Abstract
Plague, caused by the bacterium Yersinia pestis, is a mammalian vector-borne disease, transmitted by fleas that serve as the vector between rodent hosts. For many pathogens, including Y. pestis, there are strong evolutionary pressures that lead to a reduction in ‘useless genes', with only those retained that reflect function in the specific environment inhabited by the pathogen. Genetic traits critical for survival and transmission between two environments, the rodent and the flea, are conserved in epizootic/epidemic plague strains. However, there are genes that remain conserved for which no function in the flea–rodent cycle has yet been observed, indicating an additional environment may exist in the transmission cycle of plague. Here, we present evidence for highly conserved genes that suggests a role in the persistence of Y. pestis after death of its host. Furthermore, maintenance of these genes points to Y. pestis traversing a post-mortem path between, and possibly within, epizootic periods and offering insight into mechanisms that may allow Y. pestis an alternative route of transmission in the natural environment.
Keywords: Yersinia pestis, pathogen evolution, taphonomy, transmission, gene reduction/conservation
Introduction
The bacterium, Yersinia pestis, the etiological agent responsible for the Black Death (Haensch et al., 2010) and two other plague pandemics has become essentially global in its endemicity (Stenseth et al., 2008). Typically, epizootic waves of the disease occur, whereby plague sweeps through a region during a 1- or 2-year period, after which it perceptibly disappears from the landscape for years, only to re-emerge once again (Stenseth et al., 2006; Eisen and Gage, 2009). Similar re-emergence patterns were observed following the initial spread of the Black Death in Europe (Cohn, 2008). The driving force behind this dynamic is assumed to depend on the disease re-emerging in local rodent populations and their fleas passing the disease to humans. The observed periods of quiescence have prompted speculation, and investigations have aimed at determining where Y. pestis sequesters itself between epizootics. There is debate about the significance of anecdotal evidence, and much discussion of the role of the quiescent period in the life history and persistence of Y. pestis. Yet, because of the rarity of detecting Y. pestis in the environment during quiescence, it has been difficult to demonstrate a reservoir responsible for re-emergence. Here, we review information within an evolutionary context, which suggests Y. pestis may be able to affect persistence after its host has died.
Y. pestis has been intensely studied with respect to molecular mechanisms responsible for virulence and survival in the host–flea cycle. The results of research show trends in the evolution of Y. pestis and its epizootic ecology. In fact, many bacterial pathogens follow a similar evolutionary trend as that of Y. pestis (Wren, 2003), such as selection favoring functional gene reduction (Moran, 2002) and gene rearrangement (Levin and Bull, 1994; Wren, 2003). Along with this information concerning evolutionary trends, there are genetic mechanisms that enable a complete flea–rodent transmission cycle during an epizootic, namely specific genes involved in host virulence, flea colonization or core cellular function (Hinnebusch et al., 1998, 2002; Anisimov et al., 2004; Motin et al., 2004).
The trend of genome reduction appears to be associated with Y. pestis transitioning into the new environments of the host and flea, as has occurred in the evolution from Yersinia pseudotuberculosis. Adaptation allowed the pathogen to shed genes that function in the mammalian gut, serving no purpose in the mammalian blood stream (systemic infection) or the flea gut. For instance, the ability to ferment melibiose, a sugar found in the mammalian diet is converted by the gut microbiota, including most species of the genus Yersinia, into glucose and galactose before being absorbed by the body into the blood stream. However, this function has been lost in pandemic strains of Y. pestis, as the latter evolved from its enteric ancestor, Y. pseudotuberculosis (Achtman et al., 2004). The loss of genes coding for this function and of other genes reflects the superfluous nature of these genes for fitness of Y. pestis outside the mammal gut environment. Indeed, much of the genome of Y. pestis is funneled toward functionality within two environments, represented by the flea and the mammalian blood stream. If Y. pestis were obligate to both host and flea, a paradox arises with respect to certain conserved genes and their function, because they seem superfluous in both of these environments. A logical additional step in the evolution and ecology of Y. pestis can be argued, however, that conserved genes serve a function in yet a third environment, the post-mortem host.
Microbial evolution
Addition of genes for an altered ecology
Acquisition of novel traits via mobile genetic elements, is a mechanism employed by bacteria allowing relative leaps in adaptation and evolution (Frost et al., 2005). Many mobile genetic elements are readily shared among microorganisms through horizontal gene transfer, quickly becoming an intrinsic part of the recipient's expression repertoire. These genetic additions persist if they impart a selective advantage and/or facilitate survival in a novel environment (Frost et al., 2005).
