Abstract
Background.
Aging in humans is characterized by a progressive loss of muscle mass and strength known as sarcopenia. Although considered to be a normal aspect of aging, the loss of strength can have significant effects on the health, functioning, and independence of elderly individuals. Although these aspects of sarcopenia have been well studied, the molecular mechanisms leading to its development are still unclear. The nematode Caenorhabditis elegans might be a novel animal model for sarcopenia as worms experience sarcopenia during aging and mutations affecting the daf-2/insulin-like signaling pathway are able to delay this process.
Methods.
Via the use of RNA interference, we screened a total of 43 genes, most of which have been shown to be required for the enhanced longevity of daf-2 mutants, to assess for the effects of these genes on muscle function and worm mobility during aging.
Results.
We identified 17 novel genes that are essential for the delay in the onset of sarcopenia in daf-2 mutants. The identified genes include splicing factors, vacuolar sorting proteins, transcription factors, and metabolic enzymes. Using a transgenic strain that only responds to RNA interference in the body wall muscle, we also found that most of the identified genes act in muscle to prevent the onset of sarcopenia.
Conclusions.
Our results demonstrate that at least in worms, specific genetic pathways that modify the development of sarcopenia can be identified. Interestingly, almost all the identified genes also have a known human homolog, and hence, our findings may offer significant leads toward the identification of genes involved in sarcopenia in people.
Keywords: Sarcopenia, C elegans, daf-2, Mobility, Muscle, Aging
SARCOPENIA is the progressive loss of muscle mass with advancing age characterized by a decline in muscle quantity and quality (1,2). The medical importance of sarcopenia lies in the resulting reduction in strength, which serves as a risk factor for some of the impairments in mobility and functioning seen in older people (3). The decline in strength also appears to act as a risk factor for mortality (4). Although the cause of sarcopenia is not known, there is evidence from human and vertebrate animal studies for several mechanisms including disuse atrophy associated with inactivity, loss of α-motor neurons, hormonal changes, inflammatory effects, altered caloric intake, and changes muscle cell physiology (5,6). Despite this work, the primary cause of sarcopenia is unclear, and the current treatment options are rather limited. For example, a recent promising trial of testosterone was halted due to an increase in adverse events in treated participants (7). Hence, the development of new model systems could advance research in this field and hopefully lead to new treatments.
The nematode Caenorhabditis elegans has emerged as a powerful model system in multiple areas of biology. Worms are used to investigate the genetic and molecular functions of many human disease-related genes in biologic processes such as cell death and cell fate determination or to study pathological processes such as neurodegeneration and microbial pathogenesis (8). Furthermore, there are several experimental advantages to using C elegans such as a completely sequenced genome that shares significant homology with that of mammals, the ability to easily and inexpensively cultivate animals in the lab, the short life span of 2–3 weeks, and the ability to manipulate the genome by generating mutant and transgenic animals. Caenorhabditis elegans also offers the ability to complement standard mechanistic experiments with unbiased forward genetic screens. Finally, a major strength of the worm system is the ability to use RNA interference (RNAi) to manipulate gene expression and function by feeding the worm Escherichia coli bacteria producing the desired double-stranded RNA as food (9).
Like humans, C elegans also develops sarcopenia during aging (10). These muscle losses start in midlife, magnify with increasing age, and are associated with reductions in mobility (10,11). These changes in mobility are not due to neuropathology during aging as microscopic examination of worms have shown that the nervous system remains intact in old worms, whereas the myofibrils of old worms become progressively disorganized and lose myosin filaments (10,11). In addition to myofibril loss, there are also changes in nuclear architecture with aging (10).
Mutations in the daf-2/insulin-like signaling pathway in worms result in up to 100% increases in life span (12). This pathway comprises the insulin/insulin-like growth factor-1 receptor DAF-2 that signals through a conserved PI3-kinase/AKT pathway to phosphorylate and inhibit DAF-16, a forkhead family transcription factor (12). In daf-2 mutants, daf-16 is nuclear localized and activates a significant number of other target genes (12). In addition to increased longevity, daf-2 mutants also show increased resistance to oxidative stress, hypoxia, heat stress, heavy metals, and bacterial pathogens (12).
Worms with mutations in the daf-2 pathway are also resistant to the development of sarcopenia and associated declines in mobility during aging (10,11,13). Additionally, genes in the epidermal growth factor pathway and the Rag GTPase (raga-1) pathway also act to preserve mobility and muscle function (14,15). Together, these findings suggest that sarcopenia is a malleable phenotype and likely amenable to study.
This preservation of muscle mass and mobility in worms with mutations affecting the daf-2 pathway suggests that activation of daf-2 target genes either directly or indirectly affects the resistance of muscle to the development of sarcopenia. We hypothesized that specific genes acting downstream of daf-2 may be required for the delay in sarcopenia. To test this hypothesis, we investigated the effect of knockdown of a total of 43 genes, most of which have been shown to affect this pathway (16), to screen for genes, which might be required to preserve mobility and muscle function in daf-2 mutant worms.
