Abstract
Purpose
Atypical protein kinase Cι (PKCι) is an oncogene in non – small cell lung cancer (NSCLC). Here, we identify four functional gene targets of PKCι in lung adenocarcinoma (LAC), the most prominent form of NSCLC.
Experimental Design
Three independent public domain gene expression data sets were interrogated to identify genes coordinately expressed with PKCι in primary LAC tumors. Results were validated by QPCR in an independent set of primary LAC tumors. RNAi-mediated knockdown of PKCι and the target genes was used to determine whether expression of the identified genes was regulated by PKCι, and whether these target genes play a role in anchorage-independent growth and invasion of LAC cells.
Results
Meta-analysis identified seven genes whose expression correlated with PKCι in primary LAC. Subsequent QPCR analysis confirmed coordinate overexpression of four genes (COPB2, ELF3, RFC4, and PLS1) in an independent set of LAC samples. RNAi-mediated knockdown showed that PKCι regulates expression of all four genes in LAC cells, and that the four PKCι target genes play an important role in the anchorage-independent growth and invasion of LAC cells. Meta-analysis of gene expression data sets from lung squamous cell, breast, colon, prostate, and pancreas carcinomas, as well as glioblastoma, revealed that a subset of PKCι target genes, particularly COPB2 and RFC4, correlate with PKCι expression in many tumor types.
Conclusion
Meta-analysis of public gene expression data are useful in identifying novel gene targets of oncogenic PKCι signaling. Our data indicate that both common and cell type – specific signaling mechanisms contribute to PKCι-dependent transformation.
Lung cancer is the leading cause of cancer death in the United States. In 2007, there were an estimated 213,380 new cases and 160,390 deaths from lung cancer (1). The vast majority (~90%) of lung cancer is related to tobacco use (2), the predominant risk factor for this disease. Over the past 30 years, the 5-year survival rate of lung cancer patients has only increased from 7% to 15%, indicating a dire need for more effective means of prevention, diagnosis, prognosis, and treatment. Approximately 80% of lung cancer is classified as NSCLC, which is subclassified into squamous cell carcinoma, lung adenocarcinoma (LAC), and large cell carcinoma. LAC is the most prevalent subclass of lung cancer, accounting for ~50% of non–small cell lung cancer (NSCLC) cases and >40% of all lung cancers (3). LACs diagnosed at an early stage are often treated with surgery and radiotherapy, whereas patients with unresectable metastatic disease often receive combination chemotherapy (4). Unfortunately, LACs often become resistant to chemotherapy, relapse, and become fatal, underscoring the need for a better understanding of oncogenic signaling mechanisms underlying LAC and the identification of new therapeutic targets for treatment of this disease.
We recently showed that the atypical protein kinase C isozyme PKCι, is an oncogene in human NSCLC (5). PKCι is overexpressed in the majority of primary NSCLC tumors and PKCι expression is predictive of poor clinical outcome independent of tumor stage (5). The PKCι gene, PRKCI, is a target for tumor-specific gene amplification in a significant subset of NSCLC tumors, and gene amplification drives PKCι expression in these tumors. PKCι plays a requisite role in NSCLC cell transformation (5, 6). Genetic disruption of PKCι expression inhibits anchorage-independent growth and invasion of NSCLC cells in vitro, and tumorigenicity in vivo (6). PKCι is a critical downstream effector of oncogenic K-ras, the major oncogenic lesion in NSCLC (5–7).
Given its importance in oncogenic PKCι signaling in lung cancer, we initiated a study to identify downstream targets of PKCι that play a functional role in NSCLC transformation. In the present study, we used a meta-analysis of gene expression data of primary LACs to identify genes whose expression correlates with that of PKCι. Our data show the utility of this approach to identify novel genes involved in oncogenic PKCι signaling and NSCLC biology. These genes also represent potential therapeutic targets.
Materials and Methods
Data collection
Gene expression data from three independent published microarray analyses of LAC tumors were obtained from public sources. The three data sets were as follows: the Michigan data set from Beer et. al. (8) included 86 primary LACs, the Harvard data set from Bhattacharjee et. al. (9) included 136 primary LACs, and the Stanford data set from Garber et. al. (10) included 35 primary LACs. The primary data files were downloaded and rank ordered into tertiles based on PKCι expression. The expression data were categorized into three tertiles: LACs with High, Medium, and Low PKCι expression. We defined genes as coordinately expressed with PKCι if their expression was significantly increased in the High PKCι expression tertile compared with the Low PKCι expression tertile, based on two criteria: (a) Student’s t test, P < 0.05, and (b) ≥1.4- or <0.6-fold difference in mean gene expression levels between the two tertiles. Because three different expression platforms were used for analysis of these samples, we did not attempt to reanalyze the data but rather worked directly from the processed expression data within the public domain.
