Abstract
Adelgids (Insecta: Hemiptera: Adelgidae) are known as severe pests of various conifers in North America, Canada, Europe and Asia. Here, we present the first molecular identification of bacteriocyte-associated symbionts in these plant sap-sucking insects. Three geographically distant populations of members of the Adelges nordmannianae/piceae complex, identified based on coI and ef1alpha gene sequences, were investigated. Electron and light microscopy revealed two morphologically different endosymbionts, coccoid or polymorphic, which are located in distinct bacteriocytes. Phylogenetic analyses of their 16S and 23S rRNA gene sequences assigned both symbionts to novel lineages within the Gammaproteobacteria sharing <92% 16S rRNA sequence similarity with each other and showing no close relationship with known symbionts of insects. Their identity and intracellular location were confirmed by fluorescence in situ hybridization, and the names ‘Candidatus Steffania adelgidicola' and ‘Candidatus Ecksteinia adelgidicola' are proposed for tentative classification. Both symbionts were present in all individuals of all investigated populations and in different adelgid life stages including eggs, suggesting vertical transmission from mother to offspring. An 85 kb genome fragment of ‘Candidatus S. adelgidicola' was reconstructed based on a metagenomic library created from purified symbionts. Genomic features including the frequency of pseudogenes, the average length of intergenic regions and the presence of several genes which are absent in other long-term obligate symbionts, suggested that ‘Candidatus S. adelgidicola' is an evolutionarily young bacteriocyte-associated symbiont, which has been acquired after diversification of adelgids from their aphid sister group.
Keywords: evolution, genome reduction, insects, symbiosis, Buchnera
Introduction
The presence of heritable bacterial endosymbionts in insects is widespread in nature (Buchner, 1953). Their roles range from obligate mutualists with essential nutritional functions and facultative mutualists necessary for host protection to parasites manipulating the host's reproductive system (Moran et al., 2008). Thus, symbiosis with bacteria has a great impact on the ecology and the evolution of many insect hosts.
A group of insects comparatively well studied with respect to their symbionts are plant sap-sucking members of the suborder Sternorrhyncha including aphids, psyllids, whiteflies and mealybugs, with phloem-feeding aphids as a prime example. Most aphids harbor the gammaproteobacterial endosymbiont Buchnera aphidicola. Buchnera is located in specialized cells in the insect body cavity termed ‘bacteriocytes', and is vertically transmitted from mother to offspring (Baumann, 2005; Moran et al., 2008). The major function of this obligate (primary) symbiont is to convert unusable nutrition to utilizable compounds, that is, to supply its host with essential amino acids lacking in phloem sap (Shigenobu et al., 2000; Moran et al., 2003; Zientz et al., 2004; Thomas et al., 2009). Buchnera are highly specialized bacteria well adapted to symbiosis with their aphid hosts through >160 millions of years (Moran et al., 1993). As obligate intracellular bacteria, they show highly reduced genomes compared with free-living bacteria and lack many essential biosynthetic pathways (Moran et al., 2008). In addition to Buchnera, many aphids harbor additional, phylogenetically distinct facultative (secondary) symbionts such as ‘Candidatus Serratia symbiotica' (Lamelas et al., 2008), ‘Candidatus Hamiltonella defensa' (Degnan et al., 2009) and ‘Candidatus Regiella insecticola' (Moran et al., 2005; Scarborough et al., 2005).
In contrast to the well-studied aphids, our knowledge about bacterial symbionts of their sister group, the adelgids, is very scarce. Adelgids are plant sap-sucking insects comprising ∼65 highly host-specific species, some of which represent severe pests of various conifers especially in North America, Canada, Europe and Asia (Steffan, 1972; Blackman and Eastop, 1994; Havill and Foottit, 2007). Adelgids feed mainly on phloem or parenchyma cells and perform mostly a complex sexual life cycle consisting of multiple generations with host alternations on conifers by switching from the primary (spruce (Picea spp.)) to the secondary host tree (Abies, Larix, Pseudotsuga, Tsuga or Pinus); however, asexual reproduction has also been reported (Steffan, 1972; Havill and Foottit, 2007).
Early morphological and histological studies of adelgids have reported the occurrence of bacteria within bacteriocytes, between bacteriocytes and oenocytes and free in the hemocoel (Profft, 1936; Buchner, 1953; Steffan, 1968; Shields and Hirth, 2005). Rod-shaped, coccoid and polymorphic bacteria were observed in different adelgid species and sometimes within the same adelgid host (Buchner, 1953; Steffan, 1968). However, to our knowledge, no published sequences for the identification of these bacterial symbionts on the molecular level are available.
In this study, we investigated whether adelgids, like their aphid sister group, harbor Buchnera-related symbionts or whether they acquired different endosymbionts after the divergence from aphids. We analyzed adelgids of the Adelges nordmannianae/piceae complex. Both insects, the silver fir woolly adelgid A. nordmannianae (Eckstein, 1890; a.k.a. Dreyfusia nordmannianae) and the balsam woolly adelgid A. piceae (Ratzeburg, 1844; a.k.a Dreyfusia piceae), are a very closely related species pair with highly similar morphological and genetic characteristics (Eichhorn, 1967; Havill and Foottit, 2007; Havill et al., 2007). Although A. nordmannianae shows a sexual life cycle including host alternation between spruce and fir trees (Picea orientalis and Abies spp. respectively), A. piceae has an asexual life cycle and feeds on fir trees only ( Bryant, 1971; Eichhorn, 1973; Binazzi, 2000). Both are a severe pest on young firs and dreaded in the Christmas tree industry (Balch, 1952). A. nordmannianae primarily attacks young sprouts and needles and older branches but rarely the bark of the trunk. A. piceae can be located on any part of the tree depending on the host tree species (Steffan, 1972). The stylets of A. piceae pierce the bark tissue and can reach the phloem, but do not enter it. In contrast to aphids, these adelgids thus feed on cortical parenchyma cells and on the phelloderm, a layer of the periderm (Balch, 1952).
Using 16S rRNA sequence analysis and fluorescence in situ hybridization (FISH) with symbiont-specific oligonucleotide probes, we identified the bacteriocyte-associated symbionts of adelgids of the A. nordmannianae/piceae complex as previously unrecognized Gammaproteobacteria that are present in different host populations and life stages. Metagenomic analysis suggested that one of the symbionts was acquired after diversification of aphids and adelgids, recently relative to other insect symbiont lineages.
Materials and methods
Organisms
Adelgids of the A. nordmannianae/piceae complex were collected from silver fir (Abies alba Mill) and Nordmann fir (Abies nordmanniana (Steven) Spach) trees at three different sampling sites (Table 1). Infested branches were cut from the trees and stored at 4 °C until collection of the insects. The insects were used immediately, fixed for FISH and electron microscopy or stored in ethanol for DNA purification at a later time point.
