Abstract
Picocyanobacteria represented by Prochlorococcus and Synechococcus have an important role in oceanic carbon fixation and nutrient cycling. In this study, we compared the community composition of picocyanobacteria from diverse marine ecosystems ranging from estuary to open oceans, tropical to polar oceans and surface to deep water, based on the sequences of 16S-23S rRNA internal transcribed spacer (ITS). A total of 1339 ITS sequences recovered from 20 samples unveiled diverse and several previously unknown clades of Prochlorococcus and Synechococcus. Six high-light (HL)-adapted Prochlorococcus clades were identified, among which clade HLVI had not been described previously. Prochlorococcus clades HLIII, HLIV and HLV, detected in the Equatorial Pacific samples, could be related to the HNLC clades recently found in the high-nutrient, low-chlorophyll (HNLC), iron-depleted tropical oceans. At least four novel Synechococcus clades (out of six clades in total) in subcluster 5.3 were found in subtropical open oceans and the South China Sea. A niche partitioning with depth was observed in the Synechococcus subcluster 5.3. Members of Synechococcus subcluster 5.2 were dominant in the high-latitude waters (northern Bering Sea and Chukchi Sea), suggesting a possible cold-adaptation of some marine Synechococcus in this subcluster. A distinct shift of the picocyanobacterial community was observed from the Bering Sea to the Chukchi Sea, which reflected the change of water temperature. Our study demonstrates that oceanic systems contain a large pool of diverse picocyanobacteria, and further suggest that new genotypes or ecotypes of picocyanobacteria will continue to emerge, as microbial consortia are explored with advanced sequencing technology.
Keywords: cyanobacteria, Prochlorococcus, Synechococcus, diversity, global ocean, 16S-23S rRNA ITS
Introduction
Marine picocyanobacteria of the genera Prochlorococcus and Synechococcus represent the most abundant phytoplankton in the world's oceans, and are important contributors to the global primary production and carbon cycle (Li, 1994; Liu et al., 1997; Veldhuis et al., 1997). Prochlorococcus typically dominates tropical and subtropical oceans between the latitudes of 45°N and 40°S (Campbell et al., 1994; Partensky et al., 1999), whereas Synechococcus inhabits much broader marine environments ranging from the equatorial to polar regions, and from coastal to open oceans (Liu et al., 2002; Zwirglmaier et al., 2008). In general, Prochlorococcus is more abundant in warm oligotrophic waters and is absent in eutrophic coastal waters, whereas Synechococcus dominates the picocyanobacterial communities in eutrophic coastal and mesotrophic open ocean waters (Partensky et al., 1999). Remarkably, diverse Prochlorococcus and Synechococcus genotypes/ecotypes in the sea show different geographical preference and niche adaptation (see a summary in Table 1 and the description below).
Table 1. Biogeography of Prochlorococcus and Synechococcus lineages in the sea.
