Abstract
Cystic fibrosis (CF) is a common fatal genetic disorder with mortality most often resulting from microbial infections of the lungs. Culture-independent studies of CF-associated microbial communities have indicated that microbial diversity in the CF airways is much higher than suggested by culturing alone. However, these studies have relied on indirect methods to sample the CF lung such as expectorated sputum and bronchoalveolar lavage (BAL). Here, we characterize the diversity of microbial communities in tissue sections from anatomically distinct regions of the CF lung using barcoded 16S amplicon pyrosequencing. Microbial communities differed significantly between different areas of the lungs, and few taxa were common to microbial communities in all anatomical regions surveyed. Our results indicate that CF lung infections are not only polymicrobial, but also spatially heterogeneous suggesting that treatment regimes tailored to dominant populations in sputum or BAL samples may be ineffective against infections in some areas of the lung.
Keywords: cystic fibrosis, amplicon sequencing, microbial communities
Cystic fibrosis (CF) is one of the most common heritable genetic diseases in Caucasian populations, affecting over 70 000 individuals worldwide (Cystic Fibrosis Foundation, 2011). Most mortality in CF is due to microbial infections of the lungs, which lead to eventual respiratory failure (Cystic Fibrosis Foundation, 2008). Pseudomonas aeruginosa, Staphylococcus aureus and Burkholderia cepacia are common pathogens cultured from adult CF patients, but recent studies have demonstrated that microbial communities in the CF airways are highly diverse (Armougom et al., 2009; Rogers et al., 2009; Cox et al., 2010; Guss et al., 2011). Culture-based analysis of bronchoalveolar lavage (BAL) and autopsy samples indicates that microbial populations vary significantly in different spatial regions of the CF lung (Smith et al., 1998; Gutierrez et al., 2001; Gilchrist et al., 2011). Erb-Downward et al. (2011) recently demonstrated spatial heterogeneity in microbial communities from the ex-planted lung tissue of COPD patients using culture-independent methods, but no similar study has been conducted in CF patients. Here, we present the first culture-independent characterization of microbial communities isolated directly from CF lung tissue.
To characterize microbial communities, we obtained lung tissue sections from the ex-planted lungs of a transplant patient, as well as tissue sections and a tracheal mucus plug from a deceased CF patient following autopsy. Patient clinical data are presented in the Supplementary Materials and methods. Microbial communities were profiled by high-throughput barcoded amplicon sequencing of the 16S rDNA gene and were analyzed using the QIIME pipeline (Supplementary Materials and methods) (Caporaso et al., 2010).
Microbial community richness was greater in the ex-plant lungs than in the postmortem lungs. At 97% similarity, there were 498 operational taxonomic units (OTUs) corresponding to 20 phyla in the ex-plant lungs, as compared with 106 OTUs comprising nine phyla in the postmortem lungs (Figure 1; Supplementary Tables 1 and 2). Rarefaction curves and Chao1 estimates indicated that expected OTU richness by lobe was an order of magnitude higher in the ex-plant lungs (Supplementary Figure 1; Supplementary Table 1). Both sets of lungs exhibited pathology characteristic of late-stage CF respiratory disease, but the ex-plant lungs maintained some areas of normal tissue, while the postmortem lungs showed near-complete destruction of the peripheral airways in all areas (Supplementary Materials and methods). This may be correlated with reduced richness as previous studies of microbial diversity in CF and COPD have demonstrated a negative relationship between microbial diversity and disease severity (Erb-Downward et al., 2011; van der Gast et al., 2011).
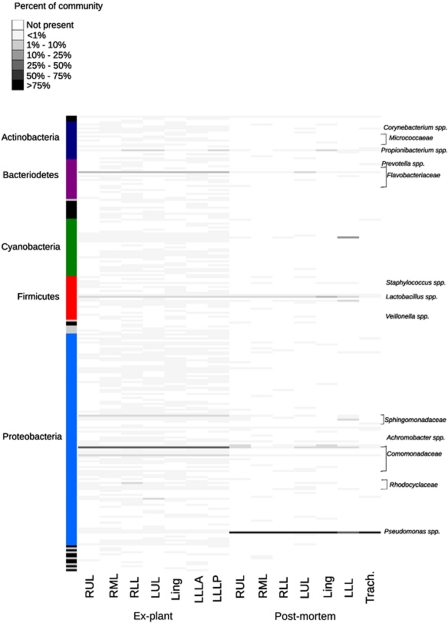
Figure 1.
