Summary
Phosphatidylinositide-3-kinases (PI3K) initiate a number of signaling pathways by recruiting other kinases, such as Akt, to the plasma membrane. One of the isoforms, PI3Kα , is an oncogene frequently mutated in several cancer types. These mutations increase PI3K kinase activity, leading to increased cell survival, cell motility, cell metabolism, and cell cycle progression. The structure of the complex between the catalytic subunit of PI3Kα , p110α , and a portion of its regulatory subunit, p85α reveals that the majority of the oncogenic mutations occur at the interfaces between p110 domains and between p110 and p85 domains. At these positions, mutations disrupt interactions resulting in changes in the kinase domain that may increase enzymatic activity. The structure also suggests that interaction with the membrane is mediated by one of the p85 domains (iSH2). These findings may provide novel structural loci for the design of new anti-cancer drugs.
Keywords: PIK3CA, PI3K, p110α, p110α /p85α, somatic mutation, hot spot, drug design
Introduction
Phosphoinositide 3-kinases (PI3Ks) are lipid kinases that phosphorylate inositol phospholipids at the 3-position of the inositol head groups 1, 2. Class IA PI3Ks are obligate heterodimers of a catalytic subunit (p110α, p110β, or p110δ; collectively called p110) and a regulatory subunit (p85α, p55α, p50α, p85β, or p55γ; collectively called p85)3. Upon activation by receptor tyrosine kinases, class IA PI3Ks preferentially use phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2; also abbreviated PIP2] as their substrate to generate phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3; also PIP3], which recruits PIP3 binding proteins to the membrane by binding to their pleckstrin homology (PH) domains. The recruited proteins include, among others, the AKT serine/threonine kinase (also known as protein kinase B or PKB) and 3-phosphoinositide-dependent protein kinase-1 (PDK1). AKT, once at the cell membrane, becomes activated and phosphorylates multiple downstream targets involved in cell survival, cell cycle progression, cell motility and metabolism 4.
PI3Ks have been implicated in cancer since phosphatidylinositol kinase (PIK) activities were identified in preparations of the oncoproteins pp60v-src and polyoma virus middle T antigen 5–7. It was subsequently shown that the oncoprotein-associated PIK activity was mediated by the p110α catalytic subunit and produced phosphatidylinositol-3-phosphate 8–10. The role of p110α in carcinogenesis was further substantiated by the discovery that avian the sarcoma virus 16 genome encodes an oncogene derived from p110α 11. Furthermore, PTEN (phosphatase and tensin homolog), which reverses the reaction catalyzed by PI3Ks by dephosphorylating 3-position on inositol head groups, was found to be a tumor suppressor gene commonly mutated in human tumors 11–13.
More recently, sequencing of PI3K genes in a variety of human cancers revealed a high frequency of mutations in the PIK3CA gene, which codes for p110α 14–22. In cancers such as colorectal, breast, and hepatocellular carcinomas these mutations occur in up to 30% of all tumors examined, making PIK3CA one of the two most frequently mutated oncogenes togehter with K-Ras 23. In vitro and in vivo studies show that most cancer-associated PIK3CA mutations lead to enhanced enzymatic activity, up-regulation of the signaling cascade, and oncogenic transformation of cells 24–28. Due to the importance of the PI3K-AKT pathway in tumorigenesis and the high frequency of p110α mutations in human cancers, small molecule inhibition of PI3Kα is regarded as a promising strategy for cancer treatment 29–31.
p110α is composed of five domains: an adaptor-binding domain (ABD), a Ras-binding domain (RBD), a C2 domain, a helical domain, and a kinase domain. The regulatory p85α also comprises five domains: an SH3 domain, a GAP domain, an N-terminal SH2 (nSH2) domain, an inter-SH2 domain (iSH2), and a C-terminal SH2 domain (cSH2). Most cancer associated mutations occur in p110α and are clustered at two small regions in the helical and kinase domains, but mutations are also found in ABD, C2, and other parts of the helical and kinase domains. The location and the effect of these mutations on the catalytic activity of the enzyme have been the subject of intense investigation. The structure of a protein homologous to p110α , the class IB catalytic subunit p110γ 32–34, was determined by X-ray crystallography, but the sequence identity between p110α and p110γ is only 35% and most p110α residues mutated in cancers are not conserved in p110γ. Other attempts to address these issues include the determination that the iSH2 domain of p85α interacts with the ABD of p110α 35. This interaction was demonstrated in detail in the crystal structure of a complex between the iSH2 and ABD 36 that showed that, contrary to previous belief, oncogenic mutations in ABD did not occur at the interface between ABD and iSH2, but the structure did not provide clues about how mutations might affect p110α activity.