MATERIALS AND METHODS
Caenorhabditis elegans Strains
TJ1060 (spe-9(hc88); fer-15(b26)), OH4125 (evIs82b; wrk-1(ok695)), and DR1568 (daf-2(e1371)) were obtained from the Caenorhabditis Genetics Center, which is supported by National Institutes of Health funding. GL227 (daf-2(e1371); spe-9(hc88); fer-15(b26)) and ALF113 (daf-2(e1371); evIs82b) were generated by standard genetic crosses. ALF110 (rde-1(ne219); bafIs110 (myo-3p:rde-1)) was generated by integrating a myo-3p:rde-1 extrachromosomal array (17) (a gift from K. Takahashi, Tokyo University of Pharmacy and Life Science) with gamma irradiation. ALF112 (daf-2(e1371); spe-9(hc88); fer-15(b26); rde-1(ne219); baf1s110 (myo-3p:rde-1)) was then generated by standard genetic crosses. Animals were maintained at 16°C on nematode growth medium plates spotted with OP50-1 (18).
RNAi Treatment
RNAi clones were obtained from the Ahringer RNAi library (Geneservice Limited, Cambridge, UK), except for clones for F40F12.7, C38C3.5, K11D12.9, Y119C1B.8, Y61A9LA.5, and Y54G2A.31, which were purchased from Open Biosystems (Huntsville, AL,) and clones for T10B9.1, Y76A2A.2, F32D8.13, Y4B6A.13, K06H7.9, and R06C1.2, which were generated by amplifying the genomic DNA using the gene pair primers available on Wormbase (for Y4B6A.13 and R06C1.2) or self-designed primers followed by subcloning of the polymerase chain reaction product into the L4440 RNAi vector (9). The sequences of the self-designed primers used were F32D8.13 (F) TGATAGCCATATCCGGTAAAC, (R) CTGAATGTGGACTTGACGAC; K06H7.9, (F)CTCACTGCGTGGGCTCTCA, (R)GGTGGACCATAAATAAGAACA; Y76A2A.2 (F) GTGTGATGAGAGTGAGATGACGA, (R)AAAAAGAAGCATCACTTGCAATC; and T10B9.1 (F) TGGCTCAATTCCAGATTTTT, (R ) CCGGTGCTCTGTTTATTGTGT.
RNAi treatment was performed by feeding animals double-stranded RNA–producing bacteria as described previously (19) except that we used a 4× concentrated RNAi culture to spot plates. After drying, 50–60 eggs isolated by hypochlorite treatment were added to the RNAi plates and kept at 20°C until the worms reached to the L4 stage before being transferred to 25°C. This is the nonpermissive temperature for adult fertility and creates a synchronized adult population.
Mobility Assay
Worms were washed from RNAi plates in S-basal buffer and placed at the center of two 10 cm Petri dishes containing agar with a drop of 1% isoamyl alcohol placed at one edge (11). The isoamyl alcohol acts as an attractant for worms. Once the worms (∼50 per plate) are placed in the center of the plate, a second drop of isoamyl alcohol and a drop of 10% sodium azide are placed at the same spot. Sodium azide paralyzes worms that reach the chemoattractant to facilitate scoring. The plates were left at room temperature for 1 hour before the locations of the worms were marked using a permanent marker and the plates are digitally imaged for measurements. We then measured the distance from the center of the plate toward the attractant that each worm moved and determined the numbers that traveled 3–4.5 cm, 0.5–3 cm, and 0–0.5 cm as previously described (11).
SAS version 9.2 (SAS Institute, Inc., Cary, NC) was used for statistical analysis. We used descriptive statistics and a line plot to summarize distances by group (RNAi treatment) and age (2/4/6/8/10 days). We fit a two-way analysis of variance model using the SAS GLM procedure with distance as the dependent variable and group, age, and Group × Age interaction as categorical factors of interest. A two-way analysis of variance model is more appropriate for the present situation because the measurements of distance at different ages were obtained from different worm populations. Appropriately constructed contrasts were used to obtain the magnitude and statistical significance of the between-group difference at each of the five ages. We repeated the analysis with age included as a continuous variable to obtain estimates of the group-specific slopes, quantifying change in distance per each day of increasing age.
Visualization of Muscle Structure
To examine the changes in muscle morphology with aging, phalloidin staining was performed as described previously (20).
Life-Span Analysis
Life-span assays were performed at 25°C with 25–30 eggs placed on two RNAi plates spotted with 2× concentrated RNAi of the gene of interest. The eggs were grown at 20°C until the L4 stage and then transferred to 25°C to obtain a synchronized adult population. The first day of adulthood was defined as Day 1. Animals were transferred to fresh plates every 2–4 days thereafter and examined daily for survival by looking for touch-provoked movement or pharyngeal pumping until all worms were dead or censored. Animals, which crawled off the plate, burst at the vulva or died from bagging were censored. All the life-span assays were repeated twice. Survival curves were generated using Prism 5 (GraphPad Software, Inc., La Jolla, CA).
Visualization of Motor Neurons
ALF113 (daf-2[e1371]; evIs82b) worms were treated with RNAi as previously and kept at 25°C until Day 10 of adulthood. In this strain, the cholinergic DA and DB ventral nerve cord motor neurons can be visualized using an unc-129p::GFP transgene (21). The Day 10 worms were transferred to nematode growth medium plates with no food for 20 minutes prior to taking digital pictures using an Olympus BX51 upright microscope and DP70 camera.