Cell lines
Human A549, H358, and H1437 LAC cell lines were obtained from American Type Culture Collection and maintained in adherent culture as suggested by the supplier.
Lentiviral RNA interference–mediated gene knockdown
Lentiviral vectors carrying short hairpin RNA interference (RNAi) against human PKCι, COPB2, ELF3, PLS1, and RFC4 were obtained from Sigma-Aldrich Mission shRNA library. A nontarget control lentiviral vector containing a short hairpin that does not recognize any human or mouse genes (NT-RNAi) was used as a negative control in all RNAi experiments. shRNA lentiviral particles were generated as described previously (11). A549, H358, and H1437 cells were stably infected with lentiviral vectors containing short hairpin RNAi–targeting gene of interest or NT-RNAi and selected using 5 μg/mL puromycin as described previously (11). The RNAi target sequences used in this study are available upon request. Immunoblot analysis of stable transfectants for PKCα, PKCι, PKCζ, PKCδ, PKCε, and actin were done as described previously (11). Antibodies were from the following sources: Transduction Laboratories [PKCι (#610176) and PKCδ (#610397)], Santa Cruz [PKCα (#8393) and PKCε (#214)], and Cell Signaling [PKCζ (#9372) and β-actin (#4967)].
RNA isolation and QPCR
Total RNA was isolated from lung cancer cell lines and primary lung adenocarinomas using RNAqueous (Ambion). PKCι, COPB2, ELF3, PLS1, and RFC4 gene expression assays were purchased from Applied Biosystems to measure mRNA abundance. Analysis was done on an Applied Biosystems 7900 thermal cycler, and data were evaluated using the RQ Manager 1.2 software as described previously (12).
Soft agar growth assays
Anchorage-independent growth was evaluated by the ability of A549 and H358 cell transfectants to form colonies in soft agar as described previously (6). After 4 wk in culture, cells were fixed in 100% methanol and stained with Giemsa. Soft agar colonies were photographed using El Logic100 Imaging System (Eastman Kodak Co.) and quantified using Image Pro Plus 5.0 (Media Cybernetics).
Cellular invasion assay
Cell invasion was assayed using Matrigel-coated Transwell cell culture chambers (Becton Dickinson) as described previously (13). After staining, invaded cells were photographed using an Olympus microscope and quantified using Image Pro Plus 5.0 (Media Cybernetics).
Statistical analysis
The Student’s t test was used to evaluate the statistical significance of the means of two groups. Kendall Rank Correlation was used to evaluate the strength of the association between two variables. SigmaStat software was used for statistical analyses, and P values of <0.05 were considered statistically significant.
Analysis of coordinate gene expression in other tumor types
Micro-array expression data for breast, colon, and prostate cancers were downloaded from the Gene Expression Omnibus (GSE2109), including all 47 prostate adenocarcinoma samples, a randomly selected 82 sample subset of the 195 colon adenocarcinoma samples, and a randomly selected 112 sample subset of the 248 breast ductal carcinoma samples. Microarray expression data for 27 pancreatic carcinomas were downloaded from Array Express (E-MEXP-1121), and microarray expression data for 44 glioblastoma samples were downloaded from The Cancer Genome Atlas (archive files broad.mit.edu_GBU.HT_HG-U133A.1.2.0, broad.mit.edu_GBU.HT_HG-U133A.2.1.0, and broad.mit.edu_ GBU.HT_HG-U133A.3.1.0). Raw cel files were processed independently for each cancer type using RMA background correction, fastlo normalization, affinities-only PM correction, and median polish for summarization. For each tumor type, the expression data were rank-ordered based on PKCι expression, and High, Medium, and Low sample tertiles were identified. Expression levels of the four target genes (COPB2, ELF3, RFC4, and PLS1) in the PKCι High tertile were compared with the expression in the PKCι Low tertile. Significant changes in expression were measured using a two-tailed, unequal variance, t test (P < 0.05). The t test was computed using the Statistics:TTest PERL library.