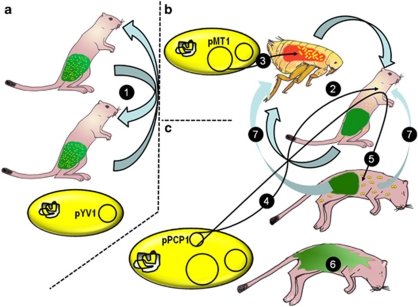
Evolution of Y. pestis from the enteric Y. pseudotuberculosis involved stepwise addition of two plasmids, pMT1 and pPCP1, respectively (Sebbane et al., 2006), within recent history, that is, >2600 years ago (Morelli et al., 2010). The pMT1 plasmid likely was responsible for the first step in the transition, as it is critical to colonization of the flea and induces septicemia at low incidence in mammals (Sebbane et al., 2006). Thus, Y. pestis can persist as a blood-borne pathogen despite a low transmission rate (Anisimov et al., 2004; Sebbane et al., 2006). Acquisition of the 9.5 kb pPCP1 plasmid increases virulence and global success of the pathogen (Figure 1). The combination of plasmids has allowed Y. pestis to flourish in the new environment of the flea and, more importantly, as a mammalian pathogen (Achtman et al., 1999; Zhou and Yang, 2009).
Figure 1.
Evolution of virulence and persistence in Y. pestis. (a) Y. pseudotuberculosis, a pathogen specialized to the mammalian gut, transmitted fecal-orally (1) first adapted into (b) flea-transmitted septicemic Y. pestis (2) (Sebbane et al., 2006), addition of pMT1 allows colonization of the flea gut (3). (c) Addition of pPCP1 both enhanced virulence and persistence. Genes pla, pst and pim on pPCP1 are expressed within the host at 37 °C. Pla, prevents fibrin formation at the site of the flea bite, allowing dissemination to lymph nodes and other tissues (4), the I259T substitution within the pla gene increases fibrinolytic activity of Pla in pandemic strains, further enhancing this virulence mechanism (Haiko et al., 2009). Pst throughout the corpse allows Y. pestis to persist longer during transmission by preventing competitive gut flora colonization of the extra-digestive tissues. Corpse acidification during decomposition should increase function of Pst from 20% (living host blood pH 7) to near 100% at pH ∼4.5 (Ferber and Brubaker, 1979) (4). Y. pestis then transmits back into the flea–rodent cycle from the corpse or soil into the host (Drancourt et al., 2006) or flea larva (Wimsatt and Biggins, 2009) (5). Typically invading gut bacteria begin corpse decomposition in the absence of Pst produced by Y. pestis (6).
Genomic reduction: the race for virulence
Signatures of selection for the specific environments occupied by obligate and semi-obligate pathogens can be observed in their gene repertoire. Genes that have specific roles in either core cell function, host invasion, or host immune evasion and transmission are maintained, since they are critical for growth and survival, while enabling exploitation of the host, a result central to the ecology of a pathogen.
The environments that Y. pestis passes through during its transmission cycle create selective pressure on maintaining function of those genes required for both short-term survival and long-term persistence and eliminating those that are not. Pathogenic bacteria tend to lose non-functional DNA (genes) quickly (Moran, 2002), especially while the pathogen adapts to a new host environment. Gene inactivation is a common feature of genes that do not contribute to fitness, yet impart some cost in transcription and translation. Y. pestis appears to have undergone a period of rapid genome reduction, as the new variant of the pathogen benefits from cessation of DNA transcription and translation, both functions no longer serving a purpose in its new environment (Straley and Perry, 1995; Hinnebusch et al., 1998; Wren, 2003). This reduction is evidenced by loss of function in 13% of the genes of Y. pestis, compared with its direct ancestor, Y. pseudotuberculosis (Achtman et al., 2004; Bearden and Brubaker, 2010).
