Abstract
Gene-modified skin grafts, produced through gene transfer to human keratinocyte stem cells, offer the possibility of therapeutic benefit for inherited skin diseases. We have previously described efficient lentiviral vector–mediated gene transfer to keratinocyte stem cells and the generation of human skin grafts for the inherited skin disease, Netherton syndrome, which arises due to mutations in serine protease inhibitor Kazal-type 5 (SPINK5). Vectors incorporating an internal murine retroviral–derived promoter [spleen focus-forming virus (SFFV)] in combination with a codon-optimized SPINK5 transgene supported high levels of reconstitution and robust correction of skin architecture. Subsequent longer-term experiments have uncovered unanticipated silencing phenomena, with loss of SPINK5 gene expression over time. The inadvertent introduction of CpG sites during codon optimization appears to have rendered vectors susceptible to silencing due to methylation across the promoter–transgene boundary. Substitution of the methylation-susceptible SFFV promoter with a 572-bp minimal human involucrin promoter (INVOp), which encodes very few CpG sites, prevented repression of the SPINK5 transgene and resulted in durable and highly compartment-specific reconstitution of lympho-epithelial Kazal-type–related inhibitor (LEKTI) in human skin grafted onto immunodeficient mice. We conclude that skin grafts modified with lentiviral vectors encoding INVOp offer a suitable platform for therapeutic gene therapy in Netherton syndrome, and our experience highlights unanticipated effects of transgene codon optimization.
Di and colleagues describe a codon-optimized lentiviral vector platform capable of driving sustained expression of SPINK5, a gene encoding the lymphoepithelial Kazal type-related inhibitor (LEKTI) protein in human skin. This approach leads to durable and highly compartment-specific reconstitution of LEKTI in human skin grafted onto immunodeficient mice, and may prove effective for genetic skin disorders such as Netherton syndrome.
Introduction
Netherton syndrome (NS) is a life-threatening and debilitating inherited skin disorder caused by defective expression of lympho-epithelial Kazal-type–related inhibitor (LEKTI), a serine protease inhibitor encoded by the SPINK5 (serine protease inhibitor Kazal-type 5) gene (Chavanas et al., 2000; Bitoun et al., 2002, 2003). LEKTI is expressed in the outer compartment of the epidermis and plays a critical role in the regulation of kallikreins 5 and 7, which hydrolyze the extracellular corneodesmosomes in the skin (Descargues et al., 2005, 2006; Ishida-Yamamoto et al., 2005). When control of kallikreins by LEKTI is lost, hyperactivated kallikreins can cause premature degradation of corneodesmosomes, resulting in overdesquamation and loss of skin-barrier function. LEKTI also inhibits elastase 2, an epidermal protease involved in profilaggrin processing and lipid lamellae structure formation of the skin, by controlling kallikrein 5–mediated cleavage of proelastase 2 (Bonnart et al., 2010). In the absence of curative therapies, we are developing ex vivo gene-therapy strategies based on lentiviral mediated gene correction of autologous keratinocyte stem cells for the generation of graftable bioengineered skin (Di et al., 2011). We have previously reported that lentiviral mediated gene expression by a spleen focus-forming virus (SFFV) internal promoter of codon-optimized SPINK5 (SPINK5co) supported sufficient LEKTI expression for architectural correction of NS skin in a skin-humanized mouse model. However, in contrast to normal LEKTI expression, transgene expression was not compartment-restricted within the epidermis, but was detectable throughout the basal and suprabasal layers. Subsequently, longer-term cultures have uncovered unanticipated silencing phenomena, which we now demonstrate are associated with methylation of CpG sites within the distal SFFV enhancer region and spreading across the transcription start site boundary within the SPINK5co transgene. We postulate that the process of codon optimization of the SPINK5 transgene, intended to improve gene expression, resulted in the inadvertent introduction of a high number of CpG sites, which then rendered the SFFV-SPINK5co configuration susceptible to methylation-mediated silencing. We show that this phenomenon can be addressed by substitution of SFFV with a 572-bp human involucrin promoter (INVOp) element (Ghazizadeh et al., 2002), which has few CpG, mediates highly compartment-specific SPINK5 expression, and is not prone to repressive methylation.
Materials and Methods
Vector generation and keratinocyte transduction
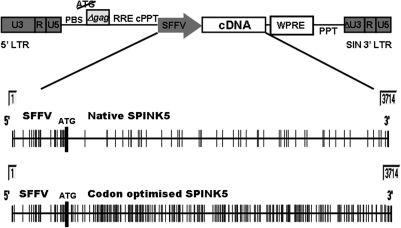
SPINK5co was synthesized by GeneArt (Regensburg, Germany) and was cloned into a previously described replication-deficient self-inactivating (SIN) HIV-1 lentiviral vector. The vector encoded the HIV-1 central polypurine tract (cPPT), start site–mutated woodchuck postregulatory element (WPRE), and SFFV promoter (Demaison et al., 2002) (Fig. 1). To generate INVOp-containing vectors, a 572-bp fragment of involucrin enhancer was cloned from the 2,500-bp full-length human INVOp (locus AF085346), using the method described by Ghazizadeh et al. (2002). Two DNA fragments containing a distal element (–2,473/–2,036) and a proximal region (–242/–1) of the INVOp were amplified from genomic DNA by PCR and cloned into pGEM-T vector (Promega, Southampton, UK). The two amplified fragments were ligated using restriction sites BglII (–2,102) and MscI (–194), and the resulting 572-bp INVOp fragment was verified by sequencing. INVOp was then cloned in place of SFFV to generate INVOp-SP/GFP and INVOp-GFP. Vector stocks were pseudotyped with vesicular stomatitis virus envelope as previously described, concentrated by ultracentrifugation, and titered on human 293T cells by flow cytometry. Keratinocytes were transduced by one round of exposure to SFFV-GFP, SFFV-SP/GFP, INVOp-GFP, or INVOp-SP/GFP vectors at a multiplicity of infection of 25.