Results and Discussion
We recently showed that PKCι is an oncogene in NSCLC (5). To identify potential downstream targets of PKCι, we did a meta-analysis of public domain gene expression data of primary human LACs. Three independent gene expression data sets (hereafter called the Michigan, Stanford, and Harvard data sets, respectively) were downloaded from public sources as described in Materials and Methods. Our strategy was to interrogate these data sets for genes whose expression is coordinately regulated with PKCι in LAC. For this purpose, we force ranked the samples from each data set based on PKCι expression, binned the samples into tertiles based on PKCι expression, and interrogated the data sets for genes whose expression correlated with PKCι (either negatively or positively). This process was repeated for all three data sets, and only genes that correlated with PKCι in all three data sets were considered further. Seven genes were identified that satisfied the inclusion criteria, COPB2, ELF3, PLS1, RFC4, ALCAM, EVPL, and TACSTD2. Table 1 summarizes the results of this analysis, including the fold-change and t test P values obtained from comparing the expression of each of these genes in the high (H) and low (L) sample set tertiles. Each gene showed a positive correlation with PKCι expression, and no genes were identified from this analysis whose expression was negatively correlated with PKCι.
Table 1.
Identification of genes coordinately expressed with PKCι in LACs
| Genes | Michigan
|
Stanford
|
Harvard
|
|||
|---|---|---|---|---|---|---|
| Gene symbol | High/Low | P | High/Low | P | High/Low | P |
| PRKCI | 3.41 | 2 × 10−9 | 5.48 | 4 × 10−11 | 2.84 | 5 × 10−14 |
| COPB2 | 1.40 | 0.006 | 1.99 | 0.002 | 1.42 | 2 × 10−4 |
| ELF3 | 1.67 | 8 × 10−4 | 3.24 | 6 × 10−7 | 1.42 | 0.010 |
| PLS1 | 1.92 | 0.007 | 2.47 | 1 × 10−4 | 1.41 | 0.008 |
| RFC4 | 1.49 | 0.037 | 4.02 | 9 × 10−7 | 1.44 | 0.007 |
| ALCAM | 1.97 | 0.016 | 1.95 | 0.024 | 1.40 | 0.023 |
| EVPL | 1.56 | 0.012 | 1.78 | 0.009 | 1.80 | 0.004 |
| TACSTD2 | 1.52 | 0.008 | 6.71 | 2 × 10−7 | 1.50 | 0.003 |
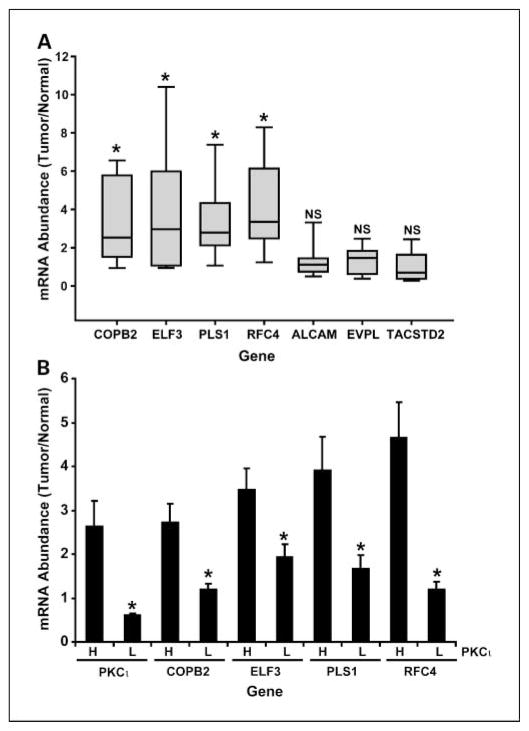
Because we were interested in identifying genes that might be functionally important in oncogenic PKCι signaling, we chose to focus our attention on genes whose expression, such as that of PKCι, was induced in LAC tumors when compared with matched normal lung tissue. For this purpose, we measured the expression of each of the 7 genes identified above in a test set of 15 primary LAC samples for which matched normal RNA was available (Fig. 1A). Four of the seven genes identified in our original analysis were significantly overexpressed in LAC tumors when compared with matched normal tissues (COPB2, ELF3, PLS1, and RFC4). Expression of ALCAM, EVPL, and TACSTD2 were not significantly elevated in LAC tumors when compared with matched normal lung tissue and therefore were not analyzed further. We further validated the coordinate overexpression of the 4 identified genes with PKCι in an expanded set of 60 primary LAC samples and corresponding matched normal lung tissues. PKCι expression was measured by QPCR, samples binned into tertiles based on PKCι expression, and expression levels of the four target genes determined by QPCR. The data were then evaluated for an association between PKCι expression and that of each of the four target genes (Fig. 1B). Our analysis revealed that expression of each of the four genes exhibited a statistically significant association with PKCι expression. Thus, four of the seven genes originally identified as being coordinately expressed with PKCι in LAC were subsequently confirmed by QPCR in an independent set of LAC tumors.
Fig. 1.