RESULTS
Screening for Genes Affecting Mobility During Aging
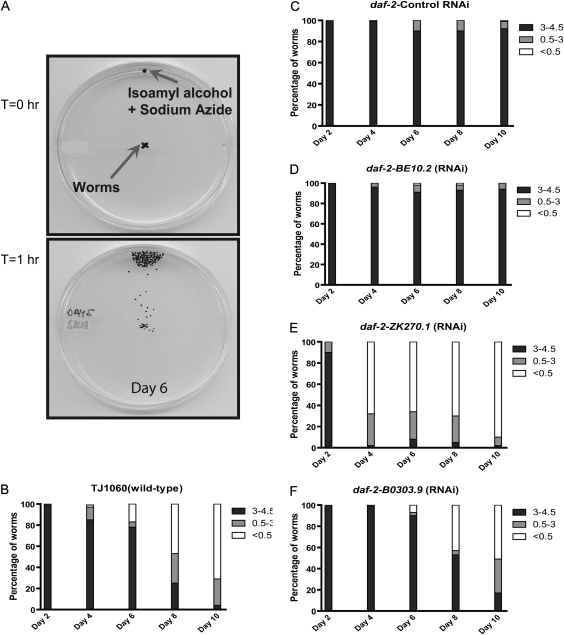
To assess muscle function, we used a chemotaxis assay that assesses the ability of worms to move between two points in response to an attractant chemical (Figure 1A) (11). This assay is very sensitive to picking up declines in muscle function and detects differences in mobility before declines in muscle structure can be observed (11).
Figure 1.
Detecting changes in mobility with a chemotaxis assay. (A) At T = 0 hour, roughly 50 worms are placed at the center of a Petri dish with a chemoattractant and sodium azide spotted at one edge. After 1 hour (T = 1 hour), the positions of the worms are marked and the plates digitally scanned. The distance traveled by each worm is then measured. Mobility data for aging (B) TJ1060 (spe-9(hc88); fer-15(b26)) worms, (C) GL227 (daf-2(e1371); spe-9(hc88); fer-15(b26)) worms treated with control RNA interference (RNAi), (D) BE10.2 RNAi, (E) ZK270.1 RNAi, and (F) B0303.9 RNAi. Shown are the percentages of worms that moved the specified distances from the starting point on each day. Black shading indicates greater distances moved and white lesser distances.
With this assay, we can reproducibly detect differences in mobility between wild-type worms and daf-2 mutants during aging. The temperature-sensitive sterile mutant TJ1060 (spe-9(hc88); fer-15(b26)) has a wild-type life span and is sterile when grown at 25°C during development, which facilitates the maintenance of a synchronized adult population (22). During aging, we observed a progressive decline in worm mobility as shown by the decrease in the number of animals moving more than 3 cm during the assay and the increase in those moving less than 0.5 cm (Figure 1B). These declines are not due to deaths or paralysis as all animals were alive and independently moving at the start of the assay. In contrast, the daf-2 mutant strain GL227 (daf-2(e1371); spe-9(hc88); fer-15(b26)), which is also sterile when grown at 25°C, exhibited enhanced mobility in this assay (Figure 1C). The differences between wild-type and daf-2 mutant worms appear as early as Day 4 of adulthood and are magnified with increasing age (Figure 1B and C). Strikingly, the GL227 strain shows no significant declines in mobility during the 10 days examined (Figure 1C).
To determine if specific genes are required for the beneficial effects of daf-2 mutations, we screened 41 genes that were previously identified as being required for the increased longevity of daf-2 mutants but not the longevity of wild-type animals (16). We also screened additional genes that were drawn from prior work showing them to be required for or affecting the daf-2 pathway (23,24). We screened these genes as we hypothesized that although required for the increased longevity of daf-2 mutants, individual genes might have a greater effect on mobility than longevity. Mobility assays were performed over a 10-day period with measurements made on Days 2, 4, 6, 8, and 10 of adulthood. The Day 2 assay was used to exclude changes in baseline mobility due to the effect of RNAi treatment instead of aging. We considered observing more than 25% of animals with less than 0.5 cm movement as meaningful as this is not observed in control RNAi experiments (Figure 1C).
From our screen, we identified 17 new genes that are required to preserve mobility in daf-2 mutants (Figure 1E and F and Tables 1 and 3), whereas two-thirds of the genes did not show any effect on mobility (Figure 1D, data not shown). For example, BE10.2 RNAi had no effect on the mobility of aging worms and, hence, is not essential for the preserved mobility and muscle function in daf-2 mutants (Figure 1D and Table 3). In contrast, both ZK270.1 and B0303.9 produced progressive declines in mobility as early as Day 4 of adulthood (Figure 1E and F and Table 3). These differences are statistically significant as shown by linear modeling and analysis of variance testing of the rates of movement decline during aging as compared with control (Table 3).
Table 1.
List of Genes Identified
| Gene | Gene Description | Human Homolog | Tissue Expressing |
| B0303.9 | Vacuolar sorting protein | VPS33A | NA |
| C29F9.1 | Uncharacterized protein | NA | NA |
| C31H2.1 | Tre-2/Bub2/cdc-16 domain | TBC1D24 | NA |
| R144.4 | WASP interacting protein | AC092898.1 | M |
| C38C3.5 | Cofilin | CFL2 | M |
| F39H11.2 | TATA binding protein-like factor | TBP | NA |
| ZK270.1 | Patched family member | NPC1 | NA |
| K11D12.9 | Golgin family member | GOLGA4 | NA |
| M01E5.5 | DNA topoisomerase I | TOP1 | I |
| C54D1.6 | β-Catenin | JUP | M, N |
| F28D1.9 | Fatty acid transport protein | SLC27A4 | M, I |
| R09B3.5 | Splicing factor | MAGOHB | NA |
| D2085.5 | Homolog of KIAA1219 | KIAA1219 | M |
| F30A10.6 | SAC1 PIP phosphatase | SACMIL | NA |
| T10B9.1 | Cytochrome P450 | CYP3A5 | NA |
| Y76A2A.2 | Copper transporting E1-E2 ATPase | ATP7A | M, I |
| R13H8.1 | daf-16, FOXO transcription factor | AFX | M, I, N |
Notes: I = intestine; M = muscle; N = nervous system; NA = not available. Annotations from Wormbase WS218.