With genome reduction potentially resulting in a higher division rate, these types of mutation would create enhanced virulence (faster exploitation of host resources) in Y. pestis, resulting in a shorter lived host. In this case, within-host intraspecific competition selects for those bacteria with the highest division rate, which translates into enhanced virulence, despite potential self-destruction beyond the immediate host (Levin and Bull, 1994; Ebert, 1998). However, this outcome is balanced against continuous culling of Y. pestis during the transmission phase. Bacterial strains that undergo optimal selection for existence within a host or vector will be stymied if a function for the next step of the transmission cycle is lost or significantly weakened. This tension between selection for increased virulence and for higher probability of transmission is created by the time limit on transmission, imposed by the death of the host (or vector), if the pathogen does not have an alternative route of transmission. A highly virulent pathogen, therefore, requires either very efficient transmission or an alternative path, in the event the host dies before it has moved on to the next host, especially if abundance of the host is too low for effective flea-borne transmission (Davis et al., 2004). Improved survival between transmission events thus becomes imperative.
Pst and Pim: without function in the flea or in the living host
After initial infection via flea bite, genes critical for virulence in Y. pestis are upregulated. This includes the upregulation of pla at 37 °C. Pla activates host plasminogen that degrades host fibrin, likely preventing the walling off of the infection, allowing the bacteria to disseminate throughout the host. During the course of an infection with Y. pestis, cells proliferate to bacteremic levels throughout the body. Lysis of Y. pestis bacterial cells during infection contributes to septic shock and subsequent death of the host (Guarner et al., 2005). Pla, pesticin (pst) and pesticin immunity protein (pim) located on the pPCP1 plasmid are all transcribed at the host core body temperature of 37 °C (Straley and Perry, 1995; Motin et al., 2004), but only Pla has been shown to have a key role in pathogenesis (Sebbane et al., 2006; Haiko et al., 2009). As expression of pst and pim are also upregulated at the host temperature, it is not surprising that the genes on this plasmid are not required for Y. pestis to colonize and block the flea (Hinnebusch et al., 1998; Bearden and Brubaker, 2010). It should also be noted that strains lacking the pPCD1 are still found in populations of Microtus spp. in Eurasia as they are able to colonize the flea and infect rodents (Anisimov, 2002). Considering the two environments of flea and rodent comprising the epizootic cycle of plague, it would appear that Y. pestis maintains, pst and pim, despite their apparent superfluous nature. It can be hypothesized, therefore, that these genes have a role in a third environment, the post-mortem host.
Taphonomy (post-mortem decay of the host)
Decay of the host is progressive and is similar among mammals, given comparable conditions. Following death, initial decay begins with cellular autolysis from accumulation of carbon dioxide that also increases acidity that, in turn, denatures and coagulates proteins (Parkinson et al., 2009). This process causes a drop in pH and in the temperature to ambient and begins a cascade effect, whereby further acidification ensues, as a result of the breakdown of organic compounds. During decomposition, bacteria from the gut invade tissues where fermentation of carbohydrates drives the pH even lower (Gill-King, 1997).
After death of the host there are many nutrient resources available to Y. pestis and other microorganisms. Given that the plague bacilli are abundant as decomposition begins, they are in an advantageous position to monopolize and exploit the resources of the dead host. Y. pestis survives and grows optimally in environments similar to the post-mortem host under acidic conditions, for example, pH 4.0 (Feodorova and Devdariani, 2001). During decomposition and acidification of the corpse, the enzymatic activity of Pst, which is most likely abundant at the time of death, increases, since Pst has been demonstrated to be most active at ca pH 4.8 (Ferber and Brubaker, 1979) and calculated by Rakin et al. (1996) to be most active at pH 5.56 (Figure 1). The only other environment that Y. pestis inhabits during its life cycle that has a similar range of pH is within the host macrophage, where the entire pPCP1 plasmid has no measurable effect on survival (Agar et al., 2009).
The decomposing corpse is, therefore, a habitat low in pH and profuse with Pst (produced while the host was alive), creating an unfavorable environment for other bacteria (Parkinson et al., 2009). In conjunction with the chemical nature of this environment, structured habitats, like soil or a corpse are more favorable for survival of colicinogenic bacteria, because colicins (antimicrobials), like Pst, function to create an effective barrier against competitive bacteria (Chao and Levin, 1981). It is likely that pesticin functions to reduce or prevent its competitors from invading the corpse from the gut (Figure 1).