Recently, we determined the crystal structure of the full-length human p110α in complex with the nSH2 and iSH2 (termed niSH2) domains of p85α 37. The p110α/niSH2 structure revealed two interdomain contacts not identified in previous structures: one between the ABD and kinase domain of p110α, and the other between the C2 of p110α and iSH2 of p85α. Several cancer-associated mutations occur at these domain interfaces. In addition, the p110α/niSH2 structure suggested a role for iSH2 in the interaction of the complex with membrane. Structural comparison of p110α/niSH2 and inhibitor-bound p110γ shows features of the active sites of the two enzymes that could be exploited for the design of isoform-specific inhibitors for cancer treatment.
Description of the p110α /niSH2 Heterodimer
General Description and Domain Structure
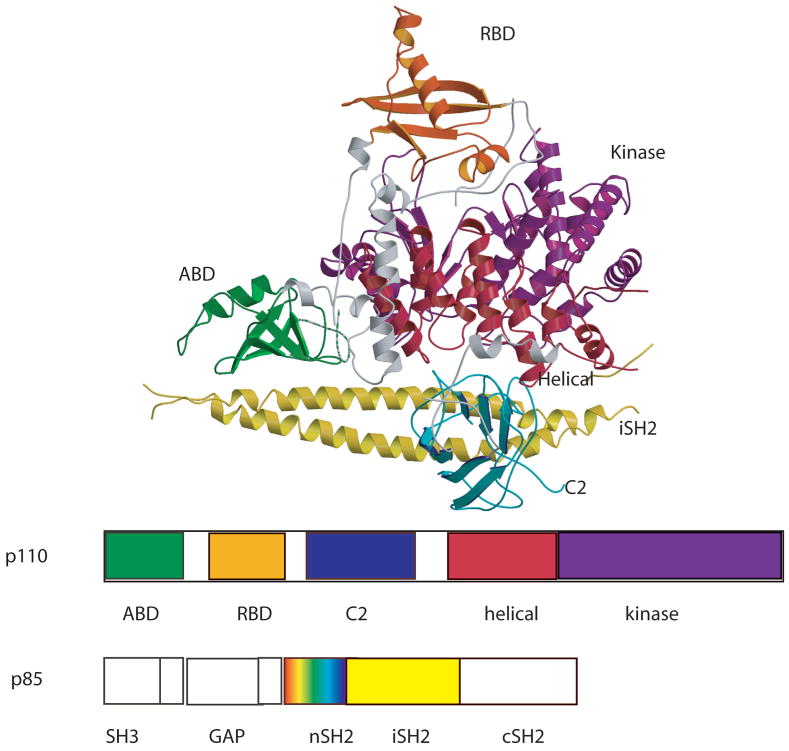
The p110α /niSH2 complex contains all five domains of the p110α subunit plus the coiled-coil (iSH2) and the nSH2 domains of p85α (Fig. 1). The structure has the overall shape of a sailing ship with the iSH2 forming the “hull” and the rest of the molecule forming a triangular “sail”. The helical and the kinase domain form the core of the molecule with the ABD at one extreme, the RBD at the very top, and the C2 domain at the center. The ABD, as expected interacts with the iSH2 of p85 (buried area 2237 Å2 (Fig. 3 a, c) but, unexpectedly so does the C2 domain (buried area 1233 Å2 (Fig. 3b,d). An additional interaction observed in this complex is between the ABD domain and the kinase domain. These domain-domain contacts are particularly important because they are the loci of most cancer associated mutations (see below).
Fig. 1. Ribbon diagram of the p110α/niSH2 heterodimer.