Table 3.
Data Summary for Identified Genes
| Mobility Decline (mm/worm/d) |
Mean Life Span (d) |
||||||||
| Gene | Whole Animal RNAi* | p | Muscle-Specific RNAi† | p | Wild-Type Animals‡ | p | Samuelson et al.§ | This Work‖ | Muscle Structure¶ |
| Control | 0.10 | 0.70 | 4.37 | 35.5–41.1 | 25.0 | +++ | |||
| daf-16 | 4.48 | <.0001 | 4.24 | <.0001 | 5.09 | .0025 | 12.7–14.2 | 12.0 | ++ |
| skn-1 | 0.34 | .3142 | ND | ND | ND | ND | ND | ND | ND |
| BE10.2 | 0.26 | .3420 | ND | ND | ND | ND | 15.9 | 12.8 | ND |
| R09B3.5 | 3.44 | <.0001 | 2.69 | .0246 | ND | ND | 25.3 | ND | + |
| ZK270.1 | 3.50 | <.0001 | 3.38 | .0024 | ND | ND | 24.0 | 22.0 | +++ |
| F39H11.2 | 4.77 | <.0001 | 4.29 | <.0001 | ND | ND | 20.1 | ND | ++ |
| K11D12.9 | 4.72 | <.0001 | 5.20 | <.0001 | ND | ND | 28.0 | ND | + |
| R144.4 | 5.02 | <.0001 | 3.10 | .0065 | 4.16 | NS | 26.0 | ND | ++ |
| F28D1.9 | 3.83 | <.0001 | 4.42 | <.0001 | 5.09 | .0025 | 11.8 | ND | + |
| M01E5.5 | 3.40 | <.0001 | 3.13 | .0059 | ND | ND | 23.5 | ND | + |
| C54D1.6# | 3.05 | <.0001 | 4.22 | <.0001 | ND | ND | ND | ND | ++ |
| C38C3.5 | 4.54 | <.0001 | 3.55 | .0013 | ND | ND | 28.0 | ND | ++ |
| F30A10.6 | 3.98 | <.0001 | 4.46 | <.0001 | ND | ND | 22.3 | ND | ++ |
| T10B9.1# | 3.09 | <.0001 | 5.50 | <.0001 | ND | ND | ND | ND | +++ |
| Y76A2A.2 | 3.95 | <.0001 | 4.48 | <.0001 | ND | ND | 11.9 | ND | ++ |
| D2085.5 | 3.10 | <.0001 | 0.72 | .9061 | ND | ND | 28.3 | ND | ++ |
| B0303.9 | 3.45 | <.0001 | 0.54 | NS | ND | ND | 27.5 | 22.0 | + |
| C29F9.1 | 3.12 | <.0001 | 0.86 | .4270 | 3.76 | NS | 25.3 | ND | + |
| C31H2.1 | 4.62 | <.0001 | 0.62 | NS | ND | ND | 25.9 | ND | ++ |
Notes: ND = not done; NS = not significant; RNAi = RNA interference.
Rate of decline of GL227 mobility.
Rate of decline of ALF112 mobility.
Rate of decline of TJ1060 mobility.
Mean life span of daf-2(e1370) treated with RNAi (16).
Mean life span of GL227 treated with RNAi.
¶Qualitative evaluation of muscle morphology where +++ represents normal morphology, ++ represents intermediate, and + represents abnormal morphology as visualized through phalloidin staining. p values were obtained on the basis of analysis of variance testing as detailed in Material and Methods.
These identified genes are involved in a wide array of processes including transcription (F39H11.2), vacuolar trafficking (B0303.9), and cytochrome P450-mediated metabolism or detoxification (T10B9.1) (Table 1). Moreover, 16 of 17 genes identified in our screen have a human homolog that makes it possible that at least some genes could be involved in mobility and sarcopenia in people.