Identification of gene coordinately expressed with PKCι in LAC. A, expression of seven genes exhibiting coordinate expression with PKCι in primary LAC tumors. Seven genes identified through meta-analysis of public domain gene expression data from LAC cases (see Table 1) were analyzed for overexpression in 15 primary LAC tumors. Four of the seven genes are significantly overexpressed in LACs when compared with matched normal controls. mRNA abundance was measured by QPCR and expressed as fold-change (tumor/normal). Boxes, upper and lower quartile values; line within each box, median value. *, significant difference between tumor and normal with a P value of <0.05. NS, not significant. B, the abundance of each of the indicated genes was determined by QPCR in 60 primary LAC and matched control samples and expressed as fold change (tumor/normal). The 60 samples were rank ordered and binned into upper tertile (H, high PKCι) and lower tertile (L, low PKCι) based on PKCι expression. The data represent the expression of each gene in the high and low PKCι groups. Columns, mean; bars, SE. *, statistically significant difference in expression of the indicated gene in the low PKCι group when compared with the high PKCι group; P < 0.05.
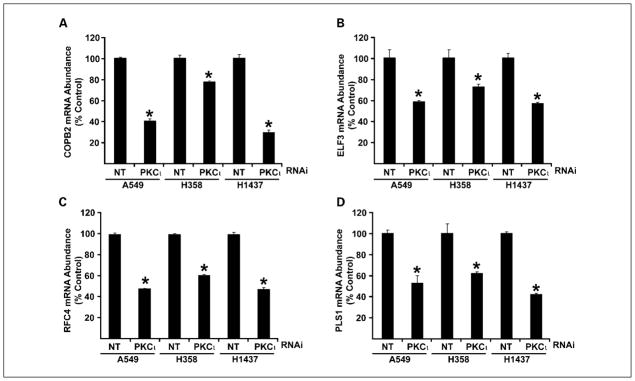
One purpose of our analysis was to identify potential downstream effectors of oncogenic PKCι signaling in LAC. Therefore, we next assessed whether the expression of any of the four PKCι-associated genes was regulated by PKCι in LAC cell lines. For this purpose, we generated three LAC cell lines in which PKCι expression was knocked down by RNAi using RNAi reagents and methods described and characterized previously (11). RNAi-mediated knockdown of PKCι caused significant inhibition of PKCι expression in each of the three LAC cell lines (Fig. 2A). Immunoblot analysis of lysates from A549, H358, and H1437 cells treated with NT and PKCι RNAi revealed a significant decrease in expression of PKCι protein but no demonstrable change in expression of PKCα, PKCζ, PKCδ, or PKCε (Fig. 2B), demonstrating specific knockdown of PKCι expression by our PKCι RNAi construct. We previously showed that PKCι is required for anchorage-independent growth of NSCLC cell lines in vitro and tumorigenicity in vivo (5, 6). Therefore, we assessed the effect of PKCι knockdown on anchorage-independent growth in soft agar (Fig. 2C). As expected, each of the three LAC cell lines in which PKCι was knocked down exhibited a significant inhibition of anchorage-independent growth when compared with nontarget control cells. Having shown effective knockdown of PKCι and consequent inhibition of PKCι-dependent transformed growth, we next assessed the effect of PKCι knockdown on the expression of the four target genes. As seen in Fig. 3, PKCι knockdown led to a significant inhibition in expression of each of the four target genes COPB2 (Fig. 3A), ELF3 (Fig. 3B), PLS1 (Fig. 3C), and RFC4 (Fig. 3D) in all three LAC cell lines. Our data indicate that PKCι regulates the expression of each of the four target genes in multiple LAC cell lines.
Fig. 2.