FIG. 1.
Schematic view of the lentiviral vector and of the SFFV promoter–transgene sites susceptible to CpG methylation. The upper panel shows SIN HIV-1-based viruses, encoding the SFFV-LTR promoter, the HIV-1 cPPT element, and a mutated WPRE. Following codon optimization, multiple novel CpG sites were introduced throughout the SPINK5co transgene compared with the native SPINK5 gene (MethylPrimer Express Software v1; Applied Biosystems, Foster City, CA).
Flow cytometry analysis
The proportion of green fluorescent protein (GFP)-positive cells and GFP intensity were monitored by flow cytometry (Becton Dickinson, San Jose, CA), and data were collected using CellQuest (Becton Dickinson; V3.0). Values were normalized against initial time points and analyzed by nonparametric tests using GraphPad Prism 5.
Skin biopsies and keratinocyte cultures
Skin biopsies were obtained by punch biopsy from a patient with NS (confirmed null homozygous mutations at c. 2200delAA in SPINK5 gene) under a protocol approved by our Local Ethics Committee and with informed consent from parents. Primary keratinocytes were isolated from skin biopsies by incubation with 0.25% trypsin-EDTA for 3 hr. Primary keratinocytes and keratinocyte cell line NTERT cells (Dickson et al., 2000) were seeded with lethally irradiated 3T3 cells and grown in the keratinocyte culture medium (Rheinwald, 1989). Transduced and untransduced control NTERT cells were treated with keratinocyte medium containing 5 μM azacitidine (5’-azacytidine; Pharmion, Hillingdon, UK), as described previously (Chien et al., 2005), with medium being replaced every 3 days for up to 40 days. During the period of 40 days, cells were passaged when they reaching confluence. Transduced and nontransduced cells without azacitidine treatment were cultured in parallel.
Pyrosequencing and methylation analysis
DNA methylation analysis was carried out by EpigenDx (Worcester, MA). In brief, genomic DNA from transduced cells cultured in the presence or absence of azacitidine was denatured and treated with sodium bisulfite using the EpiTect Bisulfite kit (Qiagen, Crawley, UK). Targeted sequences, which included a 341-bp boundary region of SFFV-SPINK5co (13 CpGs for SFFV and 8 CpGs for SPINK5co), a 246-bp boundary region of INVOp-SPINK5co (7 CpGs for INVOp and 8 CpGs for SPINK5co), a 138-bp boundary region of SFFV-eGFP (8 CpGs for SFFV and 2 CpGs for eGFP), and a 126-bp boundary region of INVOp-eGFP (3 CpGs for INVOp and 2 CpGs for eGFP), were amplified from bisulfite-treated DNA samples using specific forward and biotinylated reverse primers, followed by pyrosequencing with specific sequencing primers for each amplicon. Quantification of methylation at all individual CpG sites within the amplified PCR product was performed using PyroQ-CpG software.
Bioengineered skin preparation and grafting to immunodeficient mice
The methods for preparing and grafting bioengineered skin in nude mice have been previously described (Larcher et al., 2007). In brief, primary NS keratinocytes infected with lentiviral vectors encoding eGFP or SPINK5-eGFP cDNA linked to SFFV or INVOp were seeded on top of a fibrin matrix populated with live primary human fibroblasts (dermal equivalent). After keratinocytes reached confluence, the bioengineered skin constructs were grafted onto a dorsum of 6-week-old female immunodeficient nude mice (NMRI strain; Elevage-Janvier, Le Genest-St Isle, France). Three mice were grafted for each condition. Successfully grafted animals were anesthetized, and the presence of eGFP fluorescent grafts in the whole animals was monitored using a Kodak ISO 2000MM live imaging apparatus (Kodak, Rochester, NY) under appropriate illumination/filter pairs. Close-up photographs of fluorescent grafted areas were obtained in a stereomicroscope equipped with a GFP fluorescent illumination and digital camera (Olympus, Barcelona, Spain). Eight weeks after grafting, skin samples from grafts were taken post mortem, embedded in OCT or paraffin, and sectioned for histological and immunohistochemical examinations.
Immunostaining and immunoblotting
Immunofluorescence staining and histochemistry were performed on frozen or paraffin tissue sections (6-μm thickness) using methods previously described (Di et al., 2009). A custom-made affinity-purified rabbit polyclonal antibody (Eurogentec, Southamton, UK) directed against the C-terminus of LEKTI was used at 580 ng/ml. For immunoblotting, cells were lysed in a cooled buffer composed of 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, cocktail protease inhibitors, 1 mM phenylmethanesulfonyl fluoride (PMSF), and 1% Triton X-100 for 15 min at 4°C. Samples were then centrifuged at 12,000 rpm for 10 min to pellet the insoluble material. The total protein concentration in the supernatant was determined using the Bio-Rad protein assay kit (Bio-Rad, Hertfordshire, UK). Samples from the supernatant were further diluted in 5× sample buffer containing 100 mM dithiothreitol, 10% sodium SDS, 30% glycerol, 0.001% bromphenol blue, and 0.5 mM Tris-HCl, pH 6.8. Equal quantities of total protein were loaded in 10% SDS-PAGE. After electrophoresis, proteins were transferred to polyvinylidene difluoride membranes and incubated with LEKTI antibody overnight at room temperature with shaking. On the following day, membranes were incubated with mouse anti-rabbit IgG conjugated with horseradish peroxidase secondary antibody (Sigma, Poole, UK), and signals were detected using the ECLplus system (GE Healthcare, Buckinghamshire, UK).