The Identified Genes Have Differential Effects on daf-2 Mobility and Longevity
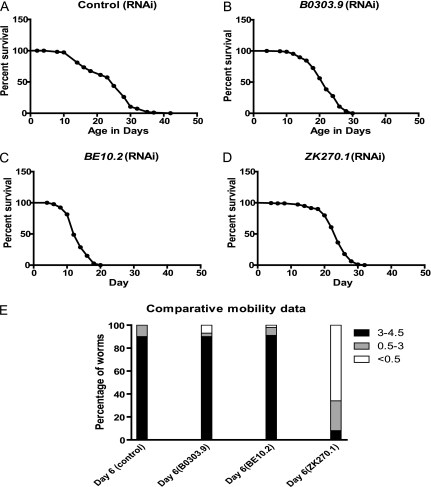
The genes we screened reduce daf-2 longevity, so we were concerned that the effects on mobility may just mirror their effects on longevity. However, we find little correlation between the magnitude of the previously published effects on life span of these genes and the magnitude of the effect we found on mobility, as our hits do not cluster among genes with the strongest effect on longevity (16) (Table 3). Consistent with differential effects on longevity and sarcopenia, we found genes with dramatic effects on life span but with no effect on mobility and other genes with lesser effects on longevity despite strong effects on mobility. For example, GL227 worms treated with B0303.9 RNAi have a mean life span of 22 days in contrast to 25 days for control RNAi (Figure 2A and B and Table 3) but declines in mobility start as early as Day 6 (Figure 1F). Similarly, GL227 worms treated with ZK270.1 RNAi have a mean life span of 22 days (Figure 2D and Table 3) but they show a progressive decline in mobility starting on Day 4 (Figure 1E). In contrast, worms treated with BE10.2 RNAi have a mean life span of only 12.8 days (Figure 2C and Table 3) but they show no declines in mobility during the first 10 days of adult life (Figure 1D). The finding that genes with strong effects on the enhanced longevity of daf-2 mutants may not necessarily impair the preserved mobility of daf-2 mutants is consistent with longevity and mobility being distinct daf-2 phenotypes.
Figure 2.
Differential effects of the identified genes on mobility and life span. Shown are Kaplan–Meier curves for the survival of GL227 (daf-2(e1371); spe-9(hc88); fer-15(b26)) worms treated with (A) control RNA interference (RNAi), (B) B0303.9 RNAi, (C) BE10.2 RNAi, and (D) ZK270.1 RNAi. In each curve, Day 1 represents the first day after the L4 larval molt. These are compared with the Day 6 mobility data of worms treated with each RNAi clone (E). Note the larger reduction in life span for worms treated with BE10.2 RNAi compared with the others and the larger mobility reduction for worms treated with ZK270.1 RNAi compared with the others. Shown is one of two trials with similar results.
Multiple Genes Act in Muscle to Preserve Mobility
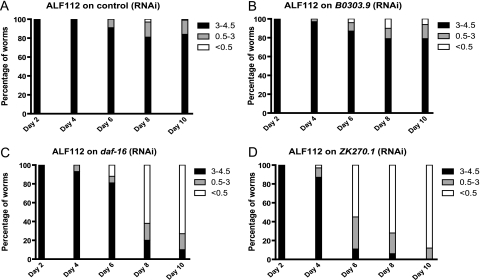
As RNAi can knock down gene expression in multiple tissues in the worm, we were unsure of where in the worms, these genes act to prevent sarcopenia. To explore this question, we used the ALF112 (daf-2(e1371), spe-9(hc88), fer-15(b26), rde-1(ne219), baf1s110 (myo-3p:rde-1)) transgenic strain that only responds to RNAi in the body wall muscle to retest the identified genes from our screen. These animals have a mutation in the rde-1 gene, which is required in a cell-autonomous manner to permit reductions in gene expression due to RNAi to occur, but the ability of these worms to respond to RNAi has been restored only in muscle by expressing rde-1 with the muscle-specific myo-3 promoter (17,25).
We performed mobility assays using the ALF112 strain treated with RNAi against the identified genes. For 13 genes, a negative effect on mobility (Figure 3C and D and Tables 2 and 3) was still observed, whereas for 4 genes, no effect on mobility was seen in this strain (Figure 3B and Tables 2 and 3). These results demonstrate that most of the genes we identified act in muscle to prevent declines in mobility during aging. This suggests that muscle plays an important role in the development or prevention of sarcopenia.
Figure 3.
Effect of muscle-specific gene knockdown on mobility of daf-2 mutants. The ALF112 (daf-2(e1371); spe-9(hc88); fer-15(b26); rde-1(ne219); bafIs110 (myo-3p:rde-1)) strain only responds to RNA interference (RNAi) in striated body wall muscle. This strain was treated with RNAi and the effects on mobility measured for (A) control RNAi, (B) B0303.9 RNAi, (C) daf-16 RNAi, and (D) ZK270.1 RNAi.
Table 2.
Classification of Genes Based on Muscle-Specific RNA interference (RNAi)
| Impair Mobility | No Effect |
| daf-16 | D2085.5 |
| R09B3.5 | B0303.9 |
| ZK270.1 | C29F9.1 |
| F39H11.2 | C31H2.1 |
| K11D12.9 | |
| R144.4 | |
| F28D1.9 | |
| M01E5.5 | |
| C54D1.6 | |
| C38C3.5 | |
| F30A10.6 | |
| T10B9.1 | |
| Y76A2A.2 |
Notes: Based on effect on mobility during aging using the ALF112 (daf-2(e1371); spe-9(hc88); fer-15(b26); rde-1(ne219); bafIs110 (myo-3p:rde-1)) strain that shows RNAi effects only in body wall muscle.