Table 1. Adelgids and their bacteriocyte-associated endosymbionts analyzed in this study.
| Adelgids | Location | Host plant |
GenBank accession number (symbionts) |
|||||
|---|---|---|---|---|---|---|---|---|
|
GenBank accession number (insect host) |
‘Candidatus Steffania adelgidicola' |
‘Candidatus Ecksteinia adelgidicola' |
||||||
| coI | ef1alpha | 16S rRNA gene | 23S rRNA gene | 16S rRNA gene | 23S rRNA gene | |||
| Adelges nordmannianae/ piceae | Gosau, Austria | Abies alba | HQ668155 | HQ668164 | HQ668158 | — | HQ668160 | — |
| Adelges nordmannianae/ piceae | Grafrath, Germany | Abies nordmanniana | HQ668157 | HQ668167 | HQ668159 | — | HQ668161 | — |
| Adelges nordmannianae/ piceae | Klausen- Leopoldsdorf, Austria | Abies alba | HQ668156 | HQ668165 HQ668166 | FR872579 | FR872579 | HQ668162 | HQ668163 |
Histology and transmission electron microscopy
Insects were prefixed in 2.5% glutaraldehyde in 0.1 sodium cacodylate buffer overnight at 4 °C and fixed in 1% osmium tetroxide in 0.1 sodium cacodylate buffer for 2 h at room temperature. Specimens were dehydrated in ethanol and embedded in Spurr's resin (Spurr, 1969). Semi-thin sections were stained with Richardson's solution (Richardson et al., 1960) and examined by light microscopy. Ultrathin sections were stained with uranyl acetate and lead citrate and examined using a Zeiss EM 902 electron microscope (Carl Zeiss, Vienna, Austria) at 80 kV.
PCR, cloning, restriction fragment length polymorphism and sequencing
Intact adelgids stored in ethanol were washed in double distilled water and DNA was purified from either single or up to 30 individuals; different life stages and eggs were used for DNA extraction using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). DNA was stored at −20 °C until further use for PCR. Adelgid (coI, ef1alpha) and bacterial (16S and 23S rRNA) genes were amplified in 35 PCR cycles using the primers and conditions listed in Supplementary Table S1. PCR reactions typically contained 2 μl template DNA, 50 pmol of each primer, 1 Unit of Taq DNA polymerase (Fermentas, St Leon-Rot, Germany), 10 × Taq buffer with KCl and 2 m MgCl2 and 0.2 m of each deoxynucleotide in a total volume of 50 μl. Both negative (no DNA added) and positive controls were included in all PCR reactions. PCR products were purified using the PCR purification kit (Qiagen) and were either sequenced directly or cloned using the TOPO TA cloning kit with the cloning vector pCR 2.1-TOPO (Invitrogen Life Technologies, Lofer, Austria) following the manufacturer's instructions. At least 20 16S and 23S rRNA gene clones each were screened by restriction fragment length polymorphism analysis using MspI (Fermentas). Nucleotide sequences were determined using the BigDye Terminator kit v3.1 (Applied Biosystems, Vienna, Austria) and an ABI 3130 XL genetic analyzer (Applied Biosystems).
Phylogenetic analysis
The program ARB (Ludwig et al., 2004) was used for phylogenetic analysis. ARB 16S and 23S rRNA databases were updated with sequences from GenBank obtained by sequence homology searches using BLASTn available at the NCBI web site (National Centre for Biotechnology Information) (Altschul et al., 1990). Databases for genes encoding cytochrome c oxidase 1 (coI) and elongation factor 1-alpha (ef1alpha), as well as a protein database RNA polymerase sigma-32 factor (RpoH) were established with representative sequences downloaded from GenBank and aligned using Mafft (Katoh et al., 2005). Phylogenetic trees were calculated using MrBayes and the maximum parsimony, distance matrix and TREEPUZZLE methods implemented in ARB (Huelsenbeck and Ronquist, 2001; Schmidt et al., 2002; Ronquist and Huelsenbeck, 2003; Ludwig et al., 2004). PhyML trees were calculated using the Mobyle portal (http://mobyle.pasteur.fr/cgi-bin/portal.py; Guindon and Gascuel, 2003). Symbiont-specific primers were designed using the probedesign/probematch tools of the ARB software package (Ludwig et al., 2004).
Fluorescence in situ hybridization
Insects were fixed in 4% paraformaldehyde for 4 h at 4 °C, crushed on a glass slide and covered with 0.2% of low melting agarose. Hybridization was performed using a standard protocol hybridization and washing buffer as described previously (Daims et al., 2005). The oligonucleotide probes used are given in Supplementary Table S1. Probe NONEUB (complementary to the bacterial probe EUB338-I) was used as negative control. Hybridized slides were examined using a confocal laser scanning microscope (LSM 510 Meta, Carl Zeiss). Symbiont-specific probes were designed using the probedesign/probematch tools of the ARB software package (Ludwig et al., 2004) and deposited at probeBase (Loy et al., 2007). Optimal hybridization conditions for symbiont-specific probes were determined in a series of hybridization experiments with increasing formamide concentrations in the hybridization buffer.
Fosmid library construction and screening
High molecular weight DNA for the construction of a fosmid library was purified from freshly collected insects and eggs from Klausen-Leopoldsdorf. To enrich bacterial symbionts, whole insects and eggs were homogenized using a Dounce tissue grinder (Wheaton, Millville, NJ, USA) in buffer A (35 m Tris-HCl, 25 m KCl, 10 m MgCl2, 250 m sucrose, pH 7.5; Ishikawa, 1982) and filtered sequentially through 53, 30 and 10 μm meshes (Eckert, Waldkirch, Germany). The remaining suspension containing both symbionts was centrifuged at 7000 r.p.m. The pellet was resuspended in TE buffer (10 m Tris-HCl, 1 m EDTA, pH 7.5), and DNA was obtained using an SDS-based DNA purification method including 1% hexadecylmethylammonium bromide, 1.5% polyvinylpyrrolidone and proteinase K (Sigma-Aldrich, Vienna, Austria) in the extraction buffer (Zhou et al., 1996). DNA was quality checked by gel electrophoresis and stored at −20 °C until further use. A large insert fosmid library was constructed using the CopyControl Fosmid Library Production Kit (Epicentre, Madison, WI, USA). A total of 14 208 fosmid clones were picked, incubated and stored in 96 MicroWell plates containing LB medium, 7% glycerol and 12.5 μg ml−1 chloramphenicol at −80 °C.
Fosmids prepared from up to 96 pooled clones served as template for the screening of the fosmid library by PCR using symbiont-specific 16S and 23S rRNA gene-targeted primers and PCR conditions described above but with 42 PCR cycles (Supplementary Table S1). PCR products from single, positive fosmid clones were sequenced directly, and the size of the inserted DNA was estimated by pulsed field gel electrophoresis. In brief, a CHEF-DR III system (Bio-Rad, Vienna, Austria) and 1% agarose gel were used at 6V cm and 14 °C with a linear switching time (0.1–5 s) for 8.4 h. Positive fosmids were end sequenced, and checked for overlapping regions using restriction fragment length polymorphism with EcoRI (Fermentas). Fosmids were sequenced using a shotgun clone library and the Sanger sequencing method by a company (LGC Genomics, Berlin, Germany). A coverage of 4 × was achieved and sequence gaps were closed by primer walking.
Annotation and comparative genome analysis
The genome fragment of ‘Candidatus Steffania adelgidicola' was automatically annotated and analyzed using the PEDANT software platform (http://pedant.gsf.de/; Frishman et al., 2001; Walter et al., 2009). Protein-coding sequences were predicted by combining intrinsic predictions from GeneMarkS (Besemer et al., 2001) and Glimmer (Delcher et al., 2007) and extrinsic information from a BLAST search against the NCBI RefSeq database (Wheeler et al., 2008). All gene models were additionally manually verified by pairwise alignments with homologous proteins found in the UniProt Knowledgebase (Consortium, 2010). Prediction of tRNA genes was performed using tRNAscan-SE (Lowe and Eddy, 1997). Pseudogene analysis was performed using a house-internal pipeline for the identification of neighboring open-reading frames with identical homologs in NCBI RefSeq. All candidates for pseudogenes were subsequently inspected manually. The NCBI COG database was used for functional classification of proteins (Tatusov et al., 2001). UniProt and the Multi-Genome Browser of the Biocyc database collection were used for synteny analysis (Karp et al., 2005; Consortium, 2010). Biochemical pathway prediction was performed using KEGG (Kyoto Encyclopedia of Genes and Genomes) (Kanehisa and Goto, 2000). Presence/absence analysis was performed by comparison with representative genomes of insect symbionts and free-living Gammaproteobacteria using the predicted protein sequences and BLASTp with an E-value cutoff of 10−9 (Altschul et al., 1990). DNAPlotter was used for generating linear DNA maps (Carver et al., 2009).