(A) Scheme of the domain organization. The same color coding is used throughout this article unless specified. Gray regions are linkers between domains. The ABD domain of p110α is green; RBD is orange; C2 is blue; helicase is red; kinase is purple. The iSH2 domain of p85α is colored yellow. The nSH2 domain of p85α (not shown; colored as a rainbow in the scheme of the construct), was not traced but it was modeled into weak density using the deposited coordinates.
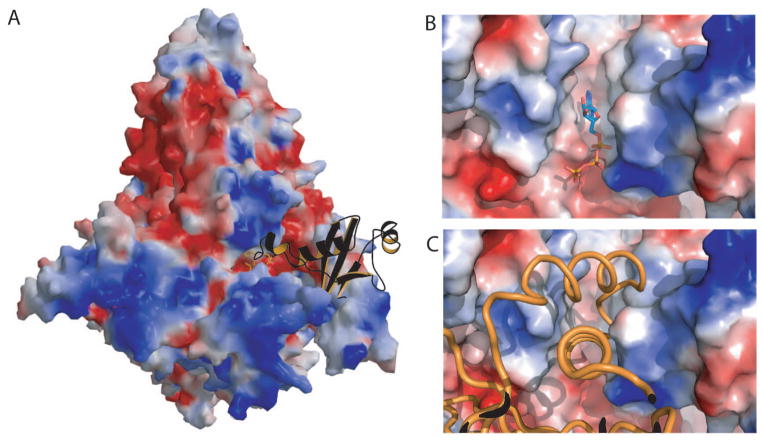
Fig. 3. Intermolecular contact in the p110α /p85 heterodimer crystal.
(A) Molecular surface of the PI3K colored as electrostatic charges showing the Ras binding domain of a neighboring molecule in the crystal (orange ribbon with black back-side) bound in the kinase domain active site. (B) Molecular surface colored as electrostatic potential showing the ATP (ball-and-stick representation) bound to PI3Kα . (C) Molecular surface colored according to the electrostatic potential showing the helix-loop-helix motif of the RBD of the neighboring molecule in the ATP binding site.
Description of the domains
The first two domains, ABD (residues 1-108) and RBD (residues 191–291), are small globular structures with an α/β topology. An 81 residue linker (109–190), containing two helices, connects the two domains. Both domains interact strongly with the kinase domain.
The RBD is followed by a linker (residues 292–329) composed by a short helix and a long coil that connects to the C2 domain (residues 330 to 480). C2 folds as a β-sandwich composed of two four-stranded antiparallel sheets and interacts not only with p110 domains but also with the iSH2 domain of p85. The main interaction of the iSH2 and the C2 domains involves H-bonds between Asp560 and Asn564 of iSH2 to Asn345 of C2 (Fig. 4b). Although the C2 domain is the least conserved of all p110 domains in class I PI3K enzymes (27% identity between α and γ), the two-sheet topology is preserved in the structures of the two isoforms 33, 37. Despite sharing the same overall fold, there are significant differences between the C2 domains of p110α and p110γ. Most differences between the C2 domains of p110α and p110γ occur in the loops connecting the β-strands. Some differences go beyond changes in conformation: the loop spanning residues 406 to 424, for example, is ten residues shorter in p110α than in p110γ. Differences in conformation between the loops of the two isoforms may be a consequence of the interaction of the C2 domain of p110α with the iSH2 domain of the regulatory subunit p85 (p85 is not the regulatory subunit of p110γ). Not only does this interaction not occur in p110γ, but the conformation that the loops adopt in p110γ makes it impossible for the iSH2 coiled-coil to fit in a position equivalent to that of the p110α /niSH2 complex.