Results
Methylation-mediated silencing of SPINK5 gene expression
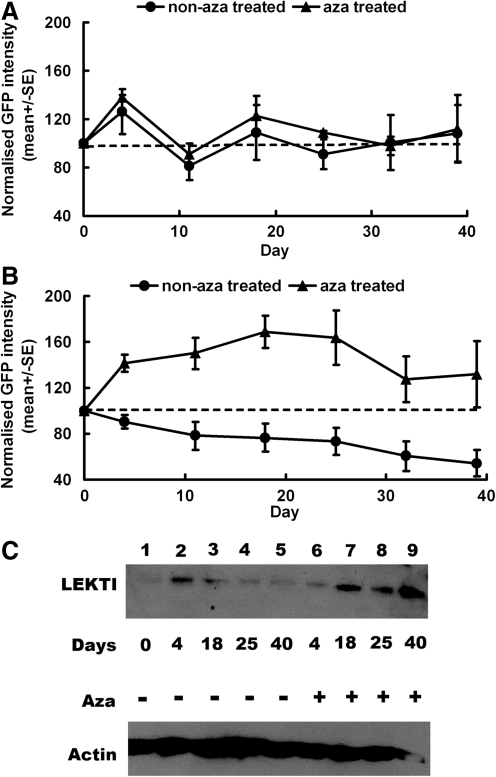
We previously reported that SIN lentiviral vectors derived from HIV-1 and incorporating an internal SFFV promoter upstream of SPINK5co/GFP (SFFV-SP/GFP) mediated high levels of LEKTI expression in primary keratinocytes, whereas protein expression from a non–codon-optimized SPINK5 transgene had been barely detectable (Di et al., 2011). We reasoned that because the native SPINK5 cDNA encodes a high proportion of rare codons and has a relatively low GC content, mRNA turnover was likely to be rapid and protein expression levels may not have been sufficient for therapeutic benefit. Therefore, gene optimization was undertaken using a proprietary algorithm (GeneArt) (Fig. 1). This removed potentially repressive sequences and increased GC content from 43% in native SPINK5 cDNA to 58% in SPINK5co. The process also included the replacement of inhibitory sites (cryptic splice sites, polyA sites, and RNA instability motifs) and the introduction of a Kozak sequence to increase translational initiation (Fath et al., 2011). Codon usage was adapted for expression in human cells, resulting in a very high “codon adaptation value” (0.93). Thereafter, vectors encoding SPINK5co mediated LEKTI expression sufficient to normalize human skin architecture in grafts derived from patients with NS (Di et al., 2011). However, we noted a decline in the expression of reporter gene eGFP in long-term cultured cells with serial propagation, suggesting repression of transgene expression. To test this, keratinocyte cell line NTERT cells were transduced with SFFV-SP/GFP and SFFV-GFP lentiviral vectors, and the intensity of eGFP expression in these cells was monitored by flow cytometry over a 40-day period. Interestingly, although eGFP intensity in cells transduced with the control SFFV-GFP vector did not change over time (Fig. 2A), it was markedly decreased in cells transduced with SFFV-SP/GFP vector (p<0.01) (Fig. 2B). Western blot analysis of LEKTI expression revealed reduction in the expression of LEKTI protein in SFFV-SP/GFP transduced cells (Fig. 2C). Quantitative PCR for integrated vector copy number confirmed the presence of stable integrants, excluding the possibility of vector-mediated toxicity, which may have resulted in the loss of gene-modified populations (data not shown). As the DNA sequences in SPINK5co had increased CpG motifs after gene codon optimization compared with those in native SPINK5 (Fig. 1), we suspected that DNA methylation might be the cause of SPINK5 transgene silencing. To investigate the possibility of methylation-mediated silencing, transduced cells were cultured in the presence of the DNA methyltransferase inhibitor, azacitidine. Flow cytometry analysis showed greater stability of GFP expression for SFFV-SP/GFP transduced populations (Fig. 2B), and western blot analysis revealed increased levels of LEKTI expression in cells cultured in the presence of azacitidine (Fig. 2C). In contrast, there were no significant changes in SFFV-GFP cells treated with azacitidine (Fig. 2A). These results supported the hypothesis that methylation-mediated silencing was compromising transgene expression from vectors expressing SP/GFP in combination with the SFFV promoter.
FIG. 2.
SFFV promoter in combination with SPINK5co results in methylation-mediated DNA silencing. NTERT keratinocytes transduced with lentiviral vectors encoding SFFV-GFP (A) or SFFV-SP/GFP (B) were cultured in the presence or absence of 5 μM azacitidine (aza) for a period of 40 days. GFP intensity in transduced cells cultured over time was monitored by flow cytometry. (A) GFP intensity in cells transduced with SFFV-GFP remained stable, and azacitidine treatment had no significant effect on these cells. (B) GFP intensity in cells transduced with SFFV-SP/GFP decreased over time and was rescued by azacitidine treatment. (C) LEKTI expression in NTERT cells transduced with SFFV-SP/GFP detected by western blot decreased over time (lanes 2–5) and were rescued by azacitidine treatment (lanes 6–9).