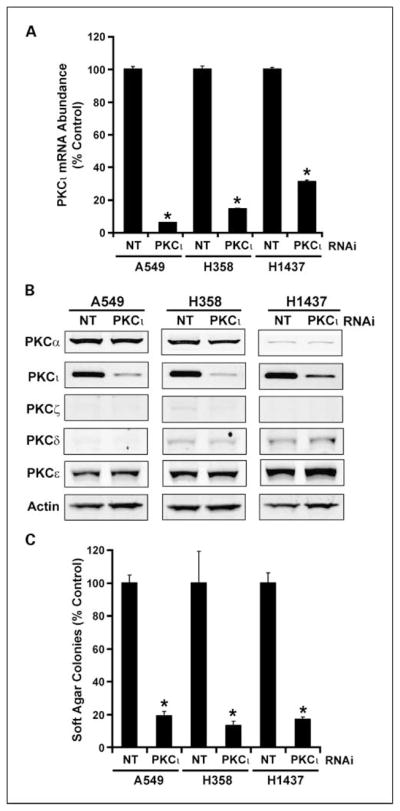
RNAi-mediated knockdown of PKCι inhibits anchorage-independent growth of LAC cell lines in soft agar. A549, H358, and H1437 LAC cell lines were stably transfected with either a nontarget (NT) lentiviral RNAi construct of a PKCι-RNAi construct. A, expression of PKCι mRNA in NT and PKCι-RNAi cells. *, statistically significant decrease in PKCι mRNA abundance when compared with NT control. Data are expressed as % NT control; n = 4; P < 0.05. B, PKCι-RNAi leads to selective knockdown of PKCι protein expression. Immunoblot analysis of total cell lysates from A549, H358, and H1437 treated with NT or PKCι-RNAi for PKCα, PKCι, PKCζ, PKCδ, PKCε, and actin. C, RNAi-mediated knockdown of PKCι inhibits anchorage-independent growth of LAC cell lines. Anchorage-independent growth in soft agar was assessed as described previously (6). Columns, mean (n = 4); bars, SE. Data are expressed as % NT control. *, statistically significant difference from NT control; P < 0.05.
Fig. 3.
PKCι regulates the expression of COPB2, ELF3, PLS1, and RFC4 in LAC cells. A549, H358, and H1437 cells transfected with either NT or PKCι-RNAi were analyzed for expression of the four genes that are coordinately overexpressed with PKCι in LAC tumors. RNAi-mediated knockdown of PKCι causes a statistically significant decrease in the expression of COPB2 (A), ELF3 (B), RFC4 (C), and PLS1 (D) in all three LAC cell lines. Data are expressed as %NT control; columns, mean (n = 4); bars, SE. *, statistically significant difference when compared with the corresponding NT control. P < 0.05.
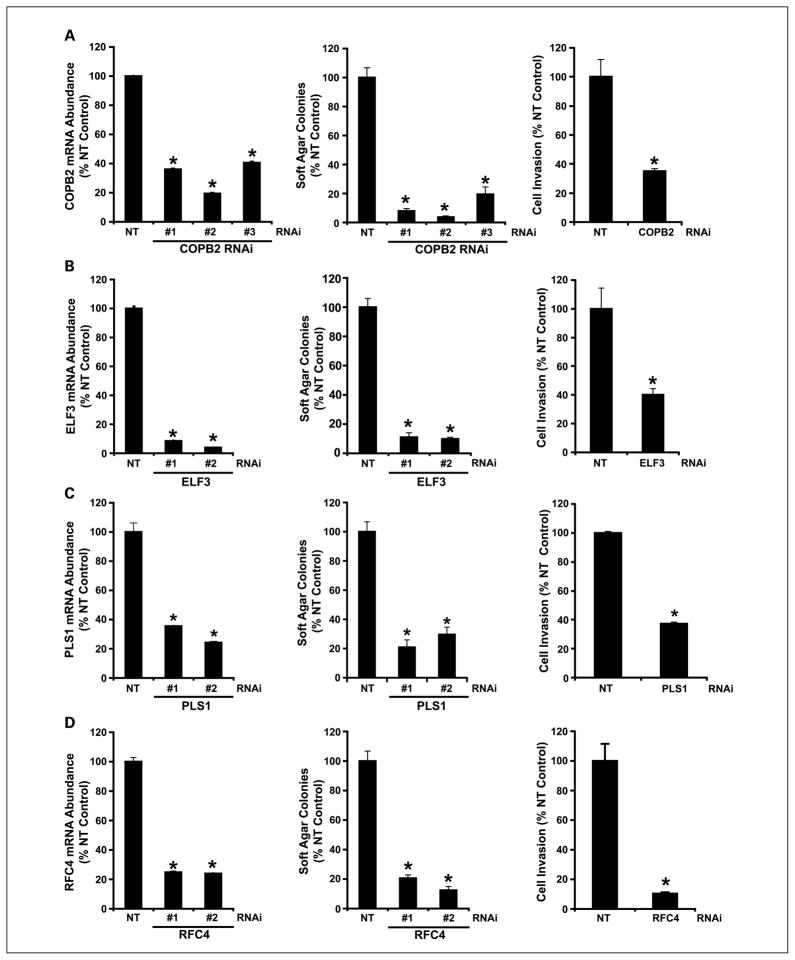
We next assessed whether the four PKCι target genes play a functional role in LAC cell transformation (Fig. 4). For this purpose, we established lentiviral RNAi reagents that target each gene. A549 cells were stably transfected with three lentiviral RNAi constructs directed against COPB2, and stably transfected cell populations were analyzed by QPCR for expression of COPB2 mRNA by QPCR (Fig. 4A). Each of the RNAi constructs significantly inhibited COPB2 mRNA abundance when compared with a nontarget RNAi control. A549/COPB2 RNAi cells were assessed for their ability to grow as colonies in soft agar (Fig. 4A). Each of the COPB2 RNAi transfectants exhibited a significant inhibition of anchorage-independent growth in soft agar when compared with nontarget control cells. The inhibition of soft agar growth was commensurate with the level of inhibition of COPB2 expression. A549 cells expressing the most effective COPB2 RNAi construct (COPB2 #2) were also assessed for ability to invade through Matrigel-coated chambers (Fig. 4A). A significant decrease in cellular invasion was observed in COPB2 knockdown cells when compared with NT control cells. We conclude that COPB2 plays a significant role in the transformed phenotype of A549 LAC cells.