Plasmid PCP1 to kill and cope
The plasmid pPCP1, is 9.5 kb in size and contains three well-defined genes, which together serve two functions: ‘to kill competing cells and to support the high invasion potential of the Y. pestis population' (Rakin et al., 1996). The pst gene encodes pesticin, which causes bacterial cell wall degradation by breaking down the glycan backbone of murein and, thereby, causing susceptible bacteria to become spheroplasts, leading to cell death. Pesticin is a unique bacteriocin with mureinidase activity (Vollmer et al., 1997) degrading murein of many Gram-negative bacteria in vivo and murein of Gram-positive bacteria in vitro (Ferber and Brubaker, 1979). The pim gene encodes the Pim protein that confers self-immunity against pesticin. A third gene on pPCP1 is pla, the product prevents the host from walling off the infection at 37 °C and allows systemic dissemination of Y. pestis from the flea bite. The LD50 of Y. pestis is only a few cells, but on loss of the pPCP1 plasmid or the pla gene, Y. pestis is significantly attenuated, as has been shown when it is administered subcutaneously, intradermally or (at least partially) intranasaly in mice, inducing no change in LD50 intraperitoneally or intravenously. The effect of loss of pPCP1 on aerosol transmission is not known, but may prove to be strain dependent (Anisimov et al., 2004; Agar et al., 2009).
A high level of virulence is required to produce sufficient bacteremia to transmit back into the flea. However, increasing virulence enhances the probability of killing the host (Engelthaler et al., 2000; Eisen et al., 2006; Bearden and Brubaker, 2010). Increased virulence in Y. pestis shortens the time frame during which the pathogen can be transmitted to uninfected fleas. Pst may aid Y. pestis in colonizing the flea as it is a regular step in the transmission cycle, although strains lacking this plasmid do not show a discernable reduction in their ability to colonize fleas (Hinnebusch et al. 1998). It is most probably not coincidental that acquisition of pPCP1 contributes to global spread of plague by simultaneously inducing increased virulence (Sebbane et al., 2006; Agar et al., 2009; Zhou and Yang, 2009; Haensch et al., 2010) and producing pesticin, the latter being useful for environmental persistence and, thereby, transmission after death of the host. While the accepted view for decades has been that Y. pestis epidemiology is a host–flea–host cycle, dominated by rodents and fleas, with aerosol transmission sporadic and local, at best, it can be argued that combined genetic and ecological evidence suggest a significant role for additional stages that include the dead host and proximate natural environment. The seemingly puzzling conservation of pesticin genes (pst and pim) is clarified if pesticin is understood as being dedicated to survival of Y. pestis between its recognized direct transmission cycles to persistence within the corpse of the host and/or the natural environment. Effecting this stage of its life cycle, by preventing invasion of competitive bacteria in the gut from migrating into tissues and fluids (Rakin et al., 1996). The recent demonstration of a viable but nonculturable stage of Y. pestis (Pawlowski et al., 2011) offers further evidence for a dormant stage of Y. pestis in the natural environment. Direct detection of Y. pestis in the natural environment employing molecular genetic methods is in progress.
Concluding perspectives
Persistence of Y. pestis during inter-epizootic periods remains enigmatic. Much of this is due to the rarity of Y. pestis in the environment between epizootics. This persistence may likely be through a series of rare infections and longevity of some fleas. For instance, various species of fleas infected with Y. pestis survive well over a year (Gage and Kosoy, 2005). Here, we connect relationships between maintained genes and habitat, which may help enable this pathogen to persist during sub-optimal conditions of low host populations (Davis et al., 2004). Pst and pim are considered most likely maintained because of a selective advantage imparted to Y. pestis strains, including those Y. pestis strains responsible for two of the most recent pandemics (Haensch et al., 2010). Before host death, expression of pst ensures an environment diffused with the antimicrobial Pst, providing Y. pestis with a survival advantage. Extended persistence between host transmission is essential and advantageous when the host population density is low and Pst/Pim increases persistence fitness creating an advantage by persisting longer between infections (Handel and Bennett, 2008).
The viable but nonculturable state has been reported for Y. pestis exposed to soil and tap water in microcosms (Suchkov et al., 1997) providing evidence that Y. pestis is capable of surviving for long periods of time outside a viable host. The viable but nonculturable (dormant) state has been demonstrated in many bacteria and proposed as a response to stress, including low temperature, limited nutrient availability, osmotic stress and altered pH of the surrounding environment. The antibacterial, pesticin, serves to protect Y. pestis from competition and facilitates long-term persistence post mortem. Subsequent introduction of viable cells into the flea–rodent cycle from the post-mortem host, thereby, allowing resuscitation of the viable but nonculturable cells via passage in the host or flea should result in replication of the bacteria.