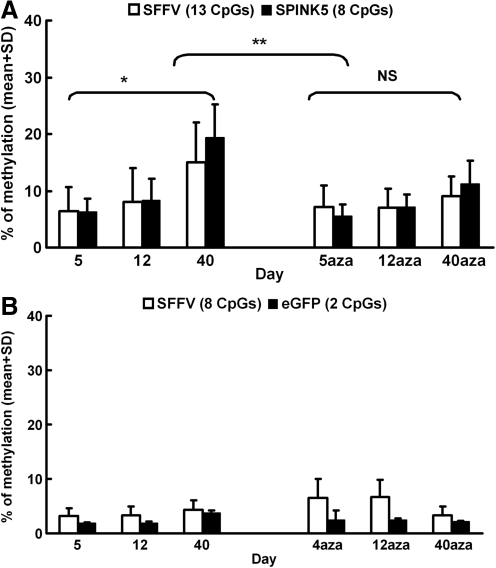
To investigate this phenomenon further, we quantified the levels of DNA methylation at individual CpG sites across the boundary region between the SFFV promoter and SPINK5co, including the transcription start site. DNA from cells transduced with the SFFV-SP/GFP and cultured in the presence or absence of azacitidine was extracted after 5, 12, and 40 days and treated with bisulfite before pyrosequencing and analysis. DNA methylation was quantified, focusing on 13 CpG sites within 174 bases of distal SFFV promoter and 8 CpG sites within 163 bases of proximal SPINK5 cDNA. The proportion of methylated CpG sites increased significantly over time in the absence of azacitidine treatment from around 6% for both SFFV and SPINK5co regions at day 5 to 15% for SFFV and 19% for SPINK5co at day 40 (p<0.001) (Fig. 3A). Moreover, by day 40, azacitidine treatment resulted in a significant reduction of DNA methylation within both SFFV and SPINK5co regions (SFFV, 9.1±3.6%; SPINK5, 11.2±4.1%; p<0.001) (Fig. 3A). Interestingly, methylation within the SFFV promoter of control groups transduced with vectors expressing eGFP alone (SFFV-GFP) was much lower (4.3±1.8% at day 40 for SFFV-GFP versus 15.1±7.0% at day 40 for SFFV-SP/GFP; p<0.001), with overall levels ranging from 1.6% to 6.6% (Fig. 3B). Thus, CpG sites within the distal region of SFFV were more susceptible to methylation when the promoter was used in combination with codon-optimized, GC-enriched SPINK5, suggesting spreading of methylation across the promoter–cDNA boundary.
FIG. 3.
Methylation analysis of the promoter–transgene boundary in NTERT cells transduced with SFFV-containing vectors. NTERT keratinocytes transduced with lentiviral vectors encoding SFFV-GFP or SFFV-SP/GFP were cultured in the presence or absence of 5 μM azacitidine (aza) for a period of 40 days. DNA was extracted after 5, 12, and 40 days and treated with bisulfite before pyrosequencing and analysis. (A) Average percentages of methylation within a 341-bp boundary region of SFFV-SPINK5co including 13 CpGs for SFFV (white bars) and 8 CpGs for SPINK5co (black bars) show increasing methylation over time (*p<0.001). Methylation was significantly reduced by day 40, following azacitidine treatment compared with cells without azacitidine treatment (**p<0.001). There was no significant (ns) change within the azacitidine-treated group over time. (B) Low methylation levels within a 138-bp boundary region including 8 CpGs for SFFV (white bars) and 2 CpGs for eGFP (black bars) were detected in cells transduced with SFFV-GFP.
The human INVOp addresses the silencing of SPINK5co
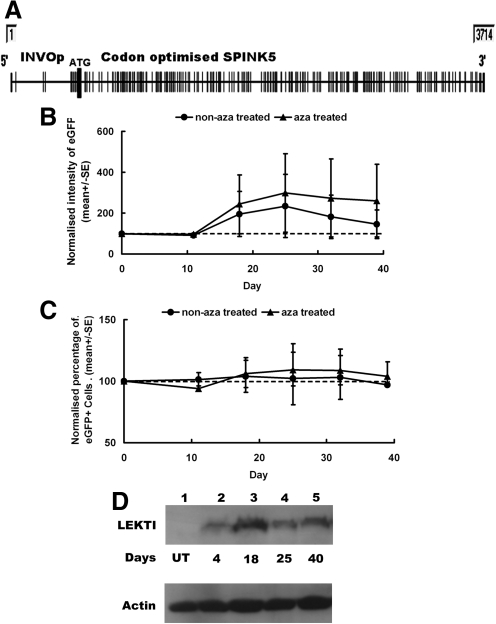
Involucrin is an epidermal protein expressed in the suprabasal layers of stratified squamous epithelium during keratinocyte differentiation. We investigated whether a 572-bp regulatory element derived from the full-length human INVOp could mediate SPINK5co transgene expression and support compartment-specific LEKTI expression. As INVOp has very few CpG sites (a total of 7 CpGs in INVOp compared with 26 CpGs within SFFV) (Fig. 4A), we hypothesized that using this promoter might prevent methylation-mediated silencing of SPINK5co transgene. We incorporated INVOp into the lentiviral constructs by substituting SFFV to generate INVOp-SP/GFP and INVOp-GFP vectors. NTERT keratinocytes were transduced and cultured with or without azacitidine, for up to 40 days. Flow cytometry analysis revealed no significant reduction in the proportion of eGFP-expressing cells or in the intensity of GFP expression in cells transduced with INVO-SP/GFP over time (Fig. 4B and C). In addition, western blot analysis showed stable LEKTI expression over the analyzed period (Fig. 4D).