Fig. 4.
COPB2, ELF3, PLS1, and RFC4 are functionally important for LAC cell transformation. A549 cells were stably transfected with multiple independent RNAi constructs targeting COPB2 (A), ELF3 (B), PLS1 (C), RFC4 (D), or a NT control RNAi construct. Target gene mRNA abundance was determined by QPCR in A549 cells expressing NT or target gene RNAi. Columns, mean (n = 4) and are presented as % NT control; bars, SE. *, statistically significant difference when compared with NT control cells. P < 0.05. The effect of target RNAi on anchorage-independent growth in soft agar and cellular invasion through Matrigel-coated chambers was assessed as previously described (13). Data are presented as % NT control; columns, mean (n = 4); bars, SE. *, statistically significant difference from NT control; P < 0.05.
A similar analysis involving RNAi-mediated knockdown of the three other PKCι regulated genes, ELF3 (Fig. 4B), PLS1 (Fig. 4C), and RFC4 (Fig. 4D), was also conducted. In each case, multiple RNAi constructs targeting the gene of interest induced significant inhibition of target gene expression and a commensurate decrease in anchorage-independent growth and cellular invasion of A549 cells through Matrigel. Taken together, these data show that each of these PKCι-regulated genes plays a functional role in NSCLC cell transformation. Similar results were obtained in H358 cells (data not shown), indicating that the observed effects are not specific to A549 cells.
Having shown the utility of using meta-analysis of gene expression data to identify relevant targets of oncogenic PKCι in LAC, we next wished to assess whether these genes are also PKCι targets in other major tumor types. Therefore, we obtained public gene expression data from the public domain for lung squamous cell carcinoma, carcinomas of the breast, colon, prostate, and pancreatic, as well as glioblastoma, and analyzed these data sets for coordinate expression of the four PKCι-regulated genes with PKCι (Table 2). Of the four genes identified in LAC, COPB2 and RFC4 showed statistically significant coordinate expression with PKCι in several other tumor types. COPB2 showed highly significant coordinate expression with PKCι in all major tumor types tested, and RFC4 showed coordinate expression in a majority of tumor types. ELF3 showed coordinate overexpression with PKCι in LAC and glioblastoma, whereas PLS1 exhibited coordinate expression with PKCι only in LAC. Taken together, these data indicate that PKCι regulates oncogenic signaling through both common and tumor type–specific mechanisms.
Table 2.
Coordinate expression of PKCι target genes in major cancer types
| Tumor type (number) | COPB2 | RFC4 | ELF3 | PLS1 |
|---|---|---|---|---|
| Lung squamous cell (129) | 2 × 10−6 | 2 × 10−8 | NS | NS |
| Breast (112) | 0.016 | 0.003 | NS | NS |
| Colon (82) | 0.008 | NS | NS | NS |
| Prostate (47) | 0.02 | NS | NS | NS |
| Pancreas (27) | 0.05 | NS | NS | NS |
| Glioblastoma (44) | 0.016 | NS | 0.04 | NS |
NOTE: P values are listed for the correlation between expression of PKCι and the indicated target gene.
Abbreviation: NS, not significant.
