Abstract
Background
After age, gender is the most important risk factor for coronary artery disease (CAD). The mechanism through which women are protected from CAD is still largely unknown, but the observed gender difference suggests the involvement of the reproductive steroid hormone signaling system. Genetic association studies of the gene encoding Estrogen Receptor alpha (ESR1) have shown conflicting results, although only a limited range of variation in the gene has been investigated.
Methods and Results
We exploited information made available by advanced new methods and resources in complex disease genetics to revisit the question of ESR1's role in risk of CAD. We performed a meta-analysis of 14 genome-wide association studies (CARDIoGRAM discovery analysis, N~87,000) to search for population-wide and gender-specific associations between CAD risk and common genetic variants throughout the coding, non-coding and flanking regions of ESR1. In additional samples from the MIGen (N~6,000), WTCCC (N~7,400) and Framingham (N~3,700) studies, we extended this search to a larger number of common and uncommon variants by imputation into a panel of haplotypes constructed using data from the 1000 Genomes project. Despite the widespread expression of ER alpha in vascular tissues, we find no evidence for involvement of common or low-frequency genetic variation throughout the ESR1 gene in modifying risk of CAD, either in the general population or as a function of gender.
Conclusions
We suggest that future research on the genetic basis of gender-related differences in CAD risk should initially prioritize other genes in the reproductive steroid hormone biosynthesis system.
Keywords: coronary artery disease; estrogen receptor alpha; menopause; polymorphism, single nucleotide; genetic association studies; meta-analysis
Introduction
After age, gender is the most important risk factor for coronary artery disease (CAD), with women aged 35–74 years having two to three times lower myocardial infarction (MI) incidence than age-matched men1. The mechanism through which women are protected from MI/CAD is still largely unknown, but the observed gender difference and the fact that CAD risk in postmenopausal women approaches that of males suggests the involvement of the sex steroid hormone system. This hypothesis was initially supported by the results of observational studies that showed lower CAD risk among post-menopausal women undergoing hormone replacement therapy (HRT)2,3. However, initial clinical trials of HRT have shown unexpected negative results4,5, even unanticipated harm, although the timing of initiation of therapy may explain these conflicting results6,7,8.
The fact that CAD clusters in families9 (estimated heritability 38–57%10) coupled with the observation of gender- and menopause-related differences in risk suggests that inter-individual variation in CAD risk may be partly mediated by population-level genetic variation in the genes that encode elements of the sex steroid hormone system. ERα is an important signaling gateway within this system, and is expressed in multiple cardiovascular tissues in both males and females11. The gene encoding ERα, ESR1, has been the subject of several candidate gene association studies in relation to CAD over the past decade with generally inconsistent results12,13,14. However, only a very limited range of the genetic variation in ESR1 has been investigated and the role of this gene in CAD risk remains to be clarified.
The last 5–7 years have seen a paradigm shift in our approach to investigating the genetic basis of complex diseases. Advanced new methods, including high-throughput genotyping15, genome-wide association studies (GWAS)16, genotype imputation17, second generation sequencing (e.g. ref 18), along with the availability of resources describing natural human genetic variation (e.g. HapMap19, 1000 Genomes Project20) allow us to explore the effect of genetic variation on phenotype more thoroughly. Also important is the manner and volume in which raw genetic data are now generated and disseminated under a model of cross-study cooperation and public data deposition, which has been key to overcoming many of the problems that limited the success of candidate gene association studies for complex diseases. While no genome-wide significant evidence for the involvement of ESR1 variation in CAD risk has been reported in recent GWAS, data from these studies may still provide important information either to support or refute this hypothesis. The fact that many robust new GWAS loci for complex diseases had previously been investigated as candidate genes (e.g. LDLR in CAD21 and several recently confirmed loci for LDL, HDL and triglycerides22) highlights the importance of revisiting the role of candidate genes in complex diseases23.
Therefore, in this paper we bring these powerful post-genomic methods and resources to bear on a classical CAD candidate gene in order to resolve a long-running unanswered question in cardiovascular genetics. For common variation in a genomic region centered on ESR1, we report the results of a large meta-analysis of GWAS of MI and CAD, and explore possible gender-specific differences. We also investigate the effect on CAD risk of low-frequency variation in this region.
Materials and Methods
Coronary Artery Disease GWAS meta-analysis
The Coronary ARtery DIsease Genome-wide Replication And Meta-analysis (CARDIoGRAM) Consortium was formed with the purpose of identifying novel susceptibility loci for CAD. Briefly, the CARDIoGRAM discovery analysis combined data from 14 published and unpublished primary GWAS, in individuals of European ancestry, including 22,233 (30.9% of which were females) cases with CAD (stable or unstable coronary events) and 64,762 controls21 (58.1% of which were females).
Each primary GWAS performed a logistic regression analysis to test for association between genotyped and imputed (using the HapMap Phase II reference panel19) SNPs and risk of CAD under an additive disease model adjusted for age and sex (see Supplementary Methods for a more detailed summary of the genotyping and quality control methods used).