FIG. 4.
INVOp supports stable LEKTI expression. (A) Schematic view of the INVOp-SPINK5co sites susceptible to CpG methylation. (B) NTERT cells transduced with INVOp-SP/GFP were cultured in the presence or absence of 5 μM azacitidine (aza) for a period of 39 days. GFP intensity did not change in the first 11 days, but was increased following day 11. This is likely to be a result of enhanced involucrin expression during keratinocyte differentiation. (C) The proportion of GFP-positive cells remained stable in the presence/absence of azacitidine treatment. (D) Western blot analysis with anti-LEKTI antibody shows stable LEKTI expression in NTERT cells transduced with INVOp-SP/GFP over time.
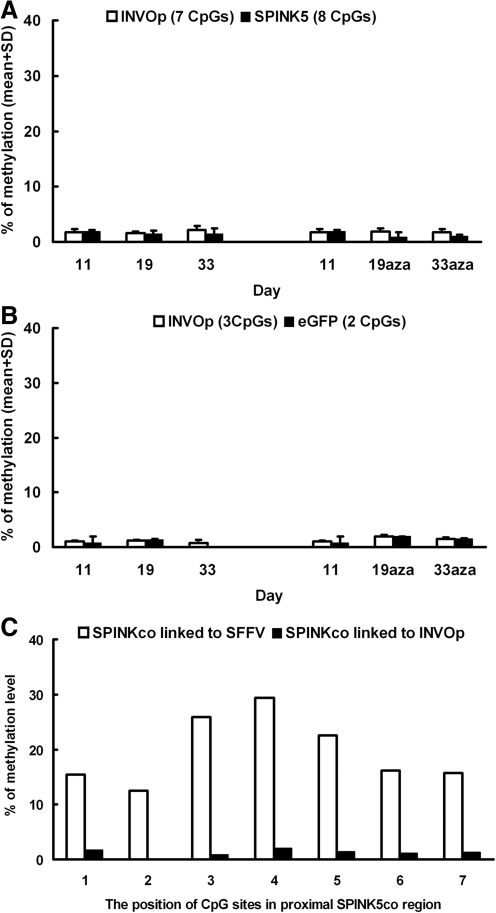
Next, to analyze the methylation status of INVOp and SPINK5co, we carried out pyrosequencing and analysis of bisulfite-treated DNA. DNA from cells transduced with the INVOp-SP/GFP and INVOp-GFP cultured in the presence or absence of azacitidine was extracted after 11, 19, and 33 days in culture and treated with bisulfite before pyrosequencing. DNA methylation was quantified, focusing on the distal region of INVOp (7 sites within 106 bp) and the proximal region of SPINK5co (8 CpG sites within 163 bp). Our results revealed low levels of methylation within the INVOp region over time in cells transduced with INVOp-SP/GFP or INVOp-GFP vectors, ranging from around 0% to 2.2% (Fig. 5A and B). Azacitidine treatment did not change the methylation status in cells transduced with either vector (Fig. 5A and B). Importantly, methylation levels within the proximal SPINK5co were significantly lower than methylation levels within the same region in vectors incorporating SFFV promoter, reaching 1.5±1.0% for INVOp-SP/GFp versus 19.4±5.9% for SFFV-SP/GFP (p<0.001). This difference was evident at each individual site analyzed (Fig. 5C). We conclude that substitution of INVOp in place of SFFV supports stable LEKTI expression in vitro and results in reduced susceptibility to transgene repression.
FIG. 5.
Methylation in vectors incorporating the INVOp. NTERT cells transduced with INVOp-SP/GFP were cultured in the presence or absence of 5 μM azacitidine (aza) for over 30 days. Methylation across the promoter–transgene boundary was quantified by pyrosequencing following bisulfite treatment of genomic DNA. (A) There was low-level methylation within the 246-bp boundary region of INVOp-SPINK5co (white bars, INVOp; black bars, transgene SPINK5co). (B) The relevant control vector configuration also showed low-level methylation within a 126-bp boundary region of INVOp-eGFP (white bars, INVOp; black bars, eGFP). (C) Methylation at seven individual CpG sites within the proximal SPINK5co region at the end of the culture period was found to be significantly lower in cells transduced with INVOp-SP vectors (black bars) compared with cells transduced with SFFV-SP constructs (white bars).
