation in the common ancestor of marsupials and eutherians (i.e. ~148 million years ago). The major difference in inactivation phenotype, and in the molecular mechanism of XCI in marsupials and eutherians (Rens et al. 2010; Chaumeil et al. 2011), suggests an ongoing sophistication of the XCI machinery on the eutherian XCR that coincided with the rise of XIST and the XIC.
This ancient mechanism was subsequently maintained in the marsupial and early eutherian lineages, but at some point before the eutherian radiation (~105 million years ago), XCI became more tightly regulated with the evolution of the XIST gene. This tighter regulation spread until it was virtually complete on the ancient eutherian X. Following the addition of XAR ~148–105 million years ago, the genes in this region came under stochastic regulation, similar to that observed on the marsupial X. In the mouse where XCR and XAR are intermixed, recruitment of the added region into tighter XIST-controlled regulation is virtually complete. In human and elephant (and presumably all structurally similar eutherian X chromosomes), however, recruitment of XAR into tight X inactivation remains incomplete.
Random monoallelic expression has been described in eutherians (Singh et al. 2003) for many autosomal loci and could be an ancient mechanism of transcriptional control (Ohlsson et al. 2001). This monoallelic expression is similar to the stochastic inactivation we observed for genes on the evolutionary distinct regions of the therian sex chromosomes (the eutherian XAR and marsupial XCR) and the independently evolved monotreme sex chromosomes. Therefore, incomplete stochastic inactivation is likely common to the initial recruitment of genes into mammalian dosage compensation systems which, in turn, may have been exapted independently (in monotreme and therian mammals) from autosomal monoallelic expression.
Electronic supplementary materials
Below is the link to the electronic supplementary material.
BACs used in this study and the genes they bear (DOC 78 kb)
Quantitative analysis of female fibroblast RNA-FISH data. Frequency of nuclei with two, one or zero signals for each X-borne locus. At least 100 nuclei were scored. Only nuclei with two signals for the autosomal control gene, and therefore diploid, were scored. Male efficiencies were used to calculate the expected frequency of nuclei with two signals, one signal and no signal for each test gene. Expected frequencies of cells with two signals were in excess of 90% for all loci, and observed frequencies were significantly different from these expected frequencies in every case (Chi-square test with 2 degrees of freedom). All data in human and mouse are collected from one cell line and in elephant from two cell lines (DOC 86 kb)
RNA-FISH results for two additional female mouse and human cell lines and one additional elephant female cell line (DOC 59 kb)
Details of BACs used in this study (DOC 49 kb)
Acknowledgements
This project was supported by an Australian Research Council discovery project (DP1094868) to PDW, JED and JMG. PDW is supported by an Australian Research Fellowship (DP0987091). TJT’s research is funded by the South African National Research Foundation. We thank Dr. Ruth Arkell and Dr. Jeff Craig for graciously providing mouse and human cell lines.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Agulnik AI, Mitchell MJ, Mattei MG, Borsani G, Avner PA, Lerner JL, Bishop CE. A novel X gene with a widely transcribed Y-linked homologue escapes X-inactivation in mouse and human. Hum Mol Genet. 1994;3(6):879–884. doi: 10.1093/hmg/3.6.879. [DOI] [PubMed] [Google Scholar]
- Al Nadaf S, Waters PD, Koina E, Deakin JE, Jordan KS, Graves JA. Activity map of the tammar X chromosome shows that marsupial X inactivation is incomplete and escape is stochastic. Genome Biol. 2010;11(12):R122. doi: 10.1186/gb-2010-11-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amar LC, Dandolo L, Hanauer A, Cook AR, Arnaud D, Mandel JL, Avner P. Conservation and reorganization of loci on the mammalian X chromosome: a molecular framework for the identification of homologous subchromosomal regions in man and mouse. Genomics. 1988;2(3):220–230. doi: 10.1016/0888-7543(88)90006-7. [DOI] [PubMed] [Google Scholar]
- Ashworth A, Rastan S, Lovell-Badge R, Kay G. X-chromosome inactivation may explain the difference in viability of XO humans and mice. Nature. 1991;351(6325):406–408. doi: 10.1038/351406a0. [DOI] [PubMed] [Google Scholar]