In this study, we meta-analyzed these study-level results using inverse-variance weighting under a fixed effects model. We performed a random effects meta-analysis for SNPs with significant between-study heterogeneity (p-heterogeneity <0.01), on the basis of Cochran’s Q-statistic. These analyses were carried out for each of 535 SNPs in a genomic region containing the entire coding and non-coding region of ESR1 (see Supplementary Table 1) and a 50kb region upstream and downstream of the gene (~547kb; Chr6: 151927808-152474406, GRCh37.p1).
Gender-stratified analysis
An equivalent analysis to that described above was performed separately for females and males in all 13 of the 14 contributing studies (CHARGE data not available), and the results were meta-analyzed in a similar way. We also formally tested for interaction between each SNP and gender by using the gender-specific effects and variances within each study to estimate those of the SNP-gender interaction term (Supplementary Methods). We then meta-analyzed the results as described for the un-stratified analysis.
Fine mapping analysis
Public GWAS data sources
To perform fine mapping studies in the region of interest, we used publicly available genotype and phenotype data from three large published GWAS: a) The Myocardial Infarction Genetics Consortium (MIGen24) is a case-control GWAS consisting of 2,967 cases of early-onset MI and 3,075 age- and sex-matched controls from six international sites in the US and Europe; b) The Wellcome Trust Case Control Consortium (WTCCC25) is a case control GWAS of CAD consisting of 1,988 cases and 5,380 controls from the UK; c) The Framingham Share Initiative dataset includes genetic data and longitudinal phenotype data, such as incidence of major cardiovascular events, for ~9,000 individuals from the Framingham Heart Study (http://www.framinghamheartstudy.org), of which we have included 3,717 in the present study (selected to maximize the number of subjects free from cardiovascular disease at baseline who had genetic data and complete follow-up data; 464 events, see below for phenotype definition; mean follow-up 13.5 years; Supplementary Appendix 1; Lluís-Ganella et al., unpublished data, 2011).
The phenotypic characteristics of these studies were as follows: MIGen cases were males aged <50 years or females aged <60 years who were diagnosed with MI on the basis of autopsy evidence, a combination of chest pain and electrocardiographic evidence, or elevation of cardiac biomarkers; WTCCC cases had a validated history of either MI or coronary revascularization (coronary artery bypass surgery or percutaneous coronary angioplasty) before their 66th birthday; in the Framingham sample, events included incident cases with MI, angina, coronary revascularization and death due to CAD.
Of the 6,042 individuals in the MIGen sample, 2,681 were previously included in the CARDIoGRAM discovery meta-analysis. All of the WTCCC cases (n~1,988) and approximately half of the controls (n~2,938) were also included in the CARDIoGRAM meta-analysis, as were many of the individuals in the Framingham sample, as part of the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium26. Genome-wide genotype data and associated phenotype data for the MIGen and Framingham samples were obtained via The Database of Genotypes and Phenotypes (dbGaP) (dbgap.ncbi.nlm.nih.gov; project number #2392). Data for the WTCCC sample were obtained from the European Genotype Archive (www.ebi.ac.uk/ega) with permission from the WTCCC Data Access Committee (www.wtccc.org.uk).
1000 Genomes Imputation in MIGen, WTCCC and Framingham
A summary of the quality control steps, imputation process, association analyses and meta-analyses performed for this analysis is shown in Supplementary Figure 1.
Imputation of un-typed genetic variants in individuals from the MIGen, WTCCC and Framingham samples was performed using IMPUTE217. Imputation was performed for SNPs in the region of interest using a reference panel of phased haplotypes (available from mathgen.stats.ox.ac.uk/impute/impute_v2.html) based on the August 2010 data release from the 1000 Genomes Project20 (1kG; 566 haplotypes from populations of European ancestry, EUR: CEU, TSI, GBR, FIN and IBS). As input for this process, we included only directly genotyped SNPs with high call rate (≥95%) and whose genotype frequencies were in HWE (p≥10−6). We carried forward to the analysis stage only those SNPs imputed with high quality (IMPUTE2 INFO metric ≥0.5).
Association and meta-analysis of genotyped and imputed SNPs in MIGen, WTCCC and Framingham
A logistic regression analysis of association between allele dosage of imputed and genotyped SNPs and MI/CAD was performed separately in the MIGen, WTCCC and Framingham samples, with adjustment for sex. Adjustment for age or other clinical covariates was not possible because no further phenotype data were available in all studies. However, the association results in the Framingham and MIGen samples were very similar after additional adjustment for age at event (data not shown), and both the MIGen and WTCCC studies were age- and sex-matched by design. To account for inter-relatedness, the analysis of the Framingham sample was also adjusted for the first two genetic principal components27. The results from these three studies were meta-analyzed as described above for the CARDIoGRAM analysis.
Statistical analysis
Apart from imputation, all analyses were performed using R version 2.11 (packages and functions indicated below by <package>::<function>). Fixed and random effects meta-analyses were performed using rmeta::meta.DSL. Association testing was performed using stats::glm for the case-control studies and survival::coxph for the cohort study.