INVOp supports robust,compartmentalized LEKTI expression in vivo
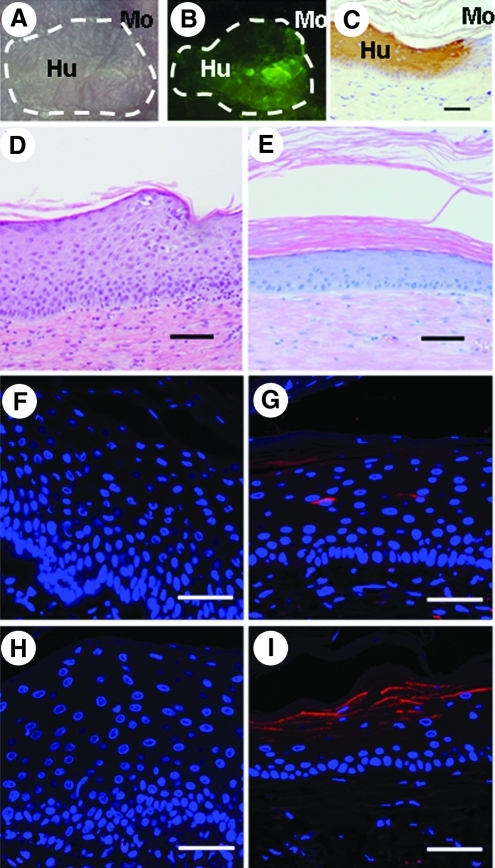
To evaluate the efficacy of the INVOp-SP/GFP combination in a skin-humanized mouse model, we transduced primary keratinocytes obtained from an NS patient with INVOp-SP/GFP and INVOp-GFP control vectors. Cells transduced with SFFV-SP/GFP and SFFV-GFP were included for comparison. Clonogenicity of keratinocytes transduced with SFFV-GFP, INVOp-SP/GFP, and INVOp-SP/GFP evaluated using colony-forming assay did not reveal significant differences among cell groups (data not shown). Transduced cells were expanded and cultured as bioengineered skin equivalents and grafted onto immunodeficient nude mice as previously described (Di et al., 2011). NS grafts transduced with INVOp-GFP showed histological features of NS, including psoriasiform changes and hypogranulosis with a parakeratotic stratum corneum and defective keratinization (Fig. 6D). LEKTI expression was not detected in grafts transduced with SFFV-GFP or INVO-GFP control vector (Fig. 6F and H). Although the initial gene-transfer efficiency monitored by flow cytometry for GFP-positive populations was lower in keratinocytes transduced with INVOp vectors (25%) compared with SFFV vectors (40%), the level of LEKTI expression and the extent of architectural correction were greatest in grafts generated from INVOp-SP/GFP (Fig. 6G and I). In addition, the localization of suprabasal LEKTI in NS grafts driven by INVOp closely mirrored the compartmentalization of native LEKTI expression (Fig. 6I). Thus, vectors incorporating INVOp supported durable SPINK5co transgene expression leading to compartment-specific LEKTI reconstitution and correction of skin architecture.
FIG. 6.
INVOp supports robust, compartment-specific LEKTI expression in NS skin grafts. NS keratinocytes were transduced with INVOp-GFP (D, H) or INVOp-SP/GFP (A-C, E, I) or SFFV-GFP (F) or SFFV-SP/GFP (G) and grafted onto nude mice. Skin grafts were examined 8 weeks after grafting. (A) Macroscopic appearances of the grafts under transmitted light. (B) Expression of eGFP within the skin grafts visible under 488-nm (blue) light. (C) Anti-human involucrin antibody staining, revealing the histological demarcation of murine (Mo) and human (Hu) skin boundary. (D, E) Histological appearance of grafts by hematoxylin and eosin staining. Results showed corrected epidermal structures with a thinner, well differentiated epidermis in the INVOp-SP/GFP grafts (E), compared with the control graft transduced with INVOp-GFP (D), which shows typical features of NS with thickened epidermis and incomplete terminal differentiation and hypogranulosis. LEKTI expression (shown in red) was absent in control groups transduced with SFFV-GFP (F) or INVOp-GFP (H), but was detectable in grafts transduced with SFFV-SP/GFP (G) and INVOp-SP/GFP (I). High-level LEKTI expression with suprabasal compartment localization was shown in the INVOp-SP/GFP graft (I), compared with the SFFV-SP/GFP graft (G) in which only limited numbers of LEKTI-positive cells were detected. Scale bar=50 μm for F–I and 200 μm for C–E.
Discussion
Gammaretroviral vectors, incorporating long terminal repeat (LTR) promoters, have been widely used in early-phase clinical studies (Qasim et al., 2009). The ability of strong enhancer elements within the LTRs to mediate transactivation of proto-oncogenes, resulting in insertional mutagenesis, has been a major concern associated with conventional retroviral configurations (Hacein-Bey-Abina et al., 2003, 2008; Ott et al., 2006; Howe et al., 2008) and, more recently, loss of transgene expression has been documented in clinical studies (Stein et al., 2010). This has been attributed to various causes, including positional variegation effects, promoter methylation, and histone modification. For example, in a phase 1 study of retroviral correction of chronic granulomatous disease, methylation of CpG sites within the SFFV promoter led to extinction of transgene expression and loss of therapeutic effects, whereas intact enhancer function mediated clonal expansion through transactivation effects. Next-generation SIN vectors, with deleted U3 LTR promoter activity, use heterologous promoters, which could address concerns relating to transactivation. However, in many preclinical studies, internal promoters derived from murine retroviral LTR sequences have remained popular, as they support high levels of transcriptional activity in a wide variety of cell types (Demaison et al., 2002). We have previously described robust and durable gene expression from SIN HIV-1 vectors incorporating an internal SFFV-derived promoter in a variety of cell types, including hematopoietic stem cells, lymphocytes (Qasim et al., 2007), fibroblasts (Fassone et al., 2010), and keratinocytes (Di et al., 2011). Evidence is accumulating that, in susceptible cell lines such as the P19 embryonic carcinoma line and hematopoietic stem cells, gene expression mediated by LTR-derived elements is rapidly repressed through CpG methylation. Notably, the SFFV-LTR includes methylation-susceptible CpG motifs within its Sp1 transcription binding site, enhancer core, and CAAT/TATA box elements (Zhang et al., 2007, 2010).