Evolution of Y. pestis, if deduced only from the known ecology of the flea–rodent cycle, creates a paradox in which PIM and PST have no, as yet, described function in either the host or the flea, yet despite having the selective advantages of genome reduction within the host, the gene pair is maintained. Logically, the paired genes would impart a selective advantage(s) to Y. pestis, if not in the flea then in the live host. Yet this is not the case, so far as current knowledge attests. A third environment offers the most credible explanation.
That Y. pestis survives post mortem from the host has been demonstrated (Eisen et al., 2008). Such post-mortem persistence may have a role in the transmission cycle, but this type of transmission has not yet been demonstrated to occur in the natural environment. It has proven difficult to demonstrate prime infection in an outbreak as having arisen from an environmental source, given the geographic range of plague outbreaks. Y. pestis has been shown to remain infectious outside both the flea and mammal host under artificial (Ayyadurai et al., 2008) and natural conditions (Drancourt et al., 2006; Eisen et al., 2008). Drancourt discusses the finding of Karimi (1963) (in French) that Y. pestis remains infectious from sealed burial chambers of burrowing rodents after 10 months and 11 months. These observations render plausible the existence of yet another stage in the natural environment.
Telluric plague, that is, plague emanating from soil is not a new concept. In 2006, Drancourt et al. renewed research on Y. pestis as a telluric bacterium, presenting possible mechanisms for reinfection of its host. The Russian research literature also explores telluric persistence of Y. pestis and mechanisms are offered for reintroduction into the plague cycle (Anisimov, 2002). Plague is infectious via the oral route and through abrasions, with the implication that transmission can occur via cannibalism (Anisimov, 2002). Wimsatt and Biggins (2009) present a plausible mechanism for Y. pestis to be transmitted from soil or a carcass into the flea–rodent cycle, whereby the flea–rodent cycle begins on infection of the larval flea, which remains infected to the adult stage, thereafter sparking a new epizootic (Wimsatt and Biggins, 2009).
In conclusion, it is necessary to understand those environments that create selective pressures shaping pathogen genomes as this allows prediction of possible environments and strategies supporting persistence. Obviously predictions for Y. pestis need to be tested and, in this context, the post-mortem environment and taphonomy of rodent corpses will prove instructive.
Acknowledgments
We would like to thank Eric De Muinck and Thomas Haverkamp for lending their knowledge during the inception of this paper. We also thank Anna Mazzarella for making Photoshop and its various layers accessible. Part of this work was funded through the US Department of Homeland Security.
References
- Achtman M, Morelli G, Zhu P, Wirth T, Diehl I, Kusecek B, et al. Microevolution and history of the plague bacillus, Yersinia pestis. Proc Natl Acad Sci USA. 2004;101:17837–17842. doi: 10.1073/pnas.0408026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agar SL, Sha J, Baze WB, Erova TE, Foltz SM, Suarez G, et al. Deletion of Braun lipoprotein gene (lpp) and curing of plasmid pPCP1 dramatically alter the virulence of Yersinia pestis CO92 in a mouse model of pneumonic plague. Microbiology. 2009;155:3247–3259. doi: 10.1099/mic.0.029124-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov AP. Factors of Yersinia pestis providing circulation and persistence of plague pathogen in ecosystems of natural foci. Communication 2. Molekulyarnaya Genetika Mikrobiologiya i Virusologiya. 2002;5:1–14. [PubMed] [Google Scholar]
- Anisimov AP, Lindler LE, Pier GB. Intraspecific diversity of Yersinia pestis. Clin Microbiol Rev. 2004;17:434–464. doi: 10.1128/CMR.17.2.434-464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayyadurai S, Houhamdi L, Lepidi H, Nappez C, Raoult D, Drancourt M. Long-term persistence of virulent Yersinia pestis in soil. Microbiology. 2008;154:2865–2871. doi: 10.1099/mic.0.2007/016154-0. [DOI] [PubMed] [Google Scholar]