To account for multiple testing, we used a Bonferroni correction based on the effective number of independent tests in the region of interest to set the threshold for declaring statistical significance (regional significance level). Since many SNPs in the region of interest were not independent, we used the technique proposed by Cheverud28 to estimate the effective number of independent tests (neff; separately for the CARDIoGRAM and fine mapping results); for this estimation, we computed pairwise LD (r2) between all pairs of SNPs in the region of interest using genotype data from the HapMapII+III CEU (344 haplotypes) or 1000 Genomes project EUR (566 haplotypes) reference panels of phased haplotypes for the CARDIoGRAM and fine mapping analyses, respectively. LD calculations in the region of interest were performed using SNPassoc::LD29.
We computed the power of each analysis to detect significant associations (Supplementary Methods). Briefly, for each SNP we computed the power of our analysis to exceed the threshold for declaring statistical significance after adjustment for multiple testing, and expressed this power in two ways: the minimum odds ratio (OR) the analysis had high or moderate power to detect (Type II error = 20% or 50%, respectively); and the power of the analysis to detect each of a series of ORs (e.g. 1.05, 1.1, etc.). We computed these values for each SNP and took the mean for all SNPs within each of a series of sub-ranges of MAF (MAF = (0,0.01], (0.01,0.02], etc.).
Results
Meta-analysis of genome-wide association studies of CAD (CARDIoGRAM)
A regional plot of global p-values from the CARDIoGRAM meta-analysis for 535 genotyped and imputed (HapMap II panel) SNPs in the region of interest is shown in Figure 1a. Considering a threshold for declaring statistically significant association of p~1.02x10−4 (neff~503), we observed no significant association between common SNPs in this gene and risk of CAD. This analysis had high power (~80%) to detect ORs of ≥1.10, ≥1.28 and ≥1.33 and moderate power (~50%) to detect ORs of ≥1.08, ≥1.23, and ≥1.26 for SNPs with MAF≥0.15, ≥0.05, and ≥0.01, respectively (Supplementary Table 2).
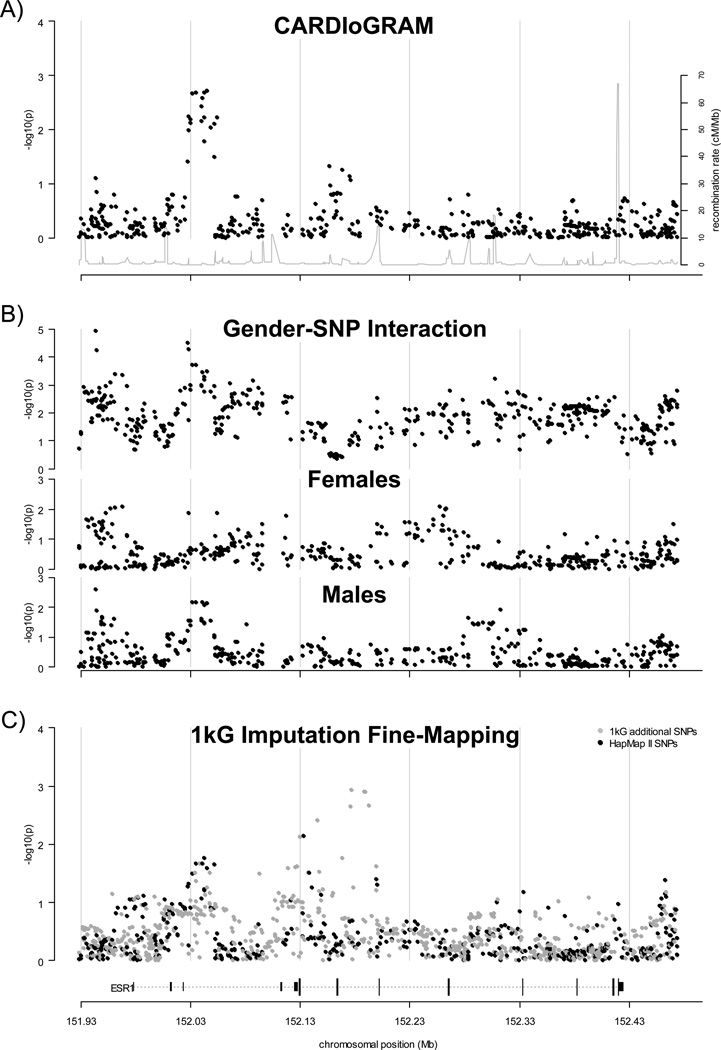
Figure 1.