- Bininda-Emonds OR, Cardillo M, Jones KE, MacPhee RD, Beck RM, Grenyer R, Price SA, Vos RA, Gittleman JL, Purvis A. The delayed rise of present-day mammals. Nature. 2007;446(7135):507–512. doi: 10.1038/nature05634. [DOI] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031):400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Carrel L, Hunt PA, Willard HF. Tissue and lineage-specific variation in inactive X chromosome expression of the murine Smcx gene. Hum Mol Genet. 1996;5(9):1361–1366. doi: 10.1093/hmg/5.9.1361. [DOI] [PubMed] [Google Scholar]
- Carrel L, Cottle AA, Goglin KC, Willard HF. A first-generation X-inactivation profile of the human X chromosome. Proc Natl Acad Sci USA. 1999;96:14440–14444. doi: 10.1073/pnas.96.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20(16):2223–2237. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Waters PD, Koina E, Gilbert C, Robinson TJ, Graves JA. Evolution from XIST-independent to XIST-controlled X-chromosome inactivation: epigenetic modifications in distantly related mammals. PLoS One. 2011;6(4):e19040. doi: 10.1371/journal.pone.0019040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DW. Directed genetic change model for X chromosome inactivation in eutherian mammals. Nature. 1971;230(5292):292–294. doi: 10.1038/230292a0. [DOI] [PubMed] [Google Scholar]
- Cooper DW, VandeBerg JL, Sharman GB, Poole WE. Phosphoglycerate kinase polymorphism in kangaroos provides further evidence for paternal X inactivation. Nat New Biol. 1971;230(13):155–157. doi: 10.1038/newbio230155a0. [DOI] [PubMed] [Google Scholar]
- Deakin JE, Hore TA, Koina E, Marshall Graves JA. The status of dosage compensation in the multiple X chromosomes of the platypus. PLoS Genet. 2008;4(7):e1000140. doi: 10.1371/journal.pgen.1000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Koina E, Waters PD, Doherty R, Patel VS, Delbridge ML, Dobson B, Fong J, Hu Y, van den Hurk C, Pask AJ, Shaw G, Smith C, Thompson K, Wakefield MJ, Yu H, Renfree MB, Graves JA. Physical map of two tammar wallaby chromosomes: a strategy for mapping in non-model mammals. Chromosome Res. 2008;16(8):1159–1175. doi: 10.1007/s10577-008-1266-y. [DOI] [PubMed] [Google Scholar]
- Deakin JE, Chaumeil J, Hore TA, Marshall Graves JA. Unravelling the evolutionary origins of X chromosome inactivation in mammals: insights from marsupials and monotremes. Chromosome Res. 2009;17(5):671–685. doi: 10.1007/s10577-009-9058-6. [DOI] [PubMed] [Google Scholar]
- Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312(5780):1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- Garrick D, Sharpe JA, Arkell R, Dobbie L, Smith AJ, Wood WG, Higgs DR, Gibbons RJ. Loss of Atrx affects trophoblast development and the pattern of X-inactivation in extraembryonic tissues. PLoS Genet. 2006;2(4):e58. doi: 10.1371/journal.pgen.0020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JAM. The evolution of mammalian sex chromosomes and the origin of sex determining genes. Philos Trans R Soc Lond B Biol Sci. 1995;350(1333):305–311. doi: 10.1098/rstb.1995.0166. [DOI] [PubMed] [Google Scholar]
- Graves JA, Gartler SM. Mammalian X chromosome inactivation: testing the hypothesis of transcriptional control. Somat Cell Mol Genet. 1986;12(3):275–280. doi: 10.1007/BF01570786. [DOI] [PubMed] [Google Scholar]
- Heard E. Delving into the diversity of facultative heterochromatin: the epigenetics of the inactive X chromosome. Curr Opin Genet Dev. 2005;15(5):482–489. doi: 10.1016/j.gde.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Hore TA, Koina E, Wakefield MJ, Marshall Graves JA. The region homologous to the X-chromosome inactivation centre has been disrupted in marsupial and monotreme mammals. Chromosome Res. 2007;15(2):147–161. doi: 10.1007/s10577-007-1119-0. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Mitchell MJ, Woods DR, Wilcox SA, Graves JA, Bishop CE. Marsupial Y chromosome encodes a homologue of the mouse Y-linked candidate spermatogenesis gene Ube1y. Nature. 1992;359(6395):528–531. doi: 10.1038/359528a0. [DOI] [PubMed] [Google Scholar]
- Mitchell MJ, Wilcox SA, Watson JM, Lerner JL, Woods DR, Scheffler J, Hearn JP, Bishop CE, Graves JAM. The origin and loss of the ubiquitin activating enzyme gene on the mammalian Y chromosome. Hum Mol Genet. 1998;7:429–434. doi: 10.1093/hmg/7.3.429. [DOI] [PubMed] [Google Scholar]
- Ohlsson R, Paldi A, Graves JA. Did genomic imprinting and X chromosome inactivation arise from stochastic expression? Trends Genet. 2001;17(3):136–141. doi: 10.1016/S0168-9525(00)02211-3. [DOI] [PubMed] [Google Scholar]