Our analysis identified four genes whose expression is regulated in human tumors by oncogenic PKCι and whose function seem to be involved in human LAC cell transformation; COPB2, RFC4, PLS1, and ELF3. Our data suggest possible key signaling pathways that may contribute to PKCι-dependent transformation. COPB2 is a subunit of the coatomer, a cytoplasmic protein complex that binds dilysine motifs and associates with Golgi nonclathrin–coated vesicles (14). The coatomer functions to mediate biosynthetic protein transport from the endoplasmic reticulum via the Golgi up to the trans-Golgi network. The coatomer is also required for Golgi membrane budding and is essential for the retrograde Golgi to endoplasmic reticulum transport of proteins. The coatomer is also involved in membrane trafficking in the exocytic pathway and for endocytic recycling of cell surface receptors. The association of the coatomer with the Golgi is regulated by binding of ADP ribosylation factors and is GTP dependent. Interestingly, COPB2 was previously identified as a binding partner for the PKCε (15). Whether COPB2 also binds PKCι remains to be determined. These data suggest that PKCι may regulate Golgi function and membrane trafficking in tumor cells. Our data are the first to our knowledge to directly implicate COPB2 in transformation.
RFC4 is a clamp loader protein that binds in a complex with proliferating cell nuclear antigen to regulate elongation of primed DNA templates by DNA polymerases (16, 17). The clamp loader also functions in unscheduled DNA repair and check point control through interactions with the RAD17 911 DNA damage checkpoint complex (18). The fact that PKCι regulates RFC4 expression suggests that PKCι may function to modulate either DNA synthesis and/or DNA damage checkpoint function in tumor cells. ELF3 is a transcriptional factor in the ETS domain family and is known to regulate expression of many growth-related genes including angiopoeitin 1, collagenase, and other transformed growth and invasion-related genes (19, 20). Interestingly, ELF3 expression has been shown to be largely restricted to epithelial cells of the lung, intestinal tract, and kidney. ELF3 has also been shown to be highly expressed in lung carcinoma tissues and in lung cancer cell lines when compared with normal lung tissue, suggesting a specific role for ELF3 in the lung (20). Our data indicate that ELF3 expression in lung tumors is regulated by oncogenic PKCι and that ELF3 plays a critical role in lung tumorigenesis. Our finding that ELF3 expression does not correlate with PKCι expression in other tumor types is consistent with the restricted expression of ELF3 to the lung observed by others. Finally, PLS1 encodes plastin, an actin-, calcium-binding protein involved in the regulation of the actin cytoskeleton (21). Interestingly, plastin has previously been reported to be overexpressed in prostate cancer cells and that genetic disruption of plastin expression with antisense mRNA inhibits invasion of prostate cancer cells (22, 23). Our data confirm the importance of plastin in tumor cell invasion and identify it as a key target of PKCι-mediated NSCLC tumor invasion. This report is the first to provide direct evidence for the involvement of most of these genes in the transformed phenotype of human tumor cells. Because so little is known about these proteins in the context of transformation, future studies will be required to elucidate the mechanisms by which these genes impinge on anchorage-independent growth and invasion. In conclusion, our results show the utility of using meta-analysis of public domain gene expression data sets to identify novel genes involved in cellular transformation downstream of a newly identified oncogene, PKCι. Because expression of these genes has been documented in primary human tumors, these genes, and the pathways in which they operate are attractive targets for development of novel prognostic markers and biomarkers, and in new therapeutic strategies for treatment of cancer.
Acknowledgments
Grant support: National Cancer Institute (CA81436), the American Lung Association/LUNGevity, and the James and Esther King Biomedical Research Program, Florida Department of Health (A.P. Fields).
We thank Dr. Eric Edell and Aaron Bungum for access to the primary NSCLC tumor tissues, Dr. Verline Justilien for immunoblot analysis, and the RNA Interference Technology Resource at the Mayo Clinic College of Medicine for production of lentiviral RNAi reagents directed against the genes of interest.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.American Cancer Society. Surveillance Research Cancer Statistics, 2004. 2004. [cited;Available from:] [Google Scholar]