Regional association results for the ESR1 gene region. Results of CARDIoGRAM global and gender-stratified meta-analysis and fine mapping analysis for a region of chromosome 6 containing the coding and non-coding exons of ESR1 and 50kb of the upstream and downstream flanking regions (−log10(p-value) shown as black points). Results shown are for a fixed or random effects meta-analysis in the absence or presence of between-study heterogeneity, respectively (see Methods). The position of the ESR1 gene is shown at the bottom of the plot (dotted line), with coding and non-coding exons shown as long and short vertical bars, respectively. Regional recombination rate (HapMap II) is shown as a grey line in plot A. A). Regional association plot of global meta-analysis results from the CARDIoGRAM study. B). Regional association plot of gender-stratified meta-analysis results from the CARDIoGRAM study. Results for the test for SNP-gender interaction are shown in the top panel; results for the association test in females and males are shown in the middle and bottom panels, respectively. C). Regional association plot of results from fine-mapping meta-analysis (MIGen, WTCCC and Framingham studies). Results for SNPs that were previously analyzed in the CARDIoGRAM study are shown as black points (i.e. SNPs that were directly genotyped or imputed using a reference panel of haplotypes generated from the Phase II HapMap CEU genotypes). Results for additional SNPs that were imputed using a reference panel of haplotypes generated using data from the 1000 Genomes project (August 2010 release) are shown as grey points.
The strongest association in this region was observed for a series of 18 SNPs lying within a ~24 Kb region of strong LD between non-coding exons E1 and T130 at the 5’ end of the gene. The direction of effect on CAD risk of the top SNP in this area (rs7749659, p= 0.0019; MAF~0. 25) was generally consistent across CARDIoGRAM studies (Supplementary Figure 2; pooled OR (95%CI) = 1.05 (1.02, 1. 08) for the G allele; range 0.85–1.21; pheterogeneity=0.28).
Gender-stratified analysis
Under the hypothesis that the effect of genetic variation in ESR1 on CAD risk differs according to gender, we analyzed data from 13 of the 14 CARDIoGRAM discovery cohorts separately in females (n= 30,615 (48.8%), of which 6,100 (19.9%) were cases) and males (n= 32,069 (51.2%), of which 13,846 (43.2%) were cases; Supplementary Figure 3). We used the same criterion for declaring statistical significance as for the un-stratified meta-analysis (p~1.02x10−4). In females we had high power (~80%) to detect ORs of ≥1.18, ≥1.47 and ≥1.58 and moderate power (~50%) to detect ORs of ≥1.15, ≥1.37, and ≥1.45 for SNPs with MAF≥0.15, ≥0.05, and ≥0.01, respectively (Supplementary Table 2). In males we had high power (~80%) to detect ORs of ≥1.15, ≥1.23 and ≥1.49 and moderate power (~50%) to detect ORs of ≥1.12, ≥1.18, and ≥1.39 for SNPs with MAF≥0.15, ≥0.05, and ≥0.01, respectively (Supplementary Table 2).
One SNP, lying ~35kb upstream of the most distal non-coding exon (Figure 1b), exceeded the threshold for regional significance in the test for interaction between gender and genotype as a predictor of CAD risk (rs9479087,MAF=0.183 in CARDIoGRAM, pint=1.2x10−5; Supplementary Figure 3). However, this variant was not significantly associated with risk in either males (p=0.0026; pooled OR (95%CI) = 1.07 (1.03, 1. 13)) or females (p=0.057; pooled OR (95%CI) = 0.94 (0.89, 1. 00)) at the regional significance level. No other regionally significant evidence for association was observed either among females (top result rs6927072 in Intron 3, p=0.0081, Figure 1b) or males (top result rs9479087, p=0.0026; Figure 1b).
Fine mapping analysis
While the density of SNP data in the HapMap II panel (CARDIoGRAM results) for this region is quite high (mean=1.15 SNPs/kb), it is possible that some stronger true association signals are not captured by these common genotyped and imputed variants. Such signals might be detected by analyzing a higher density map of common and low-frequency SNPs in this region. To explore this possibility, we imputed ~2,500 additional variants from the 1kG reference panel (~4.52 SNPs/kb), 1,451 of which were imputed with high quality in all three samples (~2.7 SNPs/kb; see Supplementary Figure 1). Imputation in the 1kG panel allowed us to test approximately ~800 additional SNPs within the region of interest that were not included in the CARDIoGRAM meta-analysis. Newly imputed SNPs had a wide range of MAF, although a large proportion had MAF in the range 0.0–0.05 (Supplementary Figure 4). After testing for association between SNPs in the 1kG panel and CAD in the MIGen, WTCCC and Framingham samples, meta-analyzing the results and correcting for multiple testing (neff~1,366; αadj~3.8x10−5), we observed no globally significant evidence for association in this region (Figure 1c). This analysis had high power (~80%) to detect ORs of ≥1.21, ≥1.44, ≥2.09 and ≥3.14 and moderate power (~50%) to detect ORs of ≥1.18, ≥1.35, ≥1.85 and ≥2.59 for SNPs with MAF≥0.15, ≥0.05, ≥0.01 and <0.01, respectively (Supplementary Table 2). The strongest association was observed for 6-152177055 (Intron 2; p=0.0012; pooled OR (95%CI) = 1.42 (1.15, 1.76) for the A allele, frequency 0.016; pheterogeneity= 0.10). We observed no significant additional gender-specific effects or gendergenotype interactions for these imputed SNPs (data not shown).