| Lineage(s) | Distribution preference or sources reported | References |
|---|---|---|
| Prochlorococcus | ||
| HLI | Upper-to-middle euphotic zone, subtropical open ocean, higher latitude | Johnson et al., 2006; Zwirglmaier et al., 2007 |
| HLII | Upper-to-middle euphotic zone, tropical open ocean, lower latitude | Johnson et al., 2006; Zwirglmaier et al., 2007 |
| HNLCs | Upper-to-middle euphotic zone, equatorial, iron-depleted ocean | Rusch et al., 2010; West et al., 2010 |
| LLI | Middle-to-lower euphotic zone, upper euphotic zone in deep mixing waters | Johnson et al., 2006; Zinser et al., 2007; Malmstrom et al., 2010 |
| LLII/III | Lower euphotic zone | Malmstrom et al., 2010 |
| LLIV | Lower euphotic zone | Johnson et al., 2006; Zinser et al., 2007 |
| LLV/VI | Oxygen-depleted intermediate depth | Lavin et al., 2010 |
| NC1 | Lower euphotic zone | Martiny et al., 2009 |
| Synechococcus | ||
| Subcluster 5.1 | ||
| I | Temperate to polar waters, co-occurrence with clade IV | Zwirglmaier et al., 2007, 2008 |
| II | Tropical/subtropical waters | Fuller et al., 2003; Zwirglmaier et al., 2008 |
| III | Oligotrophic waters | Zwirglmaier et al., 2008 |
| IV | Temperate to polar waters, co-occurrence with clade I | Zwirglmaier et al., 2007, 2008 |
| V–VII | Oceanic waters | Zwirglmaier et al., 2008 |
| VIII | Hypersaline waters | Dufresne et al., 2008 |
| IX | Rarely detected | Zwirglmaier et al., 2008 |
| XI–XIV | Gulf of Aqaba subsurface water (predominant) | Penno et al., 2006 |
| XV/XVI | Sargasso Sea (isolates and environmental sequences) | Ahlgren and Rocap, 2006 |
| CRD1/CRD2 | Costa Rica upwelling dome (predominant) | Saito et al., 2005 |
| WPC1/WPC2 | East China Sea and East Sea (isolates and environmental sequences) | Choi and Noh, 2009 |
| CB1–CB3 | Chesapeake Bay, summer | Chen et al., 2006; Cai et al., 2010 |
| Subcluster 5.2 | ||
| CB4 | Chesapeake Bay, summer | Chen et al., 2006; Cai et al., 2010 |
| CB5 | Chesapeake Bay, summer | Chen et al., 2006; Cai et al., 2010 |
| Subcluster 5.3 | Mediterranean and East Sea (three strains) and Sargasso Sea (six clones) | Ahlgren and Rocap, 2006; Dufresne et al., 2008; Choi and Noh, 2009 |
Extensive studies have delineated that Prochlorococcus is composed of two distinct ecotypes, the high-light (HL)- and low-light (LL)-adapted ecotypes. These ecotypes have remarkable correspondence with the genotypes, based on the phylogeny of the 16S rRNA gene or the 16S-23S rRNA internal transcribed spacer sequence (ITS thereafter; Moore et al., 1998; Moore and Chisholm, 1999; Rocap et al., 2002). The HL-adapted ecotype dominates the upper regions of the euphotic zone, whereas the LL-adapted ecotype is most abundant in the lower euphotic zone (Ahlgren et al., 2006; Johnson et al., 2006; Zinser et al., 2007). To date, at least 11 Prochlorococcus lineages have been identified, including the earlier six clades (HLI, HLII, LLI, LLII, LLIII and LLIV; Rocap et al., 2002), the recent three LL-adapted clades (NC1, LLV and LLVI; Martiny et al., 2009; Lavin et al., 2010), and two HL-adapted clades (HNLC1 and HNLC2) recognized in high-nutrient, low-chlorophyll (HNLC), iron-depleted waters in the Equatorial and South Pacific and tropical Indian Oceans (Rusch et al., 2010; West et al., 2010). Furthermore, HL-adapted lineages also show a complementary distribution pattern in that HLI appears to dominate in subtropical oceans (cooler, higher latitude), whereas HLII tends to dominate in tropical oceans (warmer, lower latitude; Johnson et al., 2006; Zinser et al., 2007; Zwirglmaier et al., 2008). Phylogenetic analyses, based on sequences from cultivated Prochlorococcus and clone libraries, both revealed that LL-adapted lineages harbor much larger extent of genetic variation than HL-adapted ones (Rocap et al., 2002; Ahlgren and Rocap, 2006; Zinser et al., 2006; Garczarek et al., 2007; Lavin et al., 2010).