Nucleotide sequence accession numbers
Gene sequences of symbionts and their insect hosts and the genome fragment obtained from ‘Candidatus S. adelgidicola' were deposited at the DDBJ/EMBL/GenBank databases under the accession numbers listed in Table 1.
Results and Discussion
Identification of adelgid hosts
Three adelgid populations found on Abies spp. in Austria (Gosau, Klausen-Leopoldsdorf) and Germany (Grafrath) were sampled and tentatively identified as members of the A. nordmannianae/piceae complex based on their morphology, their host plants A. alba and A. nordmanniana, as well as their location on trees. For molecular characterization of the three adelgid populations, genes encoding the cytochrome c oxidase subunit 1 (coI) and the eukaryotic elongation factor 1-alpha (ef1alpha) were partially amplified (639 nt for coI and 664 nt for ef1alpha) and sequenced from DNA extracted from up to 30 individuals. Both genes are considered useful molecular markers for inferring the phylogenetic relationships of eukaryotes, and they were used previously for analysis of adelgids (Havill et al., 2007; Foottit et al., 2009; Zurovcova et al., 2010).
Phylogenetic analyses of concatenated coI and ef1alpha data sets using neighbor joining, maximum parsimony and maximum likelihood methods confirmed the morphology-based identification of these insects and showed consistently their affiliation with the family Adelgidae in which they formed a stable, well-supported monophyletic group with A. nordmannianae and A. piceae (Supplementary Figure S1). Within this group, the concatenated data set and coI alone failed to differentiate between the species A. piceae and A. nordmannianae, which is consistent with previous studies (Havill et al., 2007; Foottit et al., 2009; Zurovcova et al., 2010). A closer inspection of the ef1alpha alignment revealed three alignment positions that differentiate A. nordmannianae from A. piceae (Supplementary Table S2). If these positions were taken as indicators for species delineation, adelgids sampled in Gosau represent A. nordmannianae and adelgids from Grafrath belong to A. piceae. Two different ef1alpha variants containing the signatures of A. nordmannianae and A. piceae, respectively, were observed for adelgids sampled in Klausen-Leopoldsdorf, which might indicate a mixed population of A. piceae and A. nordmannianae at this sampling site as observed previously on other occasions (Steffan, 1972). However, the presence of only three single-nucleotide polymorphisms in the comparatively small ef1alpha data set and inconclusive information from coI sequence data with respect to these adelgid species demonstrate that current molecular markers are not able to reliably differentiate the highly related and co-occurring species A. piceae and A. nordmannianae. This is consistent with the highly similar morphology of both insect groups (Blackman and Eastop, 1994). In concordance with previous reports, we thus refer to the adelgids investigated in this study as members of the A. nordmannianae/piceae complex.
Two morphologically different bacteriocyte-associated symbionts
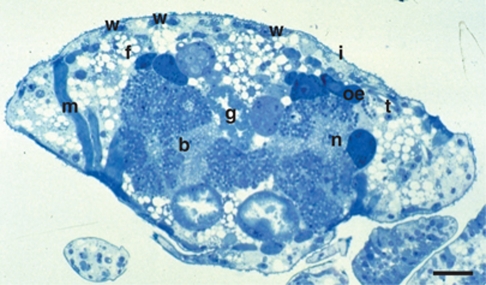
Staining of semi-thin sections of insects from the A. nordmannianae/piceae population sampled in Gosau revealed two differentially stained types of bacteriocytes in the adelgids' body cavity, which is consistent with previous studies (Profft, 1936; Buchner, 1953; Steffan, 1968). The size of the bacteriocytes increased from larval to adult stages with an average size between 30 and 50 μm in adults. In the first instar stage, the bacteriome consisted of loose-fitting cell aggregates, whereas the adult stages (apterous exulis and winged sexupara) contained a compact bacteriome (Figure 1). Similar to the situation in aphids, the bacteriome extended in two strands from the last thoracic to the seventh abdominal segment extending alongside the gut. The anterior parts of the bacteriome strands were connected to each other by bacteriocytes, ventral to the gut. In cross-sections, the bacteriome formed an H-like structure (Figure 1).
Figure 1.
The bacteriome of Adelges nordmannianae/piceae. A semi-thin cross-section through the abdomen of a sexupara (adult winged life stage) stained with Richardson's solution (Richardson et al., 1960) is shown. Bacteriocytes can be readily seen in proximity to the gut. b=bacteriocytes filled with two different types of symbionts (dark and light blue), g=gut, n=nucleus, oe=oenocytes, t=tracheole, w=wax gland plate, f=fat body cell, m=muscle, i=integument; bar represents 50 μm.
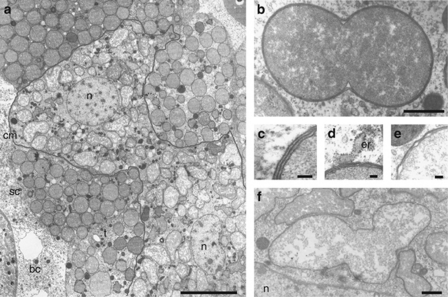
Electron microscopy confirmed the presence of two distinct types of bacteriocytes seen in histological sections. One type of bacteriocytes has single nuclei and contains coccoid, electron-dense bacteria, which divide by binary fission and are between 1 and 4 μm in diameter (Figures 2a and b). The second type of bacteriocytes is multinucleated and harbors less electron-dense, polymorphic bacteria, between 1.4 and 7 μm in size (Figures 2a and f). Both symbionts showed a Gram-negative type cell wall and were enclosed by a symbiosome membrane (Figures 2c–e). Multilamellar bodies, but no other bacterial forms, were found inside the bacteriocytes. Other tissues such as sheath cells or the hemocoel did not contain bacterial symbionts. Taken together, the association of A. nordmannianae/piceae with bacterial symbionts is very similar to the situation in aphids and other insects, which frequently contain two different kinds of bacteriocyte-associated symbionts, sometimes referred to as primary and secondary symbionts (Baumann, 2006).
Figure 2.
Ultrastructure of the bacterial symbionts of Adelges nordmannianae/piceae. (a) Electron microscopy revealed two distinct types of bacteriocytes containing different morphotypes of bacterial symbionts; bacteriocytes with electron-translucent bacteria are multinucleated; for clarity, the cell membrane of this bacteriocyte is highlighted by a gray line. (b) Coccoid symbiont (‘Candidatus Steffania adelgidicola') undergoing cell division. (c) Detail of the coccoid symbiont (‘Candidatus S. adelgidicola') showing its cell wall (two membranes) tightly surrounded by a third membrane, the symbiosome membrane, (d) which is associated with the rough endoplasmatic reticulum. (e and f) Polymorphic symbiont (‘Candidatus Ecksteinia adelgidicola') and detail view of its cell wall (two membranes) and the symbiosome membrane. Bar in panel a represents 10 μm; bars in panels b and f represent 1 μm; bars in panels c, d and e represent 90 nm. n=nucleus, t= tracheole, sc=sheath cell, bc=body cavity, cm=cell membrane of the bacteriocyte, er=rough endoplasmatic reticulum.