Discussion
In this study, we exploited post-genomic tools and resources to expand on previous candidate association studies of ESR1 in two main ways: (i) we analyzed a large number of common and uncommon genetic variants in the coding, non-coding and flanking regions of the gene, capturing a large proportion of the genetic variation throughout the gene and its regulatory regions; (ii) we performed these analyses in large samples of up to ~85,000 individuals representing multiple populations of European descent, which increases our power to detect subtle risk effects.
Despite this study's power to detect case-control differences in CAD risk of as low as 10% for a broad range of genetic variation throughout this region, we found no evidence of involvement of ESR1 in modifying CAD risk either at the population level, or as a function of gender. We consider these results surprising, given ERα's central role in estrogen and androgen signaling, its widespread expression in vascular tissues, and the importance of gender for CAD risk.
After age, male gender remains the most important independent cardiovascular risk factor (CVRF), and has a far greater impact on total risk than other important risk factors such as smoking, lipid profile, and diabetes. The physiological basis of this gender difference remains unclear, and limited research into this question has been carried out, compared to that for other risk factors, mainly because gender is non-modifiable. However, rather than considering male gender as a non-modifiable cause of increased CAD risk, it is important to remember that gender is a simple Mendelian trait determined by the presence or absence of a single gene, SRY, which is inherited on the Y chromosome in males. Since, as far as we are aware, no evidence of association between CAD and SRY has been reported it is not appropriate to consider gender as being causally associated with CAD risk. Rather, gender is a trait that is strongly associated with CAD risk via unknown and potentially modifiable factors (e.g. physiological, environmental, behavioral factors, etc.), whose effects we can partly capture by using gender as a proxy variable. It is important to identify and understand these factors because the ability to modify even a fraction of gender-associated CAD risk might have a marked impact on prevention, possibly more so than by modifying other CVRFs.
All of the loci identified by GWAS to date as being associated with CAD risk, are located on autosomes, and it seems likely that most or all of the loci that explain the remaining heritability of CAD risk will also be autosomal. Consequently, these loci are in linkage equilibrium with SRY and have equal genotype frequencies in males and females. This leads us to the simple but important conclusion that differences in CAD risk between genders can not be directly caused by genetic factors, but can only arise because of an interaction between gender and other processes associated with risk. Consequently the present study, like all association studies of primary autosomal genetic variation, does not attempt to explain differences in risk between genders. Instead we search for population-level differences in CAD risk that are driven by ESR1 variation, and whose effects may or may not be different among females compared to males (i.e. that interact with gender).
Over the past decade candidate gene association studies (e.g. 31,13) have reported generally inconsistent results regarding the role of ESR1 genetic variation in CAD risk. An initial meta-analysis including ~7,000 individuals supported association12 but this result was not upheld by two subsequent meta-analyses representing ~16,00013 and ~32,00014 individuals. However, these studies have been restricted to a very limited number of SNPs (especially rs2234693 and rs9340799, previously known as the PvuII and XbaI variants, which lie in Intron 1) out of the thousands now known to lie within the gene region. We estimate that the four most widely studied variants collectively capture (with r2 ≥ 0.8) only ~2% of the 1,450 SNPs tested in our study (data not shown). Therefore, although recent reports have found no evidence of association between ESR1 variation and CAD risk13,14, this question remains unanswered until a more complete survey of the gene is carried out. The potential gain to be made from this is illustrated by recent advances in understanding ESR1's role in modulating bone mineral density (BMD) and fracture risk, phenotypes that show intriguingly similar patterns of gender-specific and menopause-related risk to those observed for cardiovascular risk. While candidate gene studies of the role of ESR1 variation in BMD and fracture risk also examined a limited range of genetic variation and obtained similarly inconsistent results32,33, a large meta-analysis of several GWAS subsequently confirmed the involvement of ESR1 variation in modulating these phenotypes34, with highly significant evidence for association in the upstream non-coding regulatory region of the gene, in stark contrast with the lack of association we have observed for CAD.
In the discovery stage of the CARDIoGRAM study the direction of effect of the lead SNP was largely consistent across the contributing studies (Supplementary Figure 1), but fell well short of the threshold for regional statistical significance. The region of high LD containing this SNP was located within the 5' regulatory region but did not coincide with the previously reported signal for BMD and fracture risk34.
We found no broadly convincing evidence of association between ESR1 variation and CAD risk as a function of gender. Although the p-value of the gender interaction test for one SNP exceeded the significance threshold set, with opposing effects observed among males than among females, this variant was not significantly associated with CAD risk in either gender considered separately (Supplementary Figure 3). Considering the additional fact that this variant lies at a considerable distance from the regulatory (~35kb) and coding (~186kb) regions of the gene, we feel that these results do not provide strong evidence of a robust gender-specific association at this locus. In addition to gender, another potential modifier of the putative association between ESR1 variation and CAD risk is menopausal status among women. Although we were unable to investigate this issue directly, we provide some initial data on this question based on age data from the MIGen study, and we find no evidence of significantly different effects of ESR1 variation on cardiovascular risk as a function of menopausal status (see Supplementary Note).