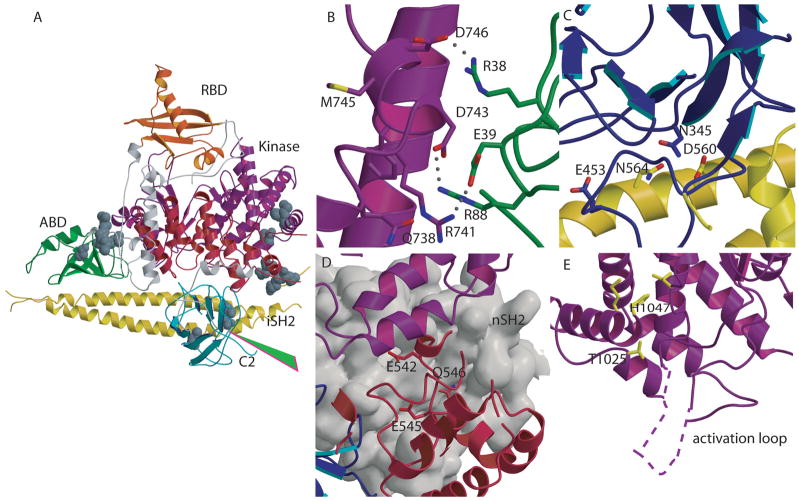
Fig. 4. Mutations in PIK3CA identified in human cancers.
(A) Location of representative mutations within p110α. Residues mutated in cancers are shown as CPK models. The start of the cancer associated truncation (residue 571 of p85) is shown by the green arrowhead. (B) Residues Arg38 and Arg88 , frequently mutated in cancers, are shown at the interface between the ABD and the kinase domains. (C) Contacts between the C2 and iSH2 domain in the p110α /p85 heterodimer. Asn345 of C2 and the residues within iSH2 (Asp560 and Asn564) with which it may interact are shown with a stick representation. (D) Residues in the helical domain commonly mutated in cancers (Glu542, Glu545, and Gln 546) are located at the interface with nSH2 (grey surface) in close proximity to the nSH2-kinase domain interface. (E) Mutations of the kinase domain, (Met 1043 and His1047), located near the C-terminal end of the activation loop, are shown in yellow. The portion of the activation loop between residues 941 to 950, not traced in the published structure, is shown as a dashed line.
A linker (residues 481 to 524) connects the C2 domain of p110α to an all α-helical domain (residues 525 to 696). The role of this helical domain is unclear, but in the structure it provides a bridge between the C2 domain and the kinase domain.
The kinase domain (residues 696 to 1068) folds as an α /β structure composed of two subdomains separated by a cleft that harbors the catalytic site of the enzyme. This architecture is reminiscent of that in other kinases. This similarity allows assignment of the catalytic and the activation loops of p110α to residues 912 to 920 and 933 to 957, respectively. The structure of this domain is highly conserved among Class I PI3Ks (rmsd between p110α and p110γ for 288 Cα atoms is 1.8 Å), especially for residues surrounding the binding pocket. The largest differences (rmsd 3.2 Å) occur in the location of the helix spanning residues 1032 to 1048. This difference is important because two positions in this helix are mutated with high frequency in cancers. The ATP binding site was identified by aligning the structure of the kinase domain of p110α with that of the same domain in the structure of the complex of p110γ with ATP. The high degree of similarity between the two structures in this region allowed an unambiguous location of the ATP. Interestingly, the ATP binding-site in the crystal structure of the p110α /niSH2 complex is occupied by a loop of the RBD domain of a neighboring molecule (residues 226 to 239) that appears to “mimic” ATP binding (Fig. 2).
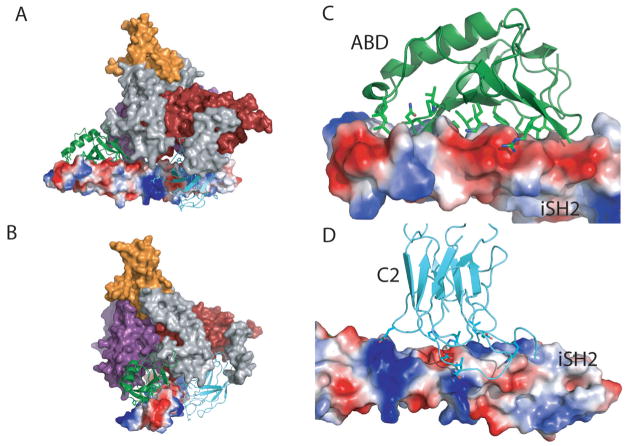
Fig. 2. Interaction between the iSH2 domain of p85 and the ABD and the C2 domain of p110.
(A) Molecular surface of the p110α /niSH2. The iSH2 is shown as a molecular surface colored according to electrostatic potential. The ABD and the C2 domain are shown as ribbons. (b) The same image rotated by 90°. (C) Close-up of the interaction between iSH2 and the ABD. (D) Close-up of the interaction between iSH2 and the C2 domain.