Novel gammaproteobacterial symbionts
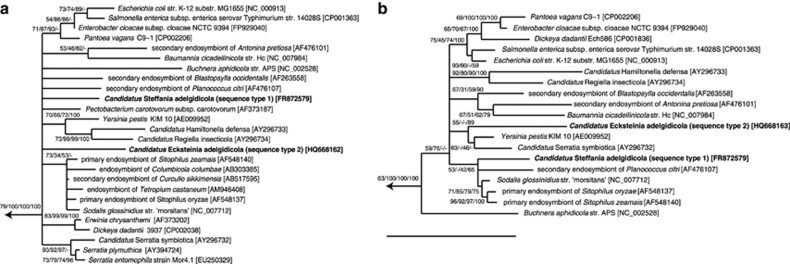
Sequencing and analysis of 16S rRNA genes amplified from up to 30 host individuals from 3 different adelgids populations revealed 2 distinct 16S rRNA sequences showing highest similarity with members of the Gammaproteobacteria. Both sequences, presumably representing the two symbiont morphotypes, were only moderately similar to each other (90.1–91.2%) and to other gammaproteobacterial sequences (∼95%). Sequence type 1 showed highest similarity to Sodalis glossinidius str. ‘morsitans', whereas sequence type 2 was most similar to Serratia plymuthica and Serratia entomophila. Both sequence types were found in all three populations and were nearly identical among the geographically distant sampling sites (99.7–99.9%). In phylogenetic trees, both sequence types established novel, deep branching lineages within the Gammaproteobacteria (Figure 3a). Depending on the method used for phylogeny inference, these lineages sometimes clustered together with other symbionts of insects, but overall comparison of different phylogenetic methods and bootstrap values did not provide support for proposing a specific affiliation of the two sequence types with other Gammaproteobacteria. The lack of phylogenetic information of rRNA genes to resolve deeply branching lineages is a well-known problem within this taxonomic clade (Williams et al., 2010).
Figure 3.
Phylogenetic relationships of the gammaproteobacterial symbionts of Adelges nordmannianae/piceae. TREEPUZZLE trees based on 16S rRNA (a) and 23S rRNA (b) genes are shown. 16S rRNA and 23S rRNA sequence types 1 represent ‘Candidatus Steffania adelgidicola' sequence types 2 represent ‘Candidatus Ecksteinia adelgidicola'. The branching order near the root of the trees could not be resolved and differs between trees obtained with MrBayes, maximum likelihood, maximum parsimony and neighbor joining methods. TREEPUZZLE support values, bootstrap values for maximum parsimony and maximum likelihood (1000 resamplings), and posterior probabilities of MrBayes are indicated at the inner nodes. GenBank/EMBL/DDBJ accession numbers are given in square brackets. Bars, 10% estimated evolutionary distance.
Consistent with the two 16S rRNA sequence types found in A. nordmannianae/piceae, we also detected two different 23S rRNA gene sequences in the insect host. 23S rRNA sequence type 1 showed highest similarity (94.2%) to the Sodalis-related primary endosymbiont of Sitophilus zeamais. Sequence type 2 was most similar to ‘Candidatus Serratia symbiotica' (90.4%). Phylogenetic analysis confirmed the affiliation of both sequences with the Gammaproteobacteria, in which they formed novel lineages. Similar to the 16S rRNA-based phylogenetic analysis, both sequences clustered in some trees with other insect symbionts, but their relationship with other Gammaproteobacteria could not be unambiguously resolved (Figure 3b).
In situ identification of ‘Candidatus S. adelgidicola' and ‘Candidatus E. adelgidicola'
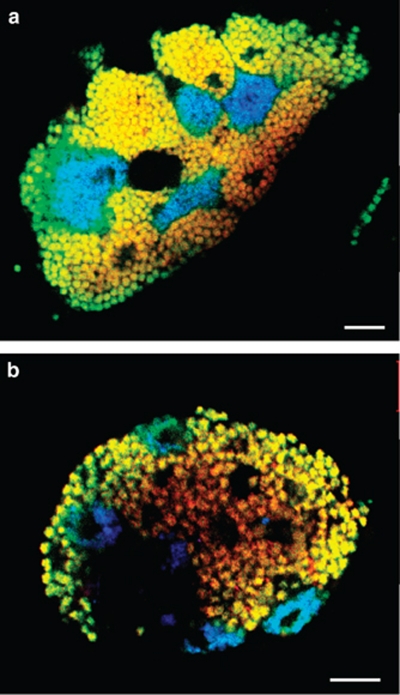
The two bacterial morphotypes observed by electron microscopy were readily visible using FISH and 16S rRNA-targeted probes in samples from all three adelgid populations (Figure 4a, Supplementary Figure S2). Probes specific for 16S and 23S rRNA sequence type 1 hybridized with the coccoid symbionts. Probes specific for sequence types 2 targeted the polymorphic symbionts, which showed an extremely low fluorescence signal that could be enhanced by application of three probes simultaneously (Supplementary Figure S3). All symbionts identified by FISH were located in bacteriocytes, which contained either one of the two symbionts. Combination with general bacterial probes demonstrated the absence of additional bacteria in this tissue.
Figure 4.
In situ identification and intracellular localization of ‘Candidatus Steffania adelgidicola' and ‘Candidatus Ecksteinia adelgidicola' in adults and eggs of A. nordmannianae/piceae. Bacterial symbionts were labeled by FISH using symbiont-specific 16S rRNA-targeted oligonucleotide probes together with a probe mix targeting all bacteria (Supplementary Table S1). Probes specific for the coccoid ‘Candidatus S. adelgidicola' were labeled with Cy3 (red); three probes specific for the polymorphic ‘Candidatus E. adelgidicola' were labeled with Cy5 (blue) and used simultaneously. Probes targeting all bacteria were labeled with FLUOS (green). The combined signal from bacterial and symbiont-specific probes appear yellow for ‘Candidatus S. adelgidicola' and blue–green for ‘Candidatus E. adelgidicola', respectively. (a) Bacteriocytes of an adult adelgid sampled in Gosau, Austria. (b) Both symbionts inside an adelgid egg. Bars represent 10 μm.
Taken together, we could show that A. nordmannianae/piceae contains two morphologically and phylogenetically distinct bacteriocyte-associated endosymbionts, which form novel evolutionary lineages within the Gammaproteobacteria. Notably, both symbionts are different from Buchnera, the obligate symbionts of most aphids which represent a sister group of the adelgids. They are also different from other known primary or secondary symbionts found in insects belonging to the order Sternorrhyncha such as mealybugs, psyllids or whiteflies (Baumann, 2006). A. nordmannianae/piceae has thus acquired its own gammaproteobacterial symbionts during evolution. The low degree of relationship with each other and with other Gammaproteobacteria justifies their taxonomic placement within two novel genera. According to Murray and Stackebrandt (1995), we propose the following provisional names.
‘Candidatus S. adelgidicola'
‘Candidatus S. adelgidicola' named in honor of the German entomologist August Wilhelm Steffan for his contributions to research on adelgids and their bacterial symbionts; ‘adelgid-icola' meaning friend or lover of the Adelgidae. This symbiont of A. nordmannianae/piceae is coccoid with a cell size between 1 and 4 μm and has a Gram-negative type cell wall. It is surrounded by a symbiosome membrane and located in single-nucleated bacteriocytes. The symbiont is not cultured in cell-free medium or a cell line. ‘Candidatus S. adelgidicola' represents a novel genus within the class Gammaproteobacteria (phylum Proteobacteria). The basis of assignment is: 16S rRNA, 23S rRNA (GenBank/EMBL/DDBJ accession numbers HQ668158, HQ668159, FR872579).