In the fine mapping analysis, imputation using data from the 1000 Genomes Project allowed us to analyze a much denser map of common variants in the region (Supplementary Figure 4), and especially to explore the role of variants with frequencies below 0.05, which are under-represented in haplotype panels based on data from the HapMap project, but which are a potentially important source of risk variance in complex diseases35,36. However, we found no additional evidence of association with CAD risk for any of these additional variants.
We highlight the fact that this study is well powered to detect genetic risk effects with sizes and frequencies that are generally plausible for common complex diseases. For example, in the CARDIoGRAM discovery analysis we have high power (~80%) to detect common variants with MAF≥0.15 that carry risk effects as low as OR ~1.1, and low frequency variants (0.01≤MAF≤0.05) that carry risk effects of OR ~1.3. In addition, the fine mapping analysis was also powered to detect associations for rare imputed variants with MAF≤0.01 effect sizes of approximately OR~3. Weaker and/or rarer risk effects than these are likely to have limited clinical relevance at the population level. In these power computations we used stringent statistical significance thresholds that account for multiple testing (see Supplementary Methods).
The most likely explanation for lack of observable association in this analysis is that no true association exists in this gene, although we note the following limitations in this study's ability to draw this conclusion:
First, this study does not address this question in populations with non-European ancestry. Second, some truly associated variants in this gene may not have been detected by this study, although these are unlikely to be simple primary sequence variants with low allelic diversity, such as common or uncommon SNPs, low-copy number polymorphisms or insertions/deletions. This analysis was also unable to detect very weak or very rare effects (Supplementary Table 2). Third, this study can not address the role of other potentially relevant forms of variation related to ERα, such as epistasis or epigenetic (e.g. promoter methylation), post-transcriptional or post-translational variation. However, if such variation exists, it is likely to be largely independent of primary sequence variation. Fourth, this study suggests that menopausal status does not modify the effect of ESR1 variation on female CAD risk, but cannot discount this possibility because of the size and imprecise design of that analysis. Fifth, these analyses were not adjusted for classical CVRFs, although a true SNP-CAD association would only be masked by confounding if the SNP had opposing effects on CAD risk and CVRF profile, which seems unlikely. Sixth, most of the studies included in these meta-analyses had a case-control design, which could lead to a bias against the discovery of variants that reduce survival.
Finally, it is important to note that we have analyzed the genetic variation in only one of the genes that encode components of the steroid sex hormone system. A more thorough exploration of this system may help to clarify the role of this system in the pathophysiology of coronary risk.
Conclusions
In conclusion, on the basis of data from a large number of subjects representing multiple samples from several populations, we find no evidence for involvement of common or uncommon genetic variation in the coding, non-coding or flanking regions of the ESR1 gene in modifying risk of CAD, irrespective of gender. However, data from observational studies and sub-analysis of clinical trials continue to support the involvement of the steroid hormone system in modulating CAD risk. Therefore, we consider that the next step in exploring the role of the sex hormone biosynthesis system in modulating CAD risk should initially be to prioritize the investigation of other genes within this system.
Supplementary Material
Acknowledgment
See supplementary appendix for full list of contributors from CARDIoGRAM Consortium. We thank all contributing members of the CARDIoGRAM Consortium for the use of cohort-level summary association results. We thank the authors of the MIGen, WTCCC and Framingham GWA studies and 1000 Genomes project for making their data publicly available, and the authors of IMPUTE2 for making 1kG-based phased haplotypes available for public use. A full list of the investigators who contributed to the generation of the WTCCC data is available from http://www.wtccc.org.uk. This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI. We also thank Ana Paula Dantas and Jana Selent for interesting discussions at the design stage.