- Bearden SW, Brubaker RR. Recent findings regarding maintenance of enzootic variants of Yersinia pestis in Sylvatic reservoirs and their significance in the evolution of epidemic plague. Vector-Borne Zoonotic Dis. 2010;10:85–92. doi: 10.1089/vbz.2009.0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao L, Levin BR. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc Natl Acad Sci USA. 1981;78:6324–6328. doi: 10.1073/pnas.78.10.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn SK. Epidemiology of the black death and successive waves of plague. Med Hist Suppl. 2008;27:74–100. [PMC free article] [PubMed] [Google Scholar]
- Davis S, Begon M, De Bruyn L, Ageyev VS, Klassovskiy NL, Pole SB, et al. Predictive thresholds for plague in Kazakhstan. Science. 2004;304:736–738. doi: 10.1126/science.1095854. [DOI] [PubMed] [Google Scholar]
- Drancourt M, Houhamdi L, Raoult D. Yersinia pestis as a telluric, human ectoparasite-borne organism. Lancet Infect Dis. 2006;6:234–241. doi: 10.1016/S1473-3099(06)70438-8. [DOI] [PubMed] [Google Scholar]
- Ebert D. Experimental evolution of parasites. Science. 1998;282:1432–1436. doi: 10.1126/science.282.5393.1432. [DOI] [PubMed] [Google Scholar]
- Eisen RJ, Bearden SW, Wilder AP, Montenieri JA, Antolin MF, Gage KL. Early-phase transmission of Yersinia pestis by unblocked fleas as a mechanism explaining rapidly spreading plague epizootics. Proc Natl Acad Sci USA. 2006;103:15380–15385. doi: 10.1073/pnas.0606831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen RJ, Gage KL. Adaptive strategies of Yersinia pestis to persist during inter-epizootic and epizootic periods. EDP Sci. 2009;40:01. doi: 10.1051/vetres:2008039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen RJ, Petersen JM, L HC, Wong D, Levy CE, Mead PS, et al. Persistence of Yersinia pestis in soil under natural conditions. Emerg Infect Dis. 2008;14:941–943. doi: 10.3201/eid1406.080029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelthaler D, Hinnebusch B, Rittner C, Gage K. Quantitative competitive PCR as a technique for exploring flea-Yersina pestis dynamics. Am J Trop Med Hyg. 2000;62:552–560. doi: 10.4269/ajtmh.2000.62.552. [DOI] [PubMed] [Google Scholar]
- Feodorova VA, Devdariani ZL. Expression of acid-stable proteins and modified lipopolysaccharide of Yersinia pestis in acidic growth medium. J Med Microbiol. 2001;50:979–985. doi: 10.1099/0022-1317-50-11-979. [DOI] [PubMed] [Google Scholar]
- Ferber DM, Brubaker RR. Mode of action of pesticin: N-acetylglucosaminidase activity. J Bacteriol. 1979;139:495–501. doi: 10.1128/jb.139.2.495-501.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost LS, Leplae R, Summers AO, Toussaint A. Mobile genetic elements: the agents of open source evolution. Nat Rev Micro. 2005;3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- Gage KL, Kosoy MY. The natural history of plague: perspectives from more than a century of research. Ann Rev Entomol. 2005;50:505–528. doi: 10.1146/annurev.ento.50.071803.130337. [DOI] [PubMed] [Google Scholar]
- Gill-King H. Forensic Taphonomy: The Postmortem Fate of Human Remains. CRC Press: Boca Raton, FL, USA, pp; 1997. pp. 94–95. [Google Scholar]
- Guarner J, Shieh W-J, Chu M, Perlman DC, Kool J, Gage KL, et al. Persistent Yersinia pestis antigens in ischemic tissues of a patient with septicemic plague. Human Pathol. 2005;36:850–853. doi: 10.1016/j.humpath.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Haensch S, Bianucci R, Signoli M, Rajerison M, Schultz M, Kacki S, et al. Distinct clones of Yersinia pestis caused the black death. PLoS Pathog. 2010;6:e1001134. doi: 10.1371/journal.ppat.1001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiko J, Kukkonen M, Ravantti JJ, Westerlund-Wikstrom B, Korhonen TK. The single substitution I259T, conserved in the plasminogen activator Pla of pandemic Yersinia pestis branches, enhances Fibrinolytic activity. J Bacteriol. 2009;191:4758–4766. doi: 10.1128/JB.00489-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel A, Bennett MR. Surviving the bottleneck: transmission mutants and the evolution of microbial populations. Genetics. 2008;180:2193–2200. doi: 10.1534/genetics.108.093013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch BJ, Fischer Elizabeth R, Schwan Tom G. Evaluation of the role of the Yersinia pestis Plasminogen activator and other plasmid encoded factors in temperature dependent blockage of the Flea. J Infect Dis. 1998;178:1406–1415. doi: 10.1086/314456. [DOI] [PubMed] [Google Scholar]