Several approaches are being investigated to address DNA methylation and histone modification, which can result in silencing of vector-mediated transgene expression. One strategy has been to incorporate chromatin insulators such as the chicken β-globin locus control region HS4 element (cHS4) into retroviral LTR regions to reduce expression variegation, but their inclusion may compromise vector titer (Gaszner and Felsenfeld, 2006), and a recent clinical report has raised questions about their inclusion in therapeutic vectors (Cavazzana-Calvo et al., 2010). Alternatively, enhancerless ubiquitous chromatin opening element (UCOE) sequences, derived from the human HNRPA2B1-CBX3 locus (A2UCOE), have been shown to confer stable transgene expression in a variety of different cell types, including hematopoietic stem cells used for the correction of murine severe combined immunodeficiency (Zhang et al., 2007, 2010). UCOEs exhibit a dominant chromatin opening capability and encode methylation-free CpG islands that potently resist DNA methylation. Although we considered substitution of SFFV with A2UCOE in vectors expressing SPINK5co, preliminary experiments using previously described A2UCOE-GFP lentiviral vectors suggested that these vectors may not support sufficiently high levels of gene expression in keratinocytes. Similarly, gene expression derived from vectors with alternative human promoter elements derived from elongation factor 1-α gene and phosphoglycerate kinase promoter were notably reduced in comparison with SFFV (data not shown). In contrast, we found that minimal (572 bp) human INVOp, with low CpG content, mediated both compartment-specific and repression-resistant transgene expression.
A general model of epigenetic repression of vector-mediated gene expression envisages methylation of enhancer-promoter elements that may spread to nearby CpG sites within the transgene (Bird and Wolffe, 1999; Curradi et al., 2002). Our observations are consistent with a model whereby, once initiated, DNA methylation is perpetuated in cis and spreads to neighboring CpG sites, breaching a threshold beyond which transcription is repressed. We conclude that juxtaposition of the SFFV and SPINK5co resulted in clustering of methylation-susceptible CpG sites, predisposing to transgene repression over time. Further investigations will be required to fully elucidate additional mechanisms, such as chromatin-mediated repression, but we have demonstrated that substituting of the CpG-rich SFFV promoter with INVOp circumvents susceptibility to methylation-mediated silencing of SPINK5co. Deployment of the INVOp is particularly attractive in the context of inherited skin diseases such as NS, where gene expression is highly compartmentalized. In the normal skin, to maintain a constant thickness of epidermis, corneocytes within the uppermost layers of the epidermis are shed by desquamation and are continuously replaced by keratinocytes undergoing differentiation. Correct epidermal assembly requires compartmentally restricted gene expression; for example, both involucrin and LEKTI proteins are expressed in the uppermost layers of the epidermis. In the absence of a well-defined promoter associated with the endogenous SPINK5 locus, INVOp provides a valuable alternative and supports LEKTI expression in suprabasal layers of the epidermis. Interestingly, essential proximal regulatory regions of human INVOp have previously been shown to encode binding sites for lens epithelium–derived growth factor (Kubo et al., 2002), a well-defined transcriptional coactivator and a critical host-cell factor that interacts with HIV-1 integrase during proviral integration. It is not know whether this association could influence vector integration or function, and detailed integration-site characterization will be required to determine whether the integration profile of lentiviral vectors encoding INVOp differs from other HIV-1-derived vectors.
The lentiviral vector system described here has addressed unanticipated problems associated with our previous proof-of-principle constructs. Further refinements are under way to produce a third-generation, four-plasmid, vector system (Dull et al., 1998) for translational studies and phase I clinical testing in a clinical trial of gene-modified skin grafts in NS.
Acknowledgments
Research at the Institute of Child Health and Great Ormond Street Hospital for Children NHS Trust benefits from R&D funding received from the UK Department of Health. This study was also supported by the Newlife Foundation for Disabled Children, Moulton Trust and Ichthyosis Support Group, and British Skin Foundation. Dr. W. Qasim is supported by Leukaemia Research and Great Ormond Street Hospital, and Prof. A. Thrasher is a Wellcome Trust Senior Fellow in Clinical Science. Drs. Del Rio and Larcher are supported by grants SAF-2010-16976 from MICINN and PI081054 from ISCIII, respectively. We are indebted to Mrs. Almudena Holguin, Blanca Duarte, and Nuria Illera for grafting experiments. We gratefully acknowledge Dr. Ayad Eddaoudi (ICH flow cytometry) and Dr. Bertrand Vernay (ICH confocal core facility) for their expertise in flow cytometry analysis and confocal microscopy support.
Author Disclosure Statement
The authors have no conflict of interest to declare.