Effects on Muscle Morphology During Aging
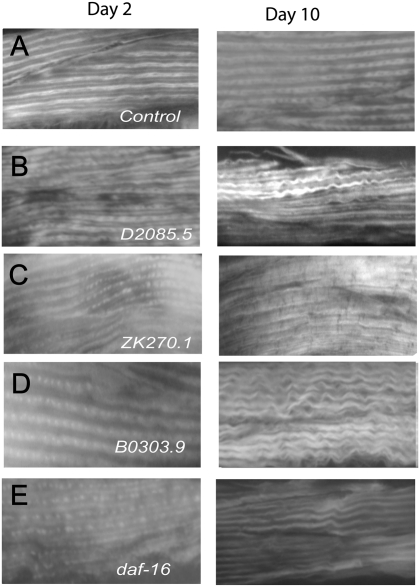
During aging in C elegans, there is a loss in the myofibril number and organization (10). We used phalloidin staining (20) to look for changes in the myofibrils of aging GL227 worms treated with control RNAi or the RNAi clones for the identified genes. We observed differences in muscle morphology for RNAi-treated Day 10 worms compared with Day 2 RNAi-treated worms or Day 10 control RNAi-treated worms for many of the identified genes (Figure 4A–E and Table 3). Often, the muscle fibers in RNAi-treated Day 10 worms are less dense and show a wavy irregular organization (Figure 4A–E). Interestingly, we found that most of the genes that act autonomously in muscle, with the exception of T10B9.1 and ZK270.1, affect muscle structure to some degree (Figure 4C and E and Table 3). Among these genes, R09B3.5, K11D12.9, F28D1.9, and M01E5.5 have the strongest effects on muscle morphology (Table 3). Surprisingly, even the genes that do not act autonomously in muscle have negative effects on muscle structure (Figure 4B and D and Table 3) These observations suggest that a large number of the identified genes affect muscle morphology and may explain the observed decline in mobility with increasing age. This finding also supports a primary effect of these genes on muscles.
Figure 4.
Effect of identified genes on muscle structure during aging. Photomicrographs (20× magnification) showing changes in muscle morphology visualized via phalloidin staining in Day 2 and Day 10 adult GL227 worms treated since egg hatching with (A) control RNA interference (RNAi), (B) D2085.5 RNAi, (C) ZK270.1 RNAi, (D) B0303.9 RNAi, and (E) daf-16 RNAi.
RNAi Treatment Does Not Affect Nervous System Structure
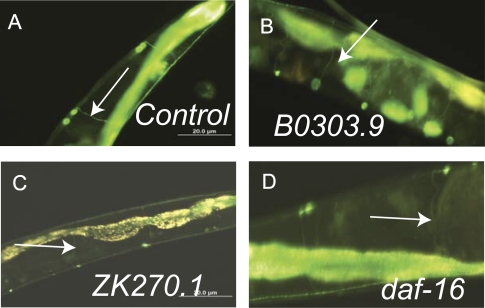
In C elegans, there are negligible changes in the structure of the nervous system during aging (10). However, we wanted to test whether knockdown of the identified genes produced adverse structural or functional changes in the nervous system. To evaluate the structure of the nervous system of aging RNAi-treated worms, we used fluorescence microscopy to visualize the ventral motor neurons using ALF113 (daf-2(e1371); evIs82b)) transgenic worms, which have unc-129:GFP reporter gene expressed in these neurons (21). Examination of Day 10 animals confirmed that these neurons are present and the axons still innervate the muscles despite the effects of both aging and RNAi treatment (Figure 5A–D). However, it is still possible that there are effects of the identified genes on the neuromuscular junction, the subcellular structure of the neurons, or aspects of neuronal function. Next, we tested the ability of RNAi-treated worms to respond to octanol (26). Caenorhabditis elegans is repelled by the odor of octanol so this assay examines the ability of the animals to detect and respond to an olfactory stimulus. We found that Day 10 RNAi-treated worms for all the genes we identified were able to respond to octanol within a 20-second time frame (data not shown). These assays confirmed that olfactory behaviors remain present and that there are no gross changes in muscle innervation in RNAi-treated animals.
Figure 5.
Motor neurons are intact in RNA interference (RNAi)-treated worms. ALF113 (daf-2(e1371); evIs82b)) worms express GFP in the DA and DB motor neurons (arrows). Shown are digital images of Day 10 ALF113 worms treated with (A) control RNAi, (B) B0303.9 RNAi, (C) ZK270.1 RNAi, and (D) daf-16 RNAi. These are representative images from pictures taken from an average of 20 treated worms.
RNAi-Treated Worms Move More Slowly
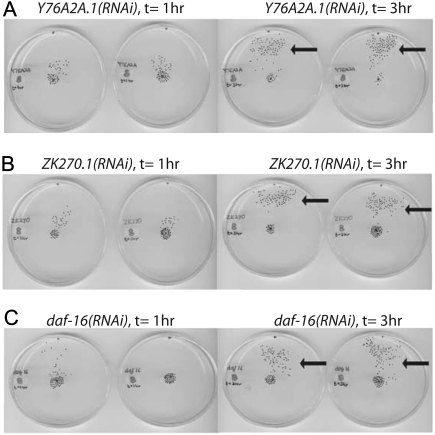
During our mobility assays, we found that when old worms were given 3 hours instead 1 hour to complete the mobility task, most of the worms were now able to reach to the end point (Figure 6A–C). This observation suggests that even older animals retain the ability to detect the attractant, but their speed of movement toward the attractant declines and accounts for the increased time required to complete the behavioral task.
Figure 6.
Loss of the identified genes makes worms move more slowly. Images of scanned Petri dishes, in duplicate, showing changes in the progress of RNA interference (RNAi)-treated GL227 worms toward the attractant when given 3 hours (right) instead of 1 hour (left). Worms were treated with (A) Y76A2A.1 RNAi, (B) ZK270.1 RNAi, and (C) daf-16 RNAi. Arrows show the increased number of worms reaching the attractant.