Relative abundance of microbial OTUs in the ex-plant and postmortem lungs. Each row represents a different OTU, and the abundance as a percentage of the total population is indicated by color. Phyla are indicated on the left, and a subset of OTU classifications is indicated on the right. A complete list of OTU classification appears in Supplementary Table 2. Lung regions are abbreviated as RUL for right upper lobe, RML for right middle lobe, RLL for right lower lobe, LUL for left upper lobe, Ling for left lingula and LLL for left lower lobe. LLLA indicates the left lower lobe anterior and LLLP indicates the left lower lobe posterior. Trach indicates the tracheal sample.
Microbial communities in both sets of lungs were dominated by a small number of highly abundant taxa. One OTU classified as Comomonadaceae at the family level comprised ∼68% of total bacterial diversity in each lobe in the ex-plant lungs (Figure 1; Supplementary Table 2). Comomonadaceae have previously been identified in CF respiratory samples, but are not commonly recognized as CF pathogens (Coenye et al., 2002; Guss et al., 2011). OTUs corresponding to Flavobacteriaceae sp. (14–23%), Diaphorobacter sp. (3–5%) and Novosphingobium sp. (2–4%) were also ubiquitous. In the postmortem lungs, Pseudomonas aeruginosa (BLASTn, 100% identity, e-value<1 × 10–180) was the most abundant taxon, comprising between 51–94% of sequences in each lobe, and 99% of the tracheal sample. Other OTUs were ubiquitous but found in lower (<3%) abundances, including phylotypes classified as Lactobacillus delbruckii, Enterococcus sp., Cloacibacterium sp. and Diaphorobacter sp.
Microbial communities in the ex-plant lungs were more similar to each other than those in the postmortem lungs when communities were compared using the weighted Unifrac distance metric (Figure 2; Supplementary Table 3). Distances were greater on average between lobes of the postmortem lungs than between lobes of the ex-plant lungs, reflecting more divergent communities (Supplementary Table 3). Less than 1% of OTUs were shared by all lobes in the postmortem lungs, while ∼60% of OTUs were identified in only one lobe (Supplementary Table 2). Several unique OTUs corresponded to potential CF pathogens, such as Xanthomonas (left upper lobe only) and Chyrseobacterium (Harrison, 2007; LiPuma, 2010). In the ex-plant lungs, ∼3% of OTUs were shared by all lobes, but most OTUs which were not universally shared occurred in low abundance (<1%). These may represent transient CF microbial populations as opposed to core CF microbiota as described in van der Gast et al. (2011).
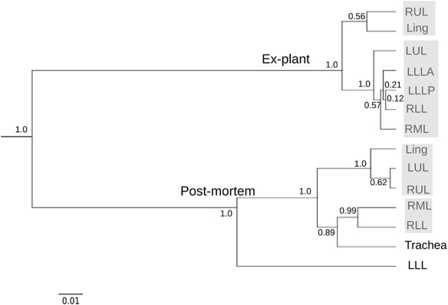
Figure 2.
Spatial clustering of microbial communities in the CF lungs. Communities were compared using the weighted Unifrac distance metric, and clustered using average linkage. Node labels indicate jackknife support values, and the bar below the tree represents a weighted Unifrac distance of 0.01. Shaded boxes indicate communities that were not significantly different from each other, but were significantly different from all other communities as determined by XIPE.
Average linkage (UMPGA) clustering based on Unifrac distances produced two main clusters corresponding to the ex-plant and postmortem lungs and several smaller clusters within each set of lungs (Figure 2). In the ex-plant lungs, microbial communities formed two significantly different clusters (Bonferroni's-corrected P-value<0.05), as determined by XIPE, which compares community distributions using non-parametric statistics (Rodriguez-Brito et al., 2006). One cluster contained the RUL and Lingula and the other contained all other lobes. This clustering was largely due to abundance differences in dominant OTUs, including Flavobacteriaceae spp. In the postmortem lungs, communities formed two clusters with the LLL as an outgroup (Figure 2). The divergence of the LLL was driven by a high abundance of chloroplast sequences, which may represent plant DNA introduced by aspiration (Bousbia et al., 2010). Aspiration may have occurred while the patient was intubated 3 weeks before death, as Bousbia et al. (2010) demonstrated the persistence of plant DNA in the lungs for several weeks in patients undergoing mechanical ventilation. Clustering of tracheal, RML and RLL communities may indicate gravity-dependent deposition of aspirated tracheal mucus, as tracheal contents would be expected to settle in the RLL and/or RML (Bittar, 2002). Despite the observed clustering, the tracheal community was significantly different from the RML and RLL, containing seven OTUs not found in the lungs, including Prevotella spp., a common constituent of oral microbiota.