References
- Bird A.P. Wolffe A.P. Methylation-induced repression—belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- Bitoun E. Chavanas S. Irvine A.D., et al. Netherton syndrome: disease expression and spectrum of SPINK5 mutations in 21 families. J. Invest. Dermatol. 2002;118:352–361. doi: 10.1046/j.1523-1747.2002.01603.x. [DOI] [PubMed] [Google Scholar]
- Bitoun E. Micheloni A. Lamant L., et al. LEKTI proteolytic processing in human primary keratinocytes, tissue distribution and defective expression in Netherton syndrome. Hum. Mol. Genet. 2003;12:2417–2430. doi: 10.1093/hmg/ddg247. [DOI] [PubMed] [Google Scholar]
- Bonnart C. Deraison C. Lacroix M., et al. Elastase 2 is expressed in human and mouse epidermis and impairs skin barrier function in Netherton syndrome through filaggrin and lipid misprocessing. J. Clin. Invest. 2010;120:871–882. doi: 10.1172/JCI41440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavazzana-Calvo M. Payen E. Negre O., et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavanas S. Bodemer C. Rochat A., et al. Mutations in SPINK5, encoding a serine protease inhibitor, cause Netherton syndrome. Nat. Genet. 2000;25:141–142. doi: 10.1038/75977. [DOI] [PubMed] [Google Scholar]
- Chien J. Staub J. Avula R., et al. Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene. 2005;24:5089–5100. doi: 10.1038/sj.onc.1208700. [DOI] [PubMed] [Google Scholar]
- Curradi M. Izzo A. Badaracco G. Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol. Cell. Biol. 2002;22:3157–3173. doi: 10.1128/MCB.22.9.3157-3173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaison C. Parsley K. Brouns G., et al. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther. 2002;13:803–813. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- Descargues P. Deraison C. Bonnart C., et al. Spink5-deficient mice mimic Netherton syndrome through degradation of desmoglein 1 by epidermal protease hyperactivity. Nat. Genet. 2005;37:56–65. doi: 10.1038/ng1493. [DOI] [PubMed] [Google Scholar]
- Descargues P. Deraison C. Prost C., et al. Corneodesmosomal cadherins are preferential targets of stratum corneum trypsin- and chymotrypsin-like hyperactivity in Netherton syndrome. J. Invest. Dermatol. 2006;126:1622–1632. doi: 10.1038/sj.jid.5700284. [DOI] [PubMed] [Google Scholar]
- Di W.L. Hennekam R.C. Callard R.E. Harper J.I. A heterozygous null mutation combined with the G1258A polymorphism of SPINK5 causes impaired LEKTI function and abnormal expression of skin barrier proteins. Br. J. Dermatol. 2009;161:404–412. doi: 10.1111/j.1365-2133.2009.09231.x. [DOI] [PubMed] [Google Scholar]
- Di W.L. Larcher F. Semenova E., et al. Ex-vivo gene therapy restores LEKTI activity and corrects the architecture of Netherton syndrome-derived skin grafts. Mol. Ther. 2011;19:408–416. doi: 10.1038/mt.2010.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickso M.A. Hahn W.C. Ino Y., et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol. Cell. Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T. Zufferey R. Kelly M., et al. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 1998;72:8463–8471. doi: 10.1128/jvi.72.11.8463-8471.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassone E. Duncan A.J. Taanman J.W., et al. FOXRED1, encoding an FAD-dependent oxidoreductase complex-I-specific molecular chaperone, is mutated in infantile-onset mitochondrial encephalopathy. Hum. Mol. Genet. 2010;19:4837–4847. doi: 10.1093/hmg/ddq414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath S. Bauer A.P. Liss M., et al. Multiparameter RNA and codon optimization: a standardized tool to assess and enhance autologous mammalian gene expression. PLoS One. 2011;6:e17596. doi: 10.1371/journal.pone.0017596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M. Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Ghazizadeh S. Doumeng C. Taichman L.B. Durable and stratum-specific gene expression in epidermis. Gene Ther. 2002;9:1278–1285. doi: 10.1038/sj.gt.3301800. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Von Kalle C. Schmidt M., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Garrigue A. Wang G.P., et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe S.J. Mansour M.R. Schwarzwaelder K., et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J. Clin. Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida-Yamamoto A. Deraison C. Bonnart C., et al. LEKTI is localized in lamellar granules, separated from KLK5 and KLK7, and is secreted in the extracellular spaces of the superficial stratum granulosum. J. Invest. Dermatol. 2005;124:360–366. doi: 10.1111/j.0022-202X.2004.23583.x. [DOI] [PubMed] [Google Scholar]
- Kubo E. Fatma N. Sharma P., et al. Transactivation of involucrin, a marker of differentiation in keratinocytes, by lens epithelium-derived growth factor (LEDGF) J. Mol. Biol. 2002;320:1053–1063. doi: 10.1016/s0022-2836(02)00551-x. [DOI] [PubMed] [Google Scholar]
- Larcher F. Dellambra E. Rico L., et al. Long-term engraftment of single genetically modified human epidermal holoclones enables safety pre-assessment of cutaneous gene therapy. Mol. Ther. 2007;15:1670–1676. doi: 10.1038/sj.mt.6300238. [DOI] [PubMed] [Google Scholar]
- Ott M.G. Schmidt M. Schwarzwaelder K., et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Qasim W. Mackey T. Sinclair J., et al. Lentiviral vectors for T-cell suicide gene therapy: preservation of T-cell effector function after cytokine-mediated transduction. Mol. Ther. 2007;15:355–360. doi: 10.1038/sj.mt.6300042. [DOI] [PubMed] [Google Scholar]
- Qasim W. Gaspar H.B. Thrasher A.J. Progress and prospects: gene therapy for inherited immunodeficiencies. Gene Ther. 2009;16:1285–1291. doi: 10.1038/gt.2009.127. [DOI] [PubMed] [Google Scholar]
- Rheinwald J.G. Methods for clonal growth and serial cultuvation of normal human epidermal keratinocytes and mesothelial cells. In: Baserga R., editor. A Practical Approach. IRL Press; Oxford: 1989. pp. 81–94. [Google Scholar]
- Stein S. Ott M.G. Schultze-Strasser S., et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nat. Med. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- Zhang F. Thornhill S.I. Howe S.J., et al. Lentiviral vectors containing an enhancer-less ubiquitously acting chromatin opening element (UCOE) provide highly reproducible and stable transgene expression in hematopoietic cells. Blood. 2007;110:1448–1457. doi: 10.1182/blood-2006-12-060814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F. Frost A.R. Blundell M.P., et al. A ubiquitous chromatin opening element (UCOE) confers resistance to DNA methylation-mediated silencing of lentiviral vectors. Mol. Ther. 2010;18:1640–1649. doi: 10.1038/mt.2010.132. [DOI] [PMC free article] [PubMed] [Google Scholar]