Cancer-associated mutations
Mutations in the ABD, C2, helical, and kinase domains of p110α (but not in p110γ) are observed with high frequency in cancers 14. It is thought that most if not all of these mutations result in enhanced enzymatic activity 14, 25. The structure of the p110α /niSH2 provides the first look at the location of the cancer-associates mutations in the relevant portions of the PI3Kα structure. Frequent mutations in the ABD domain include substitutions of Arg38 and Arg88. The most common mutations at these positions, Arg38Cys, Arg38His, and Arg88Gln, were thought to affect the interaction of the ABD with the iSH2 domain of p8536. The structures of the ABD/iSH2 and the p110α /niSH2, however, showed that residues 38 and 88 of ABD were not at the ABD/iSH2 interface (Fig. 4a,b). Instead, the structure of the p110α /niSH2 complex showed that these residues occur at an interface between the ABD and the kinase domain in which the two ABD residues are at H-bonding distance from residues in the kinase domain: Arg38 of Gln 738 and Asp743, and Arg88 of Asp746. Mutation of either arginine residue would disrupt the corresponding H-bond and possibly affect the conformation of the kinase domain in a way that changes the enzymatic activity.
Asn345 of the C2 is frequently mutated to lysine in cancers. Since C2 was thought to mediate the interaction of PI3K with the membrane 27, it was proposed that the mutation modified the interaction of the enzyme with the membrane. In the structure of the p110α /niSH2, however, Asn354 is at H-bonding distance of two residues of the iSH2 domain of p85, Asn564 and Asp560 (Fig. 4a,c). Interestingly, another common C2 mutation, Glu453Gln, is also present at the interface between C2 and iSH2. The most likely effect of these mutations appears to be the alteration of the interaction between the C2 domain of p110 and the iSH2 domain of p85 rather than changing the interaction of C2 with the membrane.
Glu542 and Glu545 of the helical domain are frequently mutated to lysine in cancers. These substitutions, which reverse the charge of the original residues, were proposed to occur in the interface between the helical domain of p110 and the nSH2 domain of p85. This proposal is supported by information from two sources. First, biochemical information suggested that Glu542 and Glu545 interact with Lys379 and Arg340 of the nSH2 of p8536, 38. Second, weak density allowed building a model of the nSH2 into the structure of the p110/niSH2 complex; these contacts were found to be present in this model (Fig. 4d). Furthermore, they occur in a region of the nSH2 domain that is also in contact with the kinase domain of p110, suggesting a mechanism by which these mutations can affect the activity of the enzyme. For example, partial disruption of the interaction between the nSH2 domain of p85 and the helical domain of p110 could reduce the inhibitory effect of the nSH2 on the kinase domain resulting in the observed increase in the activity.
One common mutation in cancers is the substitution of His1047 of the kinase domain of p110α by arginine. Interestingly, in p110γ Arg1076 is the residue equivalent to His1047 of p110α . Comparison of the two structures in this region may provide clues about the effect of this substitution. In both proteins this position is in a helix at the end of the activation loop. In p110α , His1047 forms a hydrogen bond with the carbonyl oxygen of Leu956, whereas Arg1076 in p110γ does not make an H-bond with the carbonyl of Leu987, the residue equivalent to Leu956 of p110α . Instead, it makes an H-bond with the carbonyl of Lys1000, a residue further removed along the chain. This change results in the displacement of the helix in which the residue is located by more than 3 Å despite the fact that both structures are highly similar in other regions. The change in the position of this helix, probably also present in the His1047Arg of p110α , most likely changes the conformation of the activation loop and modifies the interaction of the domain with the phosphatidylinositide substrates. Met1043Leu, a common but less frequent mutation, is located on the same helix and probably affects activity by a similar mechanism, (Fig. 4a, e).
Another mutation, T1025S, is close to the N-terminus of the catalytic loop (Fig. 4e) and may therefore alter the enzyme activity through a direct effect on the conformation of the catalytic loop.