‘Candidatus E. adelgidicola'
‘Candidatus E. adelgidicola' named in honor of the German entomologist Karl Eckstein (1859–1939) for his contributions to research on adelgids; ‘adelgid-icola' meaning friend or lover of the Adelgidae. This symbiont of A. nordmannianae/piceae is polymorphic with a cell size between 1.4 and 7 μm and has a Gram-negative type cell wall. It is surrounded by a symbiosome membrane and is located in multinucleated bacteriocytes. The symbiont is not cultured in cell-free medium or a cell line. ‘Candidatus E. adelgidicola' represents a novel genus within the class Gammaproteobacteria (phylum Proteobacteria). The basis of assignment is: 16S rRNA, 23S rRNA (GenBank/EMBL/DDBJ accession numbers HQ668160, HQ668161, HQ668162, HQ668163).
Occurrence and vertical transmission
To further investigate the occurrence of ‘Candidatus S. adelgidicola' and ‘Candidatus E. adelgidicola' within a single adelgid population, we designed PCR assays specific for the 16S rRNA gene of each of the two symbionts (Supplementary Table S1) and screened 10 individuals from the population sampled in Gosau (Supplementary Figure S4). PCR products were obtained from all individuals and 16S rRNA sequences were nearly identical (99.5%) to the previously recovered sequences of ‘Candidatus S. adelgidicola' and ‘Candidatus E. adelgidicola', suggesting that both symbionts are present in all individuals of the investigated population. We next analyzed different life stages of the adelgid host using symbiont-specific PCR assays, and we detected ‘Candidatus S. adelgidicola' and ‘Candidatus E. adelgidicola' in both sexuparae and exules (Supplementary Figure S5). In addition, we performed FISH on eggs of A. nordmannianae/piceae and could demonstrate bacteriocytes containing each of the two symbionts, respectively (Figure 4b). Taken together, these findings are strong evidence for a vertical transmission of ‘Candidatus S. adelgidicola' and ‘Candidatus E. adelgidicola' from mother to offspring, corroborating previous histological studies of A. nordmannianae/piceae and its bacterial symbionts (Profft, 1936; Buchner, 1953; Steffan, 1972). Vertical inheritance is a hallmark of obligate symbionts of insects which are well adapted to and essential for their hosts (Moran et al., 2008). Thus, ‘Candidatus S. adelgidicola' and ‘Candidatus E. adelgidicola' likely have an important role for their host's biology. This notion is consistent with the observation that the related hemlock woolly adelgid Adelges tsugae, for which morphologically similar symbionts were described, was inhibited in its development after treatment with antibiotics (Shields and Hirth, 2005).
Evidence for the presence of most biosynthetic pathways in ‘Candidatus S. adelgidicola'
Known obligate bacterial symbionts of insects mainly serve as nutrient providers complementing their hosts' diet, in some cases as the result of synergistic functions of two different symbionts (Wu et al., 2006; McCutcheon and Moran, 2007; Gosalbes et al., 2008); other symbionts can protect their hosts against parasites or heat (Montllor et al., 2002; Oliver et al., 2003; Moran et al., 2005; Vorburger et al., 2010) or they may help to expand their host's host range (Tsuchida et al., 2004). To get first insights into the genetic basis and the role of ‘Candidatus S. adelgidicola' and ‘Candidatus E. adelgidicola' for their insect host, we generated a metagenomic fosmid library from purified bacterial symbionts. In total, 14 208 fosmid clones were screened using 16S and 23S rRNA gene-specific primers for both symbionts. Although no clones containing rRNA genes of ‘Candidatus E. adelgidicola' were identified, eight clones containing rRNA genes of ‘Candidatus S. adelgidicola' were found. On the basis of restriction fragment length polymorphism analysis for estimation of the extent of overlap between these clones, we selected two clones containing the 16S rRNA gene (33 and 39 kb in size) and one clone containing the 23S rRNA gene (35 kb) for sequencing. Shotgun sequences of these three clones could be assembled into one continuous contig with a total size of 85 kb. This genome fragment of ‘Candidatus S. adelgidicola' includes 44 predicted coding sequences (CDS), 3 rRNA genes, 2 tRNAs and 5 pseudogenes (Supplementary Figure S6, Supplementary Table S3). No non-coding RNAs were detected using the Rfam database (Gardner et al., 2009).
Obligate symbionts of insects typically possess small genomes compared with free-living bacteria. As a consequence, biosynthetic pathways are frequently reduced substantially and the symbionts thus rely on the import of metabolites from their hosts (Ramsey et al., 2010). Interestingly, the genome fragment of ‘CandidatusS. adelgidicola' encodes for proteins involved in a wide range of biosynthetic pathways (Figure 5, Supplementary Table S3). This includes proteins required for glycolysis, the pentose phosphate pathway and the tricarboxylic acid cycle. Proteins involved in the synthesis of amino acids, in pyrimidine and purine metabolism, as well as in lipid and lipopolysaccharide biosynthesis are also encoded. In addition, genes coding for proteins involved in metabolism of cofactors and vitamins such as biotin and nicotinamide-adenine dinucleotide are present. Although it remains unknown whether ‘Candidatus S. adelgidicola' encodes the full gene complement for these pathways, the observed diversity of metabolic functions encoded on the genome fragment suggests that the genetic repertoire of this symbiont resembles that of most free-living bacteria or facultative symbionts (Toh et al., 2006; Degnan et al., 2009; Burke and Moran, 2011).
Figure 5.