Funding Sources: The Myocardial Infarction Genetics Consortium (MIGen)24 was funded by grant R01 HL087676 from the National Institutes of Health, USA. The Wellcome Trust Case-Control Consortium 2 was funded by the Wellcome Trust under award 085475. The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195). This work was supported by a grant from ACC1Ó (RD08-1-0024), the European Regional Development Fund (ERDF-FEDER), the Spanish Ministry of Science and Innovation through the Carlos III Health Institute [CIBER Epidemiología y Salud Pública, Red HERACLES RD06/0009, PI061254, PI09/90506] and by the Catalan Research and Technology Innovation Interdepartmental Commission [SGR 1195]. GL was funded by the Juan de la Cierva Program, Ministerio de Educación (JCI-2009-04684).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
URLs: Ensembl genome browser: www.ensembl.org; IMPUTE2 software: mathgen.stats.ox.ac.uk/impute/impute_v2.html; 1000 Genomes Project: www.1000genomes.org; HapMap: hapmap.ncbi.nlm.nih.gov/; dbSNP database: www.ncbi.nlm.nih.gov/snp/
References
- 1.Tunstall-Pedoe H, Kuulasmaa K, Mahonen M, Tolonen H, Ruokokoski E, Amouyel P. Contribution of trends in survival and coronary-event rates to changes in coronary heart disease mortality: 10-year results from 37 WHO MONICA project populations. Monitoring trends and determinants in cardiovascular disease. Lancet. 1999;353:1547–1557. doi: 10.1016/s0140-6736(99)04021-0. [DOI] [PubMed] [Google Scholar]
- 2.Barrett-Connor E, Grady D. Hormone replacement therapy, heart disease, and other considerations. Annu Rev Public Health. 1998;19:55–72. doi: 10.1146/annurev.publhealth.19.1.55. [DOI] [PubMed] [Google Scholar]
- 3.Grodstein F, Stampfer MJ, Colditz GA, Willett WC, Manson JE, Joffe M, Rosner B, Fuchs C, Hankinson SE, Hunter DJ, Hennekens CH, Speizer FE. Postmenopausal hormone therapy and mortality. N Engl J Med. 1997;336:1769–1775. doi: 10.1056/NEJM199706193362501. [DOI] [PubMed] [Google Scholar]
- 4.Rossouw JE, Anderson GL, Prentice RL, LaCroix AZ, Kooperberg C, Stefanick ML, Jackson RD, Beresford SA, Howard BV, Johnson KC, Kotchen JM, Ockene J. Risks and benefits of estrogen plus progestin in healthy postmenopausal women: principal results From the Women's Health Initiative randomized controlled trial. JAMA. 2002;288:321–333. doi: 10.1001/jama.288.3.321. [DOI] [PubMed] [Google Scholar]
- 5.Hulley S, Grady D, Bush T, Furberg C, Herrington D, Riggs B, Vittinghoff E. Randomized trial of estrogen plus progestin for secondary prevention of coronary heart disease in postmenopausal women. Heart and Estrogen/progestin Replacement Study (HERS) Research Group. JAMA. 1998;280:605–613. doi: 10.1001/jama.280.7.605. [DOI] [PubMed] [Google Scholar]
- 6.Salpeter SR, Walsh JM, Greyber E, Salpeter EE. Brief report: Coronary heart disease events associated with hormone therapy in younger and older women. A meta-analysis. J Gen Intern Med. 2006;21:363–366. doi: 10.1111/j.1525-1497.2006.00389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossouw JE, Prentice RL, Manson JE, Wu L, Barad D, Barnabei VM, Ko M, LaCroix AZ, Margolis KL, Stefanick ML. Postmenopausal hormone therapy and risk of cardiovascular disease by age and years since menopause. JAMA. 2007;297:1465–1477. doi: 10.1001/jama.297.13.1465. [DOI] [PubMed] [Google Scholar]
- 8.Dubey RK, Imthurn B, Barton M, Jackson EK. Vascular consequences of menopause and hormone therapy: importance of timing of treatment and type of estrogen. Cardiovasc Res. 2005;66:295–306. doi: 10.1016/j.cardiores.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Fox CS, Polak JF, Chazaro I, Cupples A, Wolf PA, D'Agostino RA, O'Donnell CJ. Genetic and environmental contributions to atherosclerosis phenotypes in men and women: heritability of carotid intima-media thickness in the Framingham Heart Study. Stroke. 2003;34:397–401. doi: 10.1161/01.str.0000048214.56981.6f. [DOI] [PubMed] [Google Scholar]
- 10.Zdravkovic S, Wienke A, Pedersen NL, Marenberg ME, Yashin AI, de Faire U. Heritability of death from coronary heart disease: a 36-year follow-up of 20 966 Swedish twins. J Intern Med. 2002;252:247–254. doi: 10.1046/j.1365-2796.2002.01029.x. [DOI] [PubMed] [Google Scholar]
- 11.Mendelsohn ME, Karas RH. Molecular and cellular basis of cardiovascular gender differences. Science. 2005;308:1583–1587. doi: 10.1126/science.1112062. [DOI] [PubMed] [Google Scholar]
- 12.Shearman AM, Cooper JA, Kotwinski PJ, Miller GJ, Humphries SE, Ardlie KG, Jordan B, Irenze K, Lunetta KL, Schuit SC, Uitterlinden AG, Pols HA, Demissie S, Cupples LA, Mendelsohn ME, Levy D, Housman DE. Estrogen receptor alpha gene variation is associated with risk of myocardial infarction in more than seven thousand men from five cohorts. Circ Res. 2006;98:590–592. doi: 10.1161/01.RES.0000210578.62102.a6. [DOI] [PubMed] [Google Scholar]