Effects of Identified Genes on Wild-Type Animals
Although the genes identified could have effects only on the mobility of daf-2 mutants, it is also possible that at least some of the genes could affect sarcopenia in wild-type animals. To test this possibility, we treated the wild-type TJ1060 strain with the RNAi clones for several genes and assessed the effects on mobility during aging by conducting mobility assays. Of four genes tested, we found that two (daf-16 and F28D1.9) enhance the age-related mobility decline of wild-type worms (Table 3). This suggests that at least some of the identified genes are involved in preventing sarcopenia during normal aging.
DISCUSSION
Sarcopenia, the resulting losses in muscle strength and the associated potential for adverse effects on mobility and functioning are a significant health concern even in healthy elders. Unfortunately, the causes are unclear and the available treatments are limited. Consequently, we have explored using C elegans as a model system to study this problem. Worms are increasingly used to study the genetic and biochemical events involved in aging and these animals also develop sarcopenia during aging. In worms, genes that modify sarcopenia have been identified, suggesting that the available genetic and RNAi resources can be used to study this condition. Our results show that C elegans can successfully be used to identify downstream genetic pathways that can prevent sarcopenia during aging. Of note, the effect of these genes on mobility is not the same as their effects on longevity, suggesting that the delay in sarcopenia and preservation of mobility is a distinct daf-2 phenotype that can be studied.
From our screen, we identified genes that could provide new insights into sarcopenia. For example, the gene F30A10.6 encodes a Sac1 phosphoinositide phosphatase that regulates the levels of phosphatidylinositol 4-phosphate. In yeast, Sac1 localizes to the endoplasmic reticulum and is involved in the control of lipid storage, Golgi function, vesicle formation, and organization of the actin cytoskeleton (27). Loss of any of these functions could make muscle more susceptible to the effects of aging. Similarly, the gene C31H2.1 encodes a member of the Tre-2/Bub2/Cdc16 domain protein family. These proteins play an important role in the regulation of GLUT4 trafficking to the cell surface in response to insulin, muscle contraction, or low energy states (28). The activity of family members, such as AS160, is modulated by phosphorylation via Akt in response to insulin signaling (29). Additionally, TBC1D1 has also been shown to link insulin signaling to the modulation of mammalian target of rapamycin activity (30). Interestingly, the target of rapamycin pathway has already been linked to longevity and the activation of autophagy in worms (31). F28D1.9 encodes a fatty acid transport protein homologous to FATP4 (SLC27A4). FATP4 transports long-chain fatty acids into skin and muscle (32). In worms, this gene is expressed in the myocytes of the pharynx but not the body wall muscle myocytes. This specific expression pattern may account for the noncell autonomous role for this gene in prevention of sarcopenia. Further work will be needed to explore the molecular mechanisms involved.
The identified genes also represent an opportunity for translational research in human sarcopenia as many of the identified genes have a known human homolog. It will be interesting to investigate if any of the gene homologs have positive or negative effects on muscle mass during aging in people. Likely, alleles of these genes exist in people, and their genetic effects can be explored using existing muscle-focused cohort studies like Health ABC.
Our work suggests that C elegans can be developed into an important model system to study not only the biochemical and molecular events responsible for sarcopenia but also as a novel tool for developing new targets for pharmacological intervention that may influence the development or progression of sarcopenia. Although the effects of aging on skeletal muscle are unlikely to be completely halted or reversed, the underlying mechanisms responsible for these deleterious changes present numerous targets for drug discovery with potential opportunities to attenuate muscle mass losses, improve muscle function, and preserve the functional capabilities and independence of seniors.
Acknowledgments
This work was supported by National Institutes of Health grant AG029870 to A.L.F. L.K. was supported by an Ellison Medical Foundation/AFAR Postdoctoral Fellows in Aging Research fellowship.