- Hinnebusch BJ, Rudolph AE, Cherepanov P, Dixon JE, Schwan TG, Forsberg A. Role of Yersinia murine toxin in survival of Yersinia pestis in the midgut of the flea vector. Science. 2002;296:733–735. doi: 10.1126/science.1069972. [DOI] [PubMed] [Google Scholar]
- Karimi Y.1963. Natural preservation of plague in soil (French). Bull Soc Pathol Exot Filiales 561183–1186. [PubMed]
- Levin BR, Bull JJ. Short-sighted evolution and the virulence of pathogenic microorganisms. Trends Microbiol. 1994;2:76–81. doi: 10.1016/0966-842x(94)90538-x. [DOI] [PubMed] [Google Scholar]
- Moran NA. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. [DOI] [PubMed] [Google Scholar]
- Morelli G, Song Y, Mazzoni CJ, Eppinger M, Roumagnac P, Wagner DM, et al. Yersinia pestis genome sequencing identifies patterns of global phylogenetic diversity. Nat Genet. 2010;42:1140–1143. doi: 10.1038/ng.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motin VL, Georgescu AM, Fitch JP, Gu PP, Nelson DO, Mabery SL, et al. Temporal global changes in gene expression during temperature transition in Yersinia pestis. J Bacteriol. 2004;186:6298–6305. doi: 10.1128/JB.186.18.6298-6305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson RA, Dias K-R, Horswell J, Greenwood P, Banning N, Tibbett M, et al. Microbial community analysis of human decomposition on soil. Criminal Environ Soil Forensics. 2009;24:379–394. [Google Scholar]
- Pawlowski DR, Metzger DJ, Raslawsky A, Howlett A, Siebert G, Karalus RJ, et al. Entry of Yersinia pestis into the viable but nonculturable state in a low-temperature tap water microcosm. PLoS One. 2011;6:e17585. doi: 10.1371/journal.pone.0017585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakin A, Boolgakowa E, Heesemann J. Structural and functional organization of the Yersinia pestis bacteriocin pesticin gene cluster. Microbiology. 1996;142:3415–3424. doi: 10.1099/13500872-142-12-3415. [DOI] [PubMed] [Google Scholar]
- Sebbane F, Jarrett CO, Gardner D, Long D, Hinnebusch BJ. Role of the Yersinia pestis plasminogen activator in the incidence of distinct septicemic and bubonic forms of flea-borne plague. Proc Natl Acad Sci USA. 2006;103:5526–5530. doi: 10.1073/pnas.0509544103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenseth NC, Samia NI, Viljugrein H, Kausrud KL, Begon M, Davis S, et al. Plague dynamics are driven by climate variation. Proc Natl Acad Sci USA. 2006;103:13110–13115. doi: 10.1073/pnas.0602447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenseth NC, Atshabar BB, Begon M, Belmain SR, Bertherat E, Carniel E, et al. Plague: past, present, and future. PLoS Med. 2008;5:9–13. doi: 10.1371/journal.pmed.0050003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straley SC, Perry RD. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 1995;3:310–317. doi: 10.1016/s0966-842x(00)88960-x. [DOI] [PubMed] [Google Scholar]
- Suchkov IuG, Khudiakov IV, Emel'ianenko EN, Levi MI, Pushkareva VI, Suchko IIu, et al. The possibility of preserving the causative agent of plague in resting (nonculturable) form. Zh Mikroiol Epidemiol Immunobiol. 1997;4:42–46. [PubMed] [Google Scholar]
- Vollmer W, Pilsl H, Hantke K, Holtje J, Braun V. Pesticin displays muramidase activity. J Bacteriol. 1997;179:1580–1583. doi: 10.1128/jb.179.5.1580-1583.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimsatt J, Biggins DE. A review of plague persistence with special emphasis on fleas. Natl Inst Malaria Res. 2009;46:85–99. [PubMed] [Google Scholar]
- Wren BW. The Yersiniae a model genus to study the rapid evolution of bacterial pathogens. Nat Rev Micro. 2003;1:55–64. doi: 10.1038/nrmicro730. [DOI] [PubMed] [Google Scholar]
- Zhou D, Yang R. Molecular darwinian evolution of virulence in Yersinia pestis. Infect Immun. 2009;77:2242–2250. doi: 10.1128/IAI.01477-08. [DOI] [PMC free article] [PubMed] [Google Scholar]