- 13.Kjaergaard AD, Ellervik C, Tybjaerg-Hansen A, Axelsson CK, Gronholdt ML, Grande P, Jensen GB, Nordestgaard BG. Estrogen receptor alpha polymorphism and risk of cardiovascular disease, cancer, and hip fracture: cross-sectional, cohort, and case-control studies and a meta-analysis. Circulation. 2007;115:861–871. doi: 10.1161/CIRCULATIONAHA.106.615567. [DOI] [PubMed] [Google Scholar]
- 14.Lluis-Ganella C, Lucas G, Subirana I, Escurriol V, Tomas M, Senti M, Sala J, Marrugat J, Elosua R. Qualitative assessment of previous evidence and an updated meta-analysis confirms lack of association between the ESR1 rs2234693 (PvuII) variant and coronary heart disease in men and women. Atherosclerosis. 2009;207:480–486. doi: 10.1016/j.atherosclerosis.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 15.Ding C, Jin S. High-throughput methods for SNP genotyping. Methods Mol Biol. 2009;578:245–254. doi: 10.1007/978-1-60327-411-1_16. [DOI] [PubMed] [Google Scholar]
- 16.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–1605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metzker ML. Sequencing technologies - the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 19.International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.the CARDIoGRAM Consortium. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin CY, Jin GM, Jin KY, Lee JY, Park T, Kim K, Sim X, Twee-Hee OR, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua ZJ, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees HG, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr, Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musunuru K, Lettre G, Young T, Farlow DN, Pirruccello JP, Ejebe KG, Keating BJ, Yang Q, Chen MH, Lapchyk N, Crenshaw A, Ziaugra L, Rachupka A, Benjamin EJ, Cupples LA, Fornage M, Fox ER, Heckbert SR, Hirschhorn JN, Newton-Cheh C, Nizzari MM, Paltoo DN, Papanicolaou GJ, Patel SR, Psaty BM, Rader DJ, Redline S, Rich SS, Rotter JI, Taylor HA, Jr, Tracy RP, Vasan RS, Wilson JG, Kathiresan S, Fabsitz RR, Boerwinkle E, Gabriel SB. Candidate gene association resource (CARe): design, methods, and proof of concept. Circ Cardiovasc Genet. 2010;3:267–275. doi: 10.1161/CIRCGENETICS.109.882696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myocardial Infarction Genetics Consortium. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Psaty BM, O'Donnell CJ, Gudnason V, Lunetta KL, Folsom AR, Rotter JI, Uitterlinden AG, Harris TB, Witteman JC, Boerwinkle E. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2:73–80. doi: 10.1161/CIRCGENETICS.108.829747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheverud JM. A simple correction for multiple comparisons in interval mapping genome scans. Heredity. 2001;87:52–58. doi: 10.1046/j.1365-2540.2001.00901.x. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez JR, Armengol L, Sole X, Guino E, Mercader JM, Estivill X, Moreno V. SNPassoc: an R package to perform whole genome association studies. Bioinformatics. 2007;23:644–645. doi: 10.1093/bioinformatics/btm025. [DOI] [PubMed] [Google Scholar]
- 30.Kos M, Reid G, Denger S, Gannon F. Minireview: genomic organization of the human ERalpha gene promoter region. Mol Endocrinol. 2001;15:2057–2063. doi: 10.1210/mend.15.12.0731. [DOI] [PubMed] [Google Scholar]
- 31.Koch W, Hoppmann P, Pfeufer A, Mueller JC, Schomig A, Kastrati A. No replication of association between estrogen receptor alpha gene polymorphisms and susceptibility to myocardial infarction in a large sample of patients of European descent. Circulation. 2005;112:2138–2142. doi: 10.1161/CIRCULATIONAHA.105.545913. [DOI] [PubMed] [Google Scholar]
- 32.Ioannidis JP, Ralston SH, Bennett ST, Brandi ML, Grinberg D, Karassa FB, Langdahl B, van Meurs JB, Mosekilde L, Scollen S, Albagha OM, Bustamante M, Carey AH, Dunning AM, Enjuanes A, van Leeuwen JP, Mavilia C, Masi L, McGuigan FE, Nogues X, Pols HA, Reid DM, Schuit SC, Sherlock RE, Uitterlinden AG. Differential genetic effects of ESR1 gene polymorphisms on osteoporosis outcomes. JAMA. 2004;292:2105–2114. doi: 10.1001/jama.292.17.2105. [DOI] [PubMed] [Google Scholar]
- 33.Gennari L, Merlotti D, De PV, Calabro A, Becherini L, Martini G, Nuti R. Estrogen receptor gene polymorphisms and the genetics of osteoporosis: a HuGE review. Am J Epidemiol. 2005;161:307–320. doi: 10.1093/aje/kwi055. [DOI] [PubMed] [Google Scholar]
- 34.Genetic Factors for Osteoporosis (GEFOS) Consortium. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41:1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 36.Holm H, Gudbjartsson DF, Sulem P, Masson G, Helgadottir HT, Zanon C, Magnusson OT, Helgason A, Saemundsdottir J, Gylfason A, Stefansdottir H, Gretarsdottir S, Matthiasson SE, Thorgeirsson GM, Jonasdottir A, Sigurdsson A, Stefansson H, Werge T, Rafnar T, Kiemeney LA, Parvez B, Muhammad R, Roden DM, Darbar D, Thorleifsson G, Walters GB, Kong A, Thorsteinsdottir U, Arnar DO, Stefansson K. A rare variant in MYH6 is associated with high risk of sick sinus syndrome. Nat Genet. 2011;43:316–320. doi: 10.1038/ng.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.