References
- 1.Evans WJ. What is sarcopenia? J Gerontol A Biol Sci Med Sci. 1995;50((special issue)):5–8. doi: 10.1093/gerona/50a.special_issue.5. [DOI] [PubMed] [Google Scholar]
- 2.Roubenoff R, Hughes VA. Sarcopenia: current concepts. J Gerontol A Biol Sci Med Sci. 2000;55:M716–M724. doi: 10.1093/gerona/55.12.m716. [DOI] [PubMed] [Google Scholar]
- 3.Janssen I, Heymsfield SB, Ross R. Low relative skeletal muscle mass (sarcopenia) in older persons is associated with functional impairment and physical disability. J Am Geriatr Soc. 2002;50:889–896. doi: 10.1046/j.1532-5415.2002.50216.x. [DOI] [PubMed] [Google Scholar]
- 4.Metter EJ, Talbot LA, Schrager M, Conwit R. Skeletal muscle strength as a predictor of all-cause mortality in healthy men. J Gerontol A Biol Sci Med Sci. 2002;57:B359–B365. doi: 10.1093/gerona/57.10.b359. [DOI] [PubMed] [Google Scholar]
- 5.Volpi E, Nazemi R, Fujita S. Muscle tissue changes with aging. Curr Opin Clin Nutr Metab Care. 2004;7:405–410. doi: 10.1097/01.mco.0000134362.76653.b2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenlund LJ, Nair KS. Sarcopenia-consequences, mechanisms, and potential therapies. Mech Ageing Dev. 2003;124:287–299. doi: 10.1016/s0047-6374(02)00196-3. [DOI] [PubMed] [Google Scholar]
- 7.Basaria S, Coviello AD, Travison TG, et al. Adverse events associated with testosterone administration. N Engl J Med. 2010;363:109–122. doi: 10.1056/NEJMoa1000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baumeister R, Ge L. The worm in us—Caenorhabditis elegans as a model of human disease. Trends Biotechnol. 2002;20:147–148. doi: 10.1016/s0167-7799(01)01925-4. [DOI] [PubMed] [Google Scholar]
- 9.Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 10.Herndon LA, Schmeissner PJ, Dudaronek JM, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 11.Glenn CF, Chow DK, David L, et al. Behavioral deficits during early stages of aging in Caenorhabditis elegans result from locomotory deficits possibly linked to muscle frailty. J Gerontol A Biol Sci Med Sci. 2004;59:1251–1260. doi: 10.1093/gerona/59.12.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Antebi A. Genetics of aging in Caenorhabditis elegans. PLoS Genet. 2007;3:1565–1571. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duhon SA, Johnson TE. Movement as an index of vitality: comparing wild type and the age-1 mutant of Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci. 1995;50:B254–B261. doi: 10.1093/gerona/50a.5.b254. [DOI] [PubMed] [Google Scholar]
- 14.Iwasa H, Yu S, Xue J, Driscoll M. Novel EGF pathway regulators modulate C. elegans healthspan and lifespan via EGF receptor, PLC-gamma, and IP3R activation. Aging Cell. 2010;9:490–505. doi: 10.1111/j.1474-9726.2010.00575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreiber MA, Pierce-Shimomura JT, Chan S, Parry D, McIntire SL. Manipulation of behavioral decline in Caenorhabditis elegans with the Rag GTPase raga-1. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000972. e1000972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samuelson AV, Carr CE, Ruvkun G. Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev. 2007;21:2976–2994. doi: 10.1101/gad.1588907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki M, Sagoh N, Iwasaki H, Inoue H, Takahashi K. Metalloproteases with EGF, CUB, and thrombospondin-1 domains function in molting of Caenorhabditis elegans. BiolChem. 2004;385:565–568. doi: 10.1515/BC.2004.069. [DOI] [PubMed] [Google Scholar]
- 18.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen KY, Pan H, Lin MJ, et al. Length-dependent toxicity of untranslated CUG repeats on Caenorhabditis elegans. Biochem Biophys Res Commun. 2007;352:774–779. doi: 10.1016/j.bbrc.2006.11.102. [DOI] [PubMed] [Google Scholar]
- 21.Colavita A, Krishna S, Zheng H, Padgett RW, Culotti JG. Pioneer axon guidance by UNC-129, a C. elegans TGF-beta. Science. 1998;281:706–709. doi: 10.1126/science.281.5377.706. [DOI] [PubMed] [Google Scholar]
- 22.Fabian TJ, Johnson TE. Production of age-synchronous mass cultures of Caenorhabditis elegans. J Gerontol. 1994;49:B145–B156. doi: 10.1093/geronj/49.4.b145. [DOI] [PubMed] [Google Scholar]
- 23.Murphy CT, McCarroll SA, Bargmann CI, et al. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- 24.Szewczyk NJ, Peterson BK, Barmada SJ, Parkinson LP, Jacobson LA. Opposed growth factor signals control protein degradation in muscles of Caenorhabditis elegans. EMBO J. 2007;26:935–943. doi: 10.1038/sj.emboj.7601540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabara H, Sarkissian M, Kelly WG, et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 26.Troemel ER, Chou JH, Dwyer ND, Colbert HA, Bargmann CI. Divergent seven transmembrane receptors are candidate chemosensory receptors in C. elegans. Cell. 1995;83:207–218. doi: 10.1016/0092-8674(95)90162-0. [DOI] [PubMed] [Google Scholar]
- 27.Foti M, Audhya A, Emr SD. Sac1 lipid phosphatase and Stt4 phosphatidylinositol 4-kinase regulate a pool of phosphatidylinositol 4-phosphate that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell. 2001;12:2396–2411. doi: 10.1091/mbc.12.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakamoto K, Holman GD. Emerging role for AS160/TBC1D4 and TBC1D1 in the regulation of GLUT4 traffic. Am J Physiol Endocrinol Metab. 2008;295:E29–E37. doi: 10.1152/ajpendo.90331.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sano H, Kane S, Sano E, et al. Insulin-stimulated phosphorylation of a Rab GTPase-activating protein regulates GLUT4 translocation. J Biol Chem. 2003;278:14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- 30.Zhou QL, Jiang ZY, Holik J, et al. Akt substrate TBC1D1 regulates GLUT1 expression through the mTOR pathway in 3T3-L1 adipocytes. Biochem J. 2008;411:647–655. doi: 10.1042/BJ20071084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoSGenet. 2008;4 doi: 10.1371/journal.pgen.0040024. e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jain SS, Chabowski A, Snook LA, et al. Additive effects of insulin and muscle contraction on fatty acid transport and fatty acid transporters, FAT/CD36, FABPpm, FATP1, 4 and 6. FEBS Lett. 2009;583:2294–2300. doi: 10.1016/j.febslet.2009.06.020. [DOI] [PubMed] [Google Scholar]