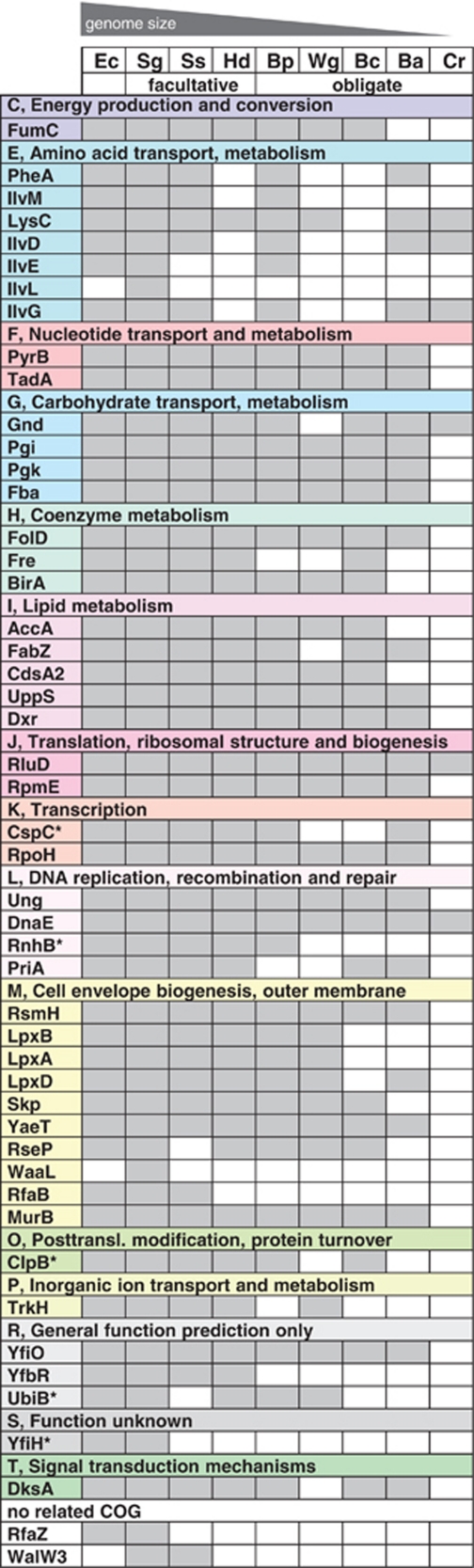
Affiliation of ‘Candidatus Steffania adelgidicola' proteins to NCBI COGs and occurrence of homologs in the genomes of E. coli and other insect symbionts. Gray boxes indicate the presence of homologs (E-value <10−9, except for pseudogenes); white indicates absence. The presence of homologs correlates with genome size, with obligate symbionts lacking many of the genes found on the ‘Candidatus S. adelgidicola' genome fragment. * Indicates pseudogenes in ‘Candidatus S. adelgidicola' it must be noted that more distant homologs of IlvL and WaaL are present in E. coli; Ec=Escherichia coli K-12 substr. MG1655 (NC_000913), Sg=Sodalis glossinidius str. ‘morsitans' (NC_007712), Ss=Serratia symbiotica str. Tucson (AENX00000000), Hd=‘Candidatus Hamiltonella defensa 5AT' (Acyrthosiphon pisum) (NC_012751), Bp=‘Candidatus Blochmannia pennsylvanicus str. BPEN' (NC_007292), Wg=Wigglesworthia glossinidia endosymbiont of Glossina brevipalpis (NC_004344), Bc=Baumannia cicadellinicola str. Hc (Homalodisca coagulata) (NC_007984), Ba=Buchnera aphidicola str. APS (Acyrthosiphon pisum) (NC_002528), Cr=‘Candidatus Carsonella ruddii PV' (NC_008512).
For example, the genome fragment of ‘CandidatusS. adelgidicola' contains genes for lipid A synthesis (lpxA, lpxB), a key lipopolysaccharide component, which are absent in the long-term insect-associated symbionts Buchnera, Baumannia cicadellinicola and ‘Candidatus Carsonella ruddii' but still present in the evolutionary younger obligate symbionts Wigglesworthia glossinidia and ‘Candidatus Blochmannia pennsylvanicus' or the facultative symbionts S. glossinidius, S. symbiotica and ‘Candidatus H. defensa' (Shigenobu et al., 2000; Akman et al., 2002; Degnan et al., 2005; Nakabachi et al., 2006; Wu et al., 2006; Moran et al., 2008).
Amino acids have a special role in many symbiotic associations between bacteria and insects. Although on the one hand, many obligate symbionts produce certain amino acids that are lacking in their hosts' diet, they possess on the other hand often only limited capabilities to synthesize other amino acids and obtain these compounds through the host metabolism or through other symbionts (Shigenobu et al., 2000; Zientz et al., 2004; Moran et al., 2008; McCutcheon et al., 2009). ‘Candidatus S. adelgidicola' encodes proteins involved in the synthesis of phenylalanine, tyrosine, tryptophan, lysine, glycine, serine, threonine, cysteine and methionine, as well as proteins for synthesis of branched-chain amino acids such as isoleucine, valine and leucine. Thus, similar to facultative symbionts like S. glossinidius (Toh et al., 2006), ‘Candidatus S. adelgidicola' might be able to generate most amino acids independent from its insect host. It is also conceivable that ‘Candidatus S. adelgidicola' has a role in essential amino acid biosynthesis for its adelgid host.
Taken together, the presence of several genes in the ‘Candidatus S. adelgidicola' genome that are also found in free-living bacteria and facultative symbionts but that are lacking in many obligate symbionts suggests that the ‘Candidatus S. adelgidicola' is more similar to known facultative symbionts with respect to its metabolic capabilities. This might indicate that ‘Candidatus S. adelgidicola' is an evolutionarily young symbiont, and that adaptation to an intracellular lifestyle is less pronounced than in long-term obligate symbionts.
‘Candidatus S. adelgidicola' is an evolutionarily young symbiont
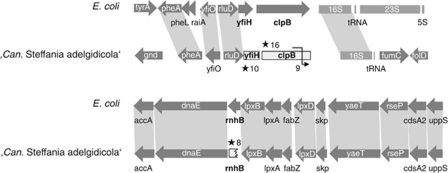
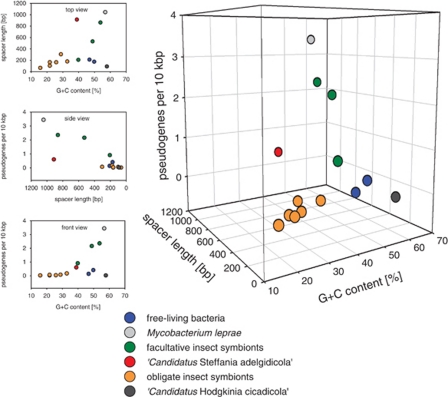
Reduction of genome size occurs through deletion of genes or genome regions. Deletion may begin with the disruption of (redundant) genes or promoter regions by point mutations, frameshifts and integration of transposable elements, which leads to the formation of pseudogenes (Mira et al., 2001). Owing to the lack of selection, these pseudogenes are degraded gradually until they have disappeared completely. This process correlates frequently with a change in the environment or lifestyle, for example, during adaptation to intracellular symbiosis with an eukaryotic host (Moran, 2002; Klasson and Andersson, 2004). The genome of the pathogen Mycobacterium leprae is a well-known example for a genome in an early stage of genome erosion (Cole et al., 2001). It contains a high number of pseudogenes (n=1116) and large intergenic spacer regions (representing strongly degraded pseudogenes, which are not recognized as such anymore). On the other end of the spectrum, the highly reduced genome of Buchnera contains only few pseudogenes and the lengths of intergenic regions are typical for more stable genomes (Mira et al., 2001). The genome fragment of ‘Candidatus S. adelgidicola' contains five pseudogenes (Supplementary Figure S6), which were formed by point mutations introducing premature stop codons and frameshifts. Three of these pseudogenes, which are absent in most obligate symbionts, are in genome regions that are syntenic with Escherichia coli (Figure 6). Extrapolated, the observed number of pseudogenes on the ‘Candidatus S. adelgidicola' genome fragment corresponds to 0.584 pseudogenes per 10 kbp, which is closer to the number of pseudogenes in facultative symbionts (0.886–2.33) than in obligate symbionts (0–0.146; Figure 7, Supplementary Table S4). Consistent with this notion, the average spacer length of ‘Candidatus S. adelgidicola' (909.3 bp) is also in the range of facultative symbionts (201.6–857.6 bp) and markedly larger than that of obligate symbionts (61.4–299.7 bp; Figure 7, Supplementary Table S4). Thus, the genome of ‘Candidatus S. adelgidicola' shows typical features of facultative symbionts and is less streamlined than the genomes of obligate symbionts.
Figure 6.
Pseudogene formation in ‘Candidatus Steffania adelgidicola'. The formation of pseudogenes (light grey boxes) in the ‘Candidatus S. adelgidicola' genome can be observed in syntenic regions with E. coli K-12 substr. MG1655. It must be noted that the genome region shown in the lower panel is also synthenic with S. glossinidius, ‘Candidatus Blochmannia pennsylvanicus' and other Gammaproteobacteria but not with Buchnera. The number of stop codons (star symbol) and frameshifts (black arrow) are noted. The rnhB gene is truncated at the 5′ end.