- 2.Carbone D. Smoking and cancer. Am J Med. 1992;93:13–7S. doi: 10.1016/0002-9343(92)90621-h. [DOI] [PubMed] [Google Scholar]
- 3.Ginsberg R, Vokes E, Rosenzweig K. Non-small cell lung cancer. In: De Vitta SHV, Rosenberg S, editors. Cancer: Principles and Practice of Oncology. Philadelphia (PA): Lippencott Williams and Wilkins; 2001. pp. 925–83. [Google Scholar]
- 4.Schiller JH. Current standards of care in small-cell and non-small-cell lung cancer. Oncology. 2001;61 (Suppl 1):3–13. doi: 10.1159/000055386. [DOI] [PubMed] [Google Scholar]
- 5.Regala RP, Weems C, Jamieson L, et al. Atypical protein kinase C ι is an oncogene in human non-small cell lung cancer. Cancer Res. 2005;65:8905–11. doi: 10.1158/0008-5472.CAN-05-2372. [DOI] [PubMed] [Google Scholar]
- 6.Regala RP, Weems C, Jamieson L, Copland JA, Thompson EA, Fields AP. Atypical protein kinase Cι plays a critical role in human lung cancer cell growth and tumorigenicity. J Biol Chem. 2005;280:31109–15. doi: 10.1074/jbc.M505402200. [DOI] [PubMed] [Google Scholar]
- 7.Murray NR, Jamieson L, Yu W, et al. Protein kinase Cι is required for Ras transformation and colon carcinogenesis in vivo. J Cell Biol. 2004;164:797–802. doi: 10.1083/jcb.200311011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–24. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci US A. 2001;98:13784–9. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frederick LA, Matthews JA, Jamieson L, et al. Matrix metalloproteinase-10 is a critical effector of protein kinase Ciota-Par6α-mediated lung cancer. Oncogene. 2008 doi: 10.1038/onc.2008.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Regala RP, Thompson EA, Fields AP. Atypical protein kinase cι expression and aurothiomalate sensitivity in human lung cancer cells. Cancer Res. 2008;68:5888–95. doi: 10.1158/0008-5472.CAN-08-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J, Anastasiadis PZ, Liu Y, Thompson EA, Fields AP. Protein kinase C βII induces cell invasion through a Ras/MEK-, PKCι/RAC 1-dependent signaling pathway. J Biol Chem. 2004;279:22118–23. doi: 10.1074/jbc.M400774200. [DOI] [PubMed] [Google Scholar]
- 14.Stenbeck G, Harter C, Brecht A, et al. β′-COP, a novel subunit of coatomer. EMBO J. 1993;12:2841–5. doi: 10.1002/j.1460-2075.1993.tb05945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Csukai M, Chen CH, De Matteis MA, Mochly-Rosen D. The coatomer protein β′-COP, a selective binding protein (RACK) for protein kinase Cε. J Biol Chem. 1997;272:29200–6. doi: 10.1074/jbc.272.46.29200. [DOI] [PubMed] [Google Scholar]
- 16.Ellison V, Stillman B. Reconstitution of recombinant human replication factor C (RFC) and identification of an RFC subcomplex possessing DNA-dependent ATPase activity. J Biol Chem. 1998;273:5979–87. doi: 10.1074/jbc.273.10.5979. [DOI] [PubMed] [Google Scholar]
- 17.Zhang G, Gibbs E, Kelman Z, O’Donnell M, Hurwitz J. Studies on the interactions between human replication factor C and human proliferating cell nuclear antigen. Proc Natl Acad Sci U S A. 1999;96:1869–74. doi: 10.1073/pnas.96.5.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffith JD, Lindsey-Boltz LA, Sancar A. Structures of the human Rad17-replication factor C and checkpoint Rad 9 – 1-1 complexes visualized by glycerol spray/low voltage microscopy. J Biol Chem. 2002;277:15233–6. doi: 10.1074/jbc.C200129200. [DOI] [PubMed] [Google Scholar]
- 19.Thomas RS, Ng AN, Zhou J, Tymms MJ, Doppler W, Kola I. The Elf group of Ets-related transcription factors. ELF3 and ELF5. Adv Exp Med Biol. 2000;480:123–8. doi: 10.1007/0-306-46832-8_15. [DOI] [PubMed] [Google Scholar]
- 20.Tymms MJ, Ng AY, Thomas RS, et al. A novel epithelial-expressed ETS gene, ELF3: human and murine cDNA sequences, murine genomic organization, human mapping to 1q32.2 and expression in tissues and cancer. Oncogene. 1997;15:2449–62. doi: 10.1038/sj.onc.1201427. [DOI] [PubMed] [Google Scholar]
- 21.Delanote V, Vandekerckhove J, Gettemans J. Plastins: versatile modulators of actin organization in (patho)physiological cellular processes. Acta Pharmacol Sin. 2005;26:769–79. doi: 10.1111/j.1745-7254.2005.00145.x. [DOI] [PubMed] [Google Scholar]
- 22.Zheng J, Rudra-Ganguly N, Miller GJ, Moffatt KA, Cote RJ, Roy-Burman P. Steroid hormone induction and expression patterns of L-plastin in normal and carcinomatous prostate tissues. Am J Pathol. 1997;150:2009–18. [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng J, Rudra-Ganguly N, Powell WC, Roy-Burman P. Suppression of prostate carcinoma cell invasion by expression of antisense L-plastin gene. Am J Pathol. 1999;155:115–22. doi: 10.1016/S0002-9440(10)65106-1. [DOI] [PMC free article] [PubMed] [Google Scholar]