Association with the lipid membrane
Phosphatidylinositides, the substrates of PI3Ks, are integral components of the plasma membrane. As part of the activation process, PI3Ks are recruited to the plasma membrane where they carry out the phosphorylation of PIP2. Since C2 domains were shown to be involved in the association of proteins with membranes, it was proposed that in p110γ loops of the C2 and the kinase domains provide the major contacts with the membrane. In p110α , the iSH2 is wedged between the C2 domain and the rest of the molecule. With this configuration, if C2 and the kinase domain interact with the membrane, iSH2 must also interact, probably contributing the major fraction of the contact surface. The two loops of the kinase domain (residues 723 to 729 and 863 to 867) and the side of the iSH2 coiled-coil proposed to interact with the membrane are lined with positively charged residues: Lys 723, Lys729, Lys 863, and Lys867 of the kinase domain of p110, and lysines 447, 448, 480, 530, 532, 551, and 561, and arginines 461, 465, 472, 480, 523, 534, 543, and 544 of the iSH2 domain of p85. It is possible that Arg349, Lys410, Arg412, Lys413, and Lys416 of the C2 domain of p110 also interact with the membrane
v-p3k
The retrovirus avian sarcoma virus 16 (ASV16) carries an oncogene v-p3k which has potent transforming activity 11. The v-p3k protein differs from its cellular counterpart, c-p3k or p110α of chicken, by a 14-amino acid N-terminal deletion and fusion to the viral Gag sequence, as well as by the following replacements: Glu52Lys, Glu116Lys, Glu469Gly, and Asn560Lys. Glu52, Glu116, and Glu469 are conserved between human and chicken p110α. An in vitro study showed elevated levels of PI(3,4)P2 and PI(3,4,5)P3 and activation of AKT in cells transformed with v-p3k 11.
One of the altered residues in v-p3k, Gly469, is located at the interface between the C2 domain and iSH2. In fact, Glu469 in the p110α/niSH2 structure may be forming a hydrogen bond to Arg481 of iSH2. The substitution of Glu469 by Gly in v-p3k might lead to effects similar to those of the C2 domain oncogenic p110α mutations observed in human cancers. Alternatively, overexpression of v-p3k may by itself lead to a high level of catalytic activity because of the lack of inhibition by the endogenous regulatory p85 subunit. It has been shown that an N-terminal extension stabilizes monomeric p110α 38 suggesting that the fusion of the N-terminus to gag in v-p3k may stabilize the protein in the absence of p85.
Drug Design Implications: p110α as a drug target
The existence of oncogenic p110α gain-of-function-mutants makes this protein an attractive therapeutic target 14. Since PI3Ks control wide range of physiological functions, ideally it would be desirable to inhibit only p110α , the isoform mutated in cancers. The availability of wide spectrum PI3K inhibitors such as wortmannin and LY294002, which inhibit PI3Kα provide a starting point 39 for the development isoform specific inhibitors. Structural differences among the isoforms can be exploited for that purpose. For example, comparison of the ATP binding sites of p110α and p110γ, shows differences in the loop spanning residues 771 and 779 ( IMSSAKRPL) in p110α and residues 803 to 811 (VMASKKKPL) in p110γ In addition, the location of the cancer associated mutations at domain interfaces such as the Glu543 “hot spot” in the helical domain points to the possibility of developing not only for inhibitors for the ATP binding site but also to compounds that disrupt protein-protein interactions between domains. Since ATP-kinase binding-sites are structurally similar, avoiding such site can maximize the likelihood of reducing cross-reactivity with kinases and other ubiquitous ATPases. The new available structure presents a new roadmap for the design of mutant specific inhibitors.
Acknowledgments
Support was provided by the Virginia and D. K. Ludwig Fund for Cancer Research. NIH grants CA 43460, GM066895 to L.M.A, and GM07309 and GM 07184 to D.M. We thank Dr. Bert Vogelstein for advice and for critical reading of the manuscript.