Our results indicate that microbial communities in the cystic fibrosis lung are spatially heterogeneous. The use of autopsy samples is a caveat to this study, as microbial community composition may be affected by aspiration at death. Despite this limitation, direct sampling of lung tissue provides a more specific characterization of microbiota than other sampling methods, which may misrepresent the true nature of CF lung disease due to a lack of spatial resolution. Spatial heterogeneities may extend further than can be resolved by 16S sequencing, including regional differences in microbial biomass, antibiotic resistance and strain level differences in highly abundant and ubiquitous populations. Treatment strategies for cystic fibrosis, which rely on antibiotics targeted at microbes of specific species with known antibiotic resistance may only be effective against pathogenic microbes in particular regions or microenvironments of the lung.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Armougom F, Bittar F, Stremler N, Rolain JM, Robert C, Dubus JC, et al. Microbial diversity in the sputum of a cystic fibrosis patient studied with 16S rDNA pyrosequencing. Eur J Clin Microbiol Infect Dis. 2009;28:1151–1154. doi: 10.1007/s10096-009-0749-x. [DOI] [PubMed] [Google Scholar]
- Bittar EE. Pulmonary Biol health Dis. Springer; 2002. [Google Scholar]
- Bousbia S, Papazian L, La Scola B, Raoult D. Detection of plant DNA in the bronchoalveolar lavage of patients with ventilator-associated pneumonia. PLoS One. 2010;5:e11298. doi: 10.1371/journal.pone.0011298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coenye T, Goris J, Spilker T, Vandamme P, LiPuma JJ. Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of inquilinus limosus gen. nov., sp. nov. J Clin Microbiol. 2002;40:2062–2069. doi: 10.1128/JCM.40.6.2062-2069.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MJ, Cox MJ, Allgaier M, Taylor B, Baek MS, Huang YJ, et al. Airway microbiota and pathogen abundance in age-stratified cystic fibrosis patients. PLoS One. 2010;5:e11044. doi: 10.1371/journal.pone.0011044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cystic Fibrosis Foundation Patient Registry 2008 Annual Report (2008
- Cystic Fibrosis Foundation 2011. Available at: http://www.cff.org ( accessed 20 February 2011).
- Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, et al. Analysis of the lung microbiome in the ‘Healthy' smoker and in COPD. PLoS One. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist FJ, Salamat S, Clayton S, Peach J, Alexander J, Lenney W, et al. 2011Bronchoalveolar lavage in children with cystic fibrosis: how many lobes should be sampled Arch Dis Child 96215–217.Available at: http://www.ncbi.nlm.nih.gov/pubmed/20930010 ( accessed 7 December 2010). [DOI] [PubMed] [Google Scholar]
- Guss AM, Roeselers G, Newton IL, Young CR, Klepac-Ceraj V, Lory S, et al. 2011Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis ISME J 520–29.Available at: http://www.ncbi.nlm.nih.gov/pubmed/20631810 ( accessed 19 September 2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez JP, Grimwood K, Armstrong DS, Carlin JB, Carzino R, Olinsky A, et al. Interlobar differences in bronchoalveolar lavage fluid from children with cystic fibrosis. Eur Respir J. 2001;17:281–286. doi: 10.1183/09031936.01.17202810. [DOI] [PubMed] [Google Scholar]
- Harrison F. Microbial ecology of the cystic fibrosis lung. Microbiology (Reading, Engl) 2007;153:917–923. doi: 10.1099/mic.0.2006/004077-0. [DOI] [PubMed] [Google Scholar]
- LiPuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev. 2010;23:299–323. doi: 10.1128/CMR.00068-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Rohwer F, Edwards RA. An application of statistics to comparative metagenomics. BMC Bioinformatics. 2006;7:162. doi: 10.1186/1471-2105-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GB, Carroll MP, Bruce KD. Studying bacterial infections through culture-independent approaches. J Med Microbiol. 2009;58:1401–1418. doi: 10.1099/jmm.0.013334-0. [DOI] [PubMed] [Google Scholar]
- Smith D, Smith E, Pitt T, Stableforth D. Regional microbiology of the cystic fibrosis lung: a post-mortem study in adults. J Infect. 1998;37:41–43. doi: 10.1016/s0163-4453(98)90475-3. [DOI] [PubMed] [Google Scholar]
- van der Gast CJ, Walker AW, Stressmann FA, Rogers GB, Scott P, Daniels TW, et al. Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J. 2011;5:780–791. doi: 10.1038/ismej.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.