Marine Synechococcus also show high genetic diversity, and have recently been subdivided into three major subclusters, 5.1, 5.2 and 5.3 (Dufresne et al., 2008; Scanlan et al., 2009). Most marine Synechococcus belong to subcluster 5.1, which contains at least 20 recognizable lineages unveiled by different gene markers (Toledo and Palenik, 1997; Rocap et al., 2002; Fuller et al., 2003; Ahlgren and Rocap, 2006; Penno et al., 2006; Choi and Noh, 2009). Similar to Prochlorococcus, Synechococcus also shows geographic niche exploitation. Synechococcus in clades I, II and IV are dominant on a global scale (Zwirglmaier et al., 2008). More specifically, clade II Synechococcus are common in subtropical/tropical open ocean waters, whereas Synechococcus in clades I and IV are largely confined to coastal and higher latitude regions (above ca. 30°N or below 30°S; Ferris and Palenik, 1998; Toledo and Palenik, 2003; Brown et al., 2005; Zwirglmaier et al., 2007, 2008). The Synechococcus found in the estuarine or coastal bays often contain unique genotypes distinct from those in the open oceans. For example, Synechococcus strains isolated from the Chesapeake Bay and the East Sea were classified into subclusters 5.2 and 5.3, respectively, (Chen et al., 2006; Choi and Noh, 2009). However, much less is known about the biogeography of subclusters 5.2 and 5.3 Synechococcus compared with subcluster 5.1.
Previous studies revealed that the cell numbers of known picocyanobacterial genotypes could not fully account for the total community abundance determined by flow cytometry, suggesting the existence of unknown lineages (Ahlgren et al., 2006; Zinser et al., 2006). In this study, we analyzed the genetic diversity of picocyanobacteria in the global oceans, including the South China Sea, Pacific and Atlantic Oceans, sub-Arctic and Arctic waters, to provide a comprehensive understanding of the diversity of marine picocyanobacteria. The ITS region of ribosomal DNA from marine picocyanobacteria was PCR amplified, cloned and sequenced. In addition, representative cyanobacterial sequences from our study were blasted against the Global Ocean Sampling (GOS) expedition database to better understand the geographic distribution of cyanobacterial genotypes in broader oceans.
Materials and Methods
Water sample collection
Five water samples were collected from the surface waters of the North Atlantic Ocean in May and June 2005 (on board the R/V Seward Johnson), and three were from the Pacific Ocean in January 2007 (on board the R/V Kilo Moana) and September 2008 (on board the R/V Tangaroa). Seawater samples were also collected from the six depths from one station in South China Sea in July 2007 (on board the R/V Dong Fang Hong II). For the above samples, 50 ml or 2 l of water was filtered through 0.22-μm pore size 47-mm diameter polycarbonate filters (Millipore, Bedford, MA, USA). Six samples were collected from the seawater below the chlorophyll maximum in the Bering Sea and the Chukchi Sea in July 2008 (on board the R/V USCGC Healy) and August 2009 (on board the R/V Alpha Helix), respectively. All samples of water (0.5 to 1.5 l) collected from the Bering Sea and Chukchi Sea were filtered onto 25-mm diameter GF/F filters (Millipore), and immediately frozen and stored at −80 °C until DNA extraction. Temperature, salinity and the concentration of chlorophyll a at all the sampling sites in this study (Figure 1) are shown in Table 2.
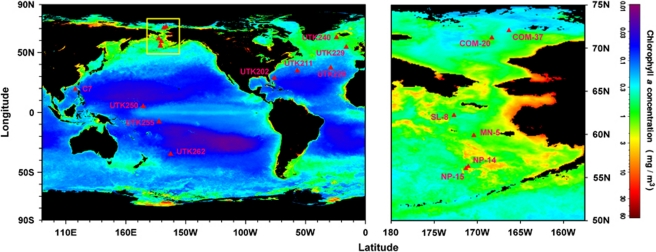
Figure 1.
Maps showing the sampling stations. Fifteen locations are indicated by red solid triangles and station names. The higher resolution map of the inset yellow box shows a precise coordinates of sampling location in the Bering Sea and the Chukchi Sea. Sampling sites were localized by using Surfer software (Golden Graphics, Golden, CO, USA). The base map was an annual composite of chlorophyll a concentration in 2005 obtained from the NASA website (http://oceancolor.gsfc.nasa.gov/).
Table 2. Environmental parameters observed and locations of stations sampled for analysis