Footnotes
The authors declare no financial or conflicts of interest in the contents of this paper
References
- 1.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 2.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 3.Geering B, Cutillas PR, Nock G, Gharbi SI, Vanhaesebroeck B. Class IA phosphoinositide 3-kinases are obligate p85-p110 heterodimers. PNAS. 2007:0700373104. doi: 10.1073/pnas.0700373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katso R, Okkenhaug K, Ahmadi K, White S, Timms J, Waterfield MD. Cellular function of phosphoinositide 3-kinases: implications for development, homeostasis, and cancer. Annu Rev Cell Dev Biol. 2001;17:615–75. doi: 10.1146/annurev.cellbio.17.1.615. [DOI] [PubMed] [Google Scholar]
- 5.Sugimoto Y, Whitman M, Cantley LC, Erikson RL. Evidence that the Rous sarcoma virus transforming gene product phosphorylates phosphatidylinositol and diacylglycerol. Proc Natl Acad Sci U S A. 1984;81:2117–21. doi: 10.1073/pnas.81.7.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whitman M, Kaplan DR, Schaffhausen B, Cantley L, Roberts TM. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature. 1985;315:239–42. doi: 10.1038/315239a0. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan DR, Whitman M, Schaffhausen B, Raptis L, Garcea RL, Pallas D, Roberts TM, Cantley L. Phosphatidylinositol metabolism and polyoma-mediated transformation. Proc Natl Acad Sci U S A. 1986;83:3624–8. doi: 10.1073/pnas.83.11.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitman M, Downes CP, Keeler M, Keller T, Cantley L. Type I phosphatidylinositol kinase makes a novel inositol phospholipid, phosphatidylinositol-3-phosphate. Nature. 1988;332:644–6. doi: 10.1038/332644a0. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter CL, Duckworth BC, Auger KR, Cohen B, Schaffhausen BS, Cantley LC. Purification and characterization of phosphoinositide 3-kinase from rat liver. J Biol Chem. 1990;265:19704–11. [PubMed] [Google Scholar]
- 10.Hiles ID, Otsu M, Volinia S, Fry MJ, Gout I, Dhand R, Panayotou G, Ruiz-Larrea F, Thompson A, Totty NF, et al. Phosphatidylinositol 3-kinase: structure and expression of the 110 kd catalytic subunit. Cell. 1992;70:419–29. doi: 10.1016/0092-8674(92)90166-a. [DOI] [PubMed] [Google Scholar]
- 11.Chang HW, Aoki M, Fruman D, Auger KR, Bellacosa A, Tsichlis PN, Cantley LC, Roberts TM, Vogt PK. Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science. 1997;276:1848–50. doi: 10.1126/science.276.5320.1848. [DOI] [PubMed] [Google Scholar]
- 12.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng DH, Tavtigian SV. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–62. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 13.Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol. 2004;22:2954–63. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 14.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, Vogelstein B, Velculescu VE. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 15.Broderick DK, Di C, Parrett TJ, Samuels YR, Cummins JM, McLendon RE, Fults DW, Velculescu VE, Bigner DD, Yan H. Mutations of PIK3CA in anaplastic oligodendrogliomas, high-grade astrocytomas, and medulloblastomas. Cancer Res. 2004;64:5048–50. doi: 10.1158/0008-5472.CAN-04-1170. [DOI] [PubMed] [Google Scholar]
- 16.Bachman KE, Argani P, Samuels Y, Silliman N, Ptak J, Szabo S, Konishi H, Karakas B, Blair BG, Lin C, Peters BA, Velculescu VE, Park BH. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther. 2004;3:772–5. doi: 10.4161/cbt.3.8.994. [DOI] [PubMed] [Google Scholar]
- 17.Lee JW, Soung YH, Kim SY, Lee HW, Park WS, Nam SW, Kim SH, Lee JY, Yoo NJ, Lee SH. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene. 2005;24:1477–80. doi: 10.1038/sj.onc.1208304. [DOI] [PubMed] [Google Scholar]
- 18.Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB, Phillips WA. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004;64:7678–81. doi: 10.1158/0008-5472.CAN-04-2933. [DOI] [PubMed] [Google Scholar]