Figure 7.
Correlation of bacterial lifestyle and general genome features for selected free-living bacteria and insect symbionts. The numbers of pseudogenes, genomic G+C content and average spacer length are displayed (for details, see Supplementary Table S4). Gammaproteobacterial obligate symbionts of insects share similar genome features and are clearly separated in this three-dimensional scatter plot. Signs for genome erosion can be observed in facultative symbionts to different degrees. The placement of ‘Candidatus Steffania adelgidicola' between these two groups suggests that ‘Candidatus S. adelgidicola' is an evolutionarily young symbiont the genome of which is in the process of size reduction. For comparison, ‘Candidatus Hodgkinia cicadicola', an alphaproteobacterial symbiont of cicadas is shown. Its genome is highly reduced but in contrast to the genomes of gammaproteobacterial insect symbionts shows a high G+C content.
Within the Gammaproteobacteria, genome reduction is also accompanied by a decrease in genomic G+C content (Moran et al., 2008). The average G+C content of the genome fragment of ‘Candidatus S. adelgidicola' is 39.1%. Although the G+C content of the closest free-living relative of ‘Candidatus S. adelgidicola' is unknown, this is in the range of the genomic G+C content of other facultative symbionts of insects but notably higher than the G+C content in highly reduced bacterial genomes from obligate symbionts such as Buchnera and Wigglesworthia (Figure 7, Supplementary Table S4; Williams et al., 2010). These genome features and the lifestyle of bacterial symbionts of insects reflect the age of their association with their insect hosts, with facultative symbionts being evolutionary younger symbionts than obligate symbionts (Supplementary Table S4; Moran et al., 2008). Taken together, if G+C content, pseudogenes and spacer length are taken as proxies for the degree of genome reduction, lifestyle and evolutionary age of the symbiosis, ‘Candidatus S. adelgidicola' is an evolutionarily young bacteriocyte-associated symbiont and its genome might still be undergoing genome reduction.
Although phylogenetic analysis of 16S and 23S rRNA genes failed to identify the closest relative of ‘Candidatus S. adelgidicola' (Figure 3), two lines of evidence suggest an affiliation with Sodalis-related symbionts of diverse insect hosts (Kaiwa et al., 2010). First, 42 out of 44 predicted proteins encoded on the ‘Candidatus S. adelgidicola' genome fragment show S. glossinidius proteins as closest homologs. Second, in phylogenetic trees based on the RNA polymerase sigma-32 factor RpoH, ‘Candidatus S. adelgidicola' formed a monophyletic group with the respective S. glossinidius protein (Supplementary Figure S7). Sodalis-related symbionts are considered evolutionary young symbionts showing less reduced genomes compared with long-term-associated obligate symbionts (Rio et al., 2003), corroborating our findings from the analysis of the ‘Candidatus S. adelgidicola' genome fragment.
Conclusions
This study shows that adelgids of the A. nordmannianae/piceae complex harbor two novel Gammaproteobacteria as bacteriocyte-associated symbionts, both of which show no close relationship with known facultative or obligate symbionts of other insects. These adelgids have thus acquired their symbionts independently from members of their sister group, the aphids. The genome of one of these symbionts, ‘Candidatus S. adelgidicola', is reduced compared with the genome of free-living Gammaproteobacteria, but it is less streamlined than the genomes of known long-term obligate insect symbionts. The symbiosis between ‘Candidatus S. adelgidicola' and its insect host thus most likely represents an evolutionary young association, which was established after diversification of the Adelgidae. We predict that other members of this insect family have acquired their symbionts independently, a notion which is also supported by evidence for a larger morphological diversity of bacterial symbionts in this insect family (Profft, 1936; Steffan, 1968). Further molecular characterization of other adelgid symbionts is required to improve our understanding of the evolutionary history of these associations and to reveal the specific functions of these intracellular symbionts for their insect hosts.
Acknowledgments
We gratefully acknowledge Marc Mußmann for advice during fosmid library preparation, and Frank Maixner and Patrick Tischler for help with sampling and annotation, respectively. This work was partially funded by the Austrian Science Fund (FWF) grants Y277-B03 and P22533-B17.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, et al. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat Genet. 2002;32:402–407. doi: 10.1038/ng986. [DOI] [PubMed] [Google Scholar]
- Altschul S, Gish W, Miller W, Myers E, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Balch RE. Studies of the balsam woolly aphid Adelges piceae (Ratz.) and its effects on balsam fir, Abies balsamea (L.) Mill. Can Dep Agric Pub. 1952;867:1–76. [Google Scholar]
- Baumann P. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- Baumann P.2006Diversity of prokaryote-insect associations within the Sternorrhyncha (psyllids, whiteflies, aphids, mealybugs)In: Bourtzis K, Miller TA (eds)Insect Symbiosis CRC Press: Boca Raton; 1–24. [Google Scholar]
- Besemer J, Lomsadze A, Borodovsky M. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucl Acids Res. 2001;29:2607–2618. doi: 10.1093/nar/29.12.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binazzi A. Notes on and key to winged forms of adelgids recorded from Italy (Homoptera, Aphidoidea, Adelgidae) Redia. 2000;83:187–215. [Google Scholar]
- Blackman RL, Eastop VF. Aphids of the World's Trees: An Identification and Information Guide. CAB International: Wallingford; 1994. [Google Scholar]
- Bryant DG. Balsam woolly aphid Adelges piceae (Homoptera: Phylloxeridae) seasonal and spatial development in crowns of balsam fir, Abies balsamea. Can Entomol. 1971;103:1411–1420. [Google Scholar]
- Buchner P.1953Endosymbiose der Tiere mit Pflanzlichen Mikroorganismenvol. 12Birkhäuser: Basel/Stuttgart [Google Scholar]
- Burke GR, Moran NA. Massive genomic decay in Serratia symbiotica, a recently evolved symbiont of aphids. Genome Biol Evol. 2011;3:195–208. doi: 10.1093/gbe/evr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- Consortium TU. The Universal Protein Resource (UniProt) in 2010. Nucl Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daims H, Stoecker K, Wagner M.2005Fluorescence in situ hybridization for the detection of prokaryotesIn: Osborn A, Smith C (eds)Advanced Methods in Molecular Microbial Ecology Bios-Garland: Abingdon, UK; 213–239. [Google Scholar]
- Degnan PH, Lazarus AB, Wernegreen JJ. Genome sequence of Blochmannia pennsylvanicus indicates parallel evolutionary trends among bacterial mutualists of insects. Genome Res. 2005;15:1023–1033. doi: 10.1101/gr.3771305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Yu Y, Sisneros N, Wing RA, Moran NA. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc Natl Acad Sci USA. 2009;106:9063–9068. doi: 10.1073/pnas.0900194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn O. On methods of differentiating the species of the harmful white woolly aphids (genus Dreyfusia Cb.=Adelges An.) on fir, and the consequences for forest protection. Tech Bull CIBC. 1967;8:53–82. [Google Scholar]
- Eichhorn O. Folgerungen aus dem Auftreten der Gallen von Dreyfusia nordmannianae Eckst. (= D. nüsslini CB) in Mitteleuropa auf ihren Generationszyklus. Z Angew Entomol. 1973;74:196–199. [Google Scholar]
- Foottit RG, Maw HEL, Havill NP, Ahern RG, Montgomery ME. DNA barcodes to identify species and explore diversity in the Adelgidae (Insecta: Hemiptera: Aphidoidea) Mol Ecol Resour. 2009;9:188–195. doi: 10.1111/j.1755-0998.2009.02644.x. [DOI] [PubMed] [Google Scholar]