- Ohno S. Sex chromosomes and sex-linked genes. New York: Springer; 1967. [Google Scholar]
- Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf JP, Renard JP, Duranthon V, Heard E. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472(7343):370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- Pask A, Renfree MB, Marshall Graves JA. The human sex-reversing ATRX gene has a homologue on the marsupial Y chromosome, ATRY: implications for the evolution of mammalian sex determination. Proc Natl Acad Sci USA. 2000;97(24):13198–13202. doi: 10.1073/pnas.230424497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrat C, Okamoto I, Diabangouaya P, Vialon V, Le Baccon P, Chow J, Heard E. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc Natl Acad Sci USA. 2009;106(13):5198–5203. doi: 10.1073/pnas.0810683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudsepp T, Lee EJ, Kata SR, Brinkmeyer C, Mickelson JR, Skow LC, Womack JE, Chowdhary BP. Exceptional conservation of horse-human gene order on X chromosome revealed by high-resolution radiation hybrid mapping. Proc Natl Acad Sci USA. 2004;101(8):2386–2391. doi: 10.1073/pnas.0308513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rens W, Wallduck MS, Lovell FL, Ferguson-Smith MA, Ferguson-Smith AC. Epigenetic modifications on X chromosomes in marsupial and monotreme mammals and implications for evolution of dosage compensation. Proc Natl Acad Sci USA. 2010;107(41):17657–17662. doi: 10.1073/pnas.0910322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez Delgado CL, Waters PD, Gilbert C, Robinson TJ, Graves JA. Physical mapping of the elephant X chromosome: conservation of gene order over 105 million years. Chromosome Res. 2009;17(7):917–926. doi: 10.1007/s10577-009-9079-1. [DOI] [PubMed] [Google Scholar]
- Sharman GB. Late DNA replication in the paternally derived X chromosome of female kangaroos. Nature. 1971;230(5291):231–232. doi: 10.1038/230231a0. [DOI] [PubMed] [Google Scholar]
- Sheardown S, Norris D, Fisher A, Brockdorff N. The mouse Smcx gene exhibits developmental and tissue specific variation in degree of escape from X inactivation. Hum Mol Genet. 1996;5(9):1355–1360. doi: 10.1093/hmg/5.9.1355. [DOI] [PubMed] [Google Scholar]
- Singh N, Ebrahimi FA, Gimelbrant AA, Ensminger AW, Tackett MR, Qi P, Gribnau J, Chess A. Coordination of the random asynchronous replication of autosomal loci. Nat Genet. 2003;33(3):339–341. doi: 10.1038/ng1102. [DOI] [PubMed] [Google Scholar]
- Sudbrak R, Wieczorek G, Nuber UA, Mann W, Kirchner R, Erdogan F, Brown CJ, Wohrle D, Sterk P, Kalscheuer VM, Berger W, Lehrach H, Ropers HH. X chromosome-specific cDNA arrays: identification of genes that escape from X-inactivation and other applications. Hum Mol Genet. 2001;10(1):77–83. doi: 10.1093/hmg/10.1.77. [DOI] [PubMed] [Google Scholar]
- Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, Grützner F, Deakin JE, Whittington CM, Schatzkamer K, Kremitzki CL, Graves T, Ferguson-Smith MA, Warren W, Graves JAM. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008;18(6):965–973. doi: 10.1101/gr.7101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters PD, Duffy B, Frost CJ, Delbridge ML, Graves JA. The human Y chromosome derives largely from a single autosomal region added to the sex chromosomes 80–130 million years ago. Cytogenet Cell Genet. 2001;92(1–2):74–79. doi: 10.1159/000056872. [DOI] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20(5):614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BACs used in this study and the genes they bear (DOC 78 kb)
Quantitative analysis of female fibroblast RNA-FISH data. Frequency of nuclei with two, one or zero signals for each X-borne locus. At least 100 nuclei were scored. Only nuclei with two signals for the autosomal control gene, and therefore diploid, were scored. Male efficiencies were used to calculate the expected frequency of nuclei with two signals, one signal and no signal for each test gene. Expected frequencies of cells with two signals were in excess of 90% for all loci, and observed frequencies were significantly different from these expected frequencies in every case (Chi-square test with 2 degrees of freedom). All data in human and mouse are collected from one cell line and in elephant from two cell lines (DOC 86 kb)
RNA-FISH results for two additional female mouse and human cell lines and one additional elephant female cell line (DOC 59 kb)
Details of BACs used in this study (DOC 49 kb)