- 19.Levine DA, Bogomolniy F, Yee CJ, Lash A, Barakat RR, Borgen PI, Boyd J. Frequent mutation of the PIK3CA gene in ovarian and breast cancers. Clin Cancer Res. 2005;11:2875–8. doi: 10.1158/1078-0432.CCR-04-2142. [DOI] [PubMed] [Google Scholar]
- 20.Saal LH, Holm K, Maurer M, Memeo L, Su T, Wang X, Yu JS, Malmstrom PO, Mansukhani M, Enoksson J, Hibshoosh H, Borg A, Parsons R. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65:2554–9. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Helland A, Holm R, Kristensen GB, Borresen-Dale AL. PIK3CA mutations in advanced ovarian carcinomas. Hum Mutat. 2005;25:322. doi: 10.1002/humu.9316. [DOI] [PubMed] [Google Scholar]
- 22.Vogt PK, Kang S, Elsliger MA, Gymnopoulos M. Cancer-specific mutations in phosphatidylinositol 3-kinase. Trends Biochem Sci. 2007;32:342–9. doi: 10.1016/j.tibs.2007.05.005. [DOI] [PubMed] [Google Scholar]
- 23.Bos JL. Ras oncogenes in human cancer: a review. Cancer Research. 1989;49:4682–9. [PubMed] [Google Scholar]
- 24.Ikenoue T. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005;65:4562–7. doi: 10.1158/0008-5472.CAN-04-4114. [DOI] [PubMed] [Google Scholar]
- 25.Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A. 2005;102:802–7. doi: 10.1073/pnas.0408864102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A. 2006;103:1475–9. doi: 10.1073/pnas.0510857103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0701005104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carson JD, Van Aller G, Lehr R, Sinnamon RH, Kirkpatrick RB, Auger KR, Dhanak D, Copeland RA, Gontarek RR, Tummino PJ, Luo L. Effects of oncogenic p110alpha subunit mutations on the lipid kinase activity of phosphoinositide 3-kinase. Biochem J. 2008;409:519–24. doi: 10.1042/BJ20070681. [DOI] [PubMed] [Google Scholar]
- 29.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 30.Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell. 2003;4:257–62. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 31.Stephens L, Williams R, Hawkins P. Phosphoinositide 3-kinases as drug targets in cancer. Curr Opin Pharmacol. 2005;5:357–65. doi: 10.1016/j.coph.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 32.Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol Cell. 2000;6:909–19. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 33.Walker EH, Perisic O, Ried C, Stephens L, Williams RL. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature. 1999;402:313–20. doi: 10.1038/46319. [DOI] [PubMed] [Google Scholar]
- 34.Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, Hawkins PT, Stephens L, Eccleston JF, Williams RL. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103:931–43. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 35.Dhand R, Hara K, Hiles I, Bax B, Gout I, Panayotou G, Fry MJ, Yonezawa K, Kasuga M, Waterfield MD. PI 3-kinase: structural and functional analysis of intersubunit interactions. Embo J. 1994;13:511–21. doi: 10.1002/j.1460-2075.1994.tb06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miled N, Yan Y, Hon W-C, Perisic O, Zvelebil M, Inbar Y, Schneidman-Duhovny D, Wolfson HJ, Backer JM, Williams RL. Mechanism of Two Classes of Cancer Mutations in the Phosphoinositide 3-Kinase Catalytic Subunit. Science. 2007;317:239–42. doi: 10.1126/science.1135394. [DOI] [PubMed] [Google Scholar]
- 37.Huang CH, Mandelker D, Schmidt-Kittler O, Samuels Y, Velculescu VE, Kinzler KW, Vogelstein B, Gabelli SB, Amzel LM. The structure of a human p110α/p85α complex elucidates the effects of oncogenic PI3Kα mutations. Science. 2007;318:1744–8. doi: 10.1126/science.1150799. [DOI] [PubMed] [Google Scholar]
- 38.Yu J, Wjasow C, Backer JM. Regulation of the p85/p110alpha phosphatidylinositol 3'-kinase. Distinct roles for the n-terminal and c-terminal SH2 domains. J Biol Chem. 1998;273:30199–203. doi: 10.1074/jbc.273.46.30199. [DOI] [PubMed] [Google Scholar]
- 39.Ward SG, Finan P. Isoform-specific phosphoinositide 3-kinase inhibitors as therapeutic agents. Curr Opin Pharmacol. 2003;3:426–34. doi: 10.1016/s1471-4892(03)00078-x. [DOI] [PubMed] [Google Scholar]