- Frishman D, Albermann K, Hani J, Heumann K, Metanomski A, Zollner A, et al. Functional and structural genomics using PEDANT. Bioinformatics. 2001;17:44–57. doi: 10.1093/bioinformatics/17.1.44. [DOI] [PubMed] [Google Scholar]
- Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, Lindgreen S, et al. Rfam: updates to the RNA families database. Nucl Acids Res. 2009;37:D136–D140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosalbes MJ, Lamelas A, Moya A, Latorre A. The striking case of tryptophan provision in the cedar aphid Cinara cedri. J Bacteriol. 2008;190:6026–6029. doi: 10.1128/JB.00525-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Havill NP, Foottit RG. Biology and evolution of Adelgidae. Annu Rev Entomol. 2007;52:325–349. doi: 10.1146/annurev.ento.52.110405.091303. [DOI] [PubMed] [Google Scholar]
- Havill NP, Foottit RG, von Dohlen CD. Evolution of host specialization in the Adelgidae (Insecta: Hemiptera) inferred from molecular phylogenetics. Mol Phylogenet Evol. 2007;44:357–370. doi: 10.1016/j.ympev.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ishikawa H. Host-symbiont interactions in the protein synthesis in the pea aphid, Acyrthosiphon pisum. Insect Biochem. 1982;12:613–622. [Google Scholar]
- Kaiwa N, Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Kimura N, et al. 2010Primary gut symbiont and secondary Sodalis-allied symbiont in the scutellerid stinkbug Cantao ocellatus Appl Environ Microbiol 763486–3494.AEM.00421-00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucl Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahrén D, et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucl Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucl Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson L, Andersson SGE. Evolution of minimal-gene-sets in host-dependent bacteria. Trends Microbiol. 2004;12:37–43. doi: 10.1016/j.tim.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Lamelas A, Perez-Brocal V, Gomez-Valero L, Gosalbes MJ, Moya A, Latorre A. Evolution of the secondary symbiont ‘Candidatus Serratia symbiotica' in aphid species of the subfamily Lachninae. Appl Environ Microbiol. 2008;74:4236–4240. doi: 10.1128/AEM.00022-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T, Eddy S. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy A, Maixner F, Wagner M, Horn M. probeBase: an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucl Acids Res. 2007;35:D800–D804. doi: 10.1093/nar/gkl856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucl Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Montllor CB, Maxmen A, Purcell AH. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol. 2002;27:189–195. [Google Scholar]
- Moran NA. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- Moran NA, Munson MA, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc R Soc B. 1993;253:167–171. [Google Scholar]
- Moran NA, Plague GR, Sandstrom JP, Wilcox JL. A genomic perspective on nutrient provisioning by bacterial symbionts of insects. Proc Natl Acad Sci USA. 2003;100 (Suppl 2:14543–14548. doi: 10.1073/pnas.2135345100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA, Russell JA, Koga R, Fukatsu T. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl Environ Microbiol. 2005;71:3302–3310. doi: 10.1128/AEM.71.6.3302-3310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray RGE, Stackebrandt E. Taxonomic note: implementation of the provisional status Candidatus for incompletely described procaryotes. Int J Syst Bacteriol. 1995;45:186–187. doi: 10.1099/00207713-45-1-186. [DOI] [PubMed] [Google Scholar]
- Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267–26. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- Oliver KM, Russell JA, Moran NA, Hunter MS. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci USA. 2003;100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Profft J. Beiträge zur Symbiose der Aphiden und Psylliden. Z Morphol Oekol Tiere. 1936;32:289–326. [Google Scholar]
- Ramsey JS, MacDonald SJ, Jander G, Nakabachi A, Thomas GH, Douglas AE. Genomic evidence for complementary purine metabolism in the pea aphid, Acyrthosiphon pisum, and its symbiotic bacterium Buchnera aphidicola. Insect Mol Biol. 2010;19:241–248. doi: 10.1111/j.1365-2583.2009.00945.x. [DOI] [PubMed] [Google Scholar]
- Richardson KC, Jarett L, Finke EH. Embedding in epoxy resins for ultrathin sectioning in electron microscopy. Stain Technol. 1960;35:313–323. doi: 10.3109/10520296009114754. [DOI] [PubMed] [Google Scholar]
- Rio RV, Lefevre C, Heddi A, Aksoy S. Comparative genomics of insect-symbiotic bacteria: influence of host environment on microbial genome composition. Appl Environ Microbiol. 2003;69:6825–6832. doi: 10.1128/AEM.69.11.6825-6832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Scarborough CL, Ferrari J, Godfray HC. Aphid protected from pathogen by endosymbiont. Science. 2005;310:1781. doi: 10.1126/science.1120180. [DOI] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Shields KS, Hirth RT.2005Bacterial Endosymbionts of Adelges Tsugae Annand: Potential Targets for Biocontrol? USDA For. Serv: Morgantown; ; http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.65 .638&rep=rep1&type=pdf#page=371 . [Google Scholar]
- Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS Nat. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- Spurr AR. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- Steffan AW. Evolution und Systematik der Adelgidae (Homoptera: Aphidina): Eine Verwandtschaftsanalyse auf vorwiegend ethologischer, zytologischer und karyologischer Grundlage. Zoologica. 1968;115:1–139. [Google Scholar]
- Steffan AW. Unterordnung Aphidina, Blattläuse. Parey: Hamburg; 1972. [Google Scholar]
- Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, et al. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucl Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Zucker J, Macdonald S, Sorokin A, Goryanin I, Douglas A. A fragile metabolic network adapted for cooperation in the symbiotic bacterium Buchnera aphidicola. BMC Syst Biol. 2009;3:24. doi: 10.1186/1752-0509-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, Weiss BL, Perkin SAH, Yamashita A, Oshima K, Hattori M, et al. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16:149–156. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchida T, Koga R, Fukatsu T. Host plant specialization governed by facultative symbiont. Science. 2004;303:1989. doi: 10.1126/science.1094611. [DOI] [PubMed] [Google Scholar]
- Vorburger C, Gehrer L, Rodriguez P. A strain of the bacterial symbiont Regiella insecticola protects aphids against parasitoids. Biol Lett. 2010;6:109–111. doi: 10.1098/rsbl.2009.0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MC, Rattei T, Arnold R, Güldener U, Münsterkötter M, Nenova K, et al. PEDANT covers all complete RefSeq genomes. Nucl Acids Res. 2009;37:D408–D411. doi: 10.1093/nar/gkn749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, et al. Database resources of the National Center for Biotechnology Information. Nucl Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KP, Gillespie JJ, Sobral BWS, Nordberg EK, Snyder EE, Shallom JM, et al. 2010Phylogeny of Gammaproteobacteria J Bacteriol 1922305–2314.JB.01480-01409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Daugherty SC, Aken SE, Pai GH, Watkins KL, Khouri H, et al. Metabolic complementarity and genomics of the dual bacterial symbiosis of sharpshooters. PLoS Biol. 2006;4:e188. doi: 10.1371/journal.pbio.0040188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zientz E, Dandekar T, Gross R. Metabolic interdependence of obligate intracellular bacteria and their insect hosts. Microbiol Mol Biol Rev. 2004;68:745–770. doi: 10.1128/MMBR.68.4.745-770.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurovcova M, Havelka J, Stary P, Vechtova P, Chundelova D, Jarosova A, et al. ‘DNA barcoding' is of limited value for identifying adelgids (Hemiptera: Adelgidae) but supports traditional morphological taxonomy. Eur J Entomol. 2010;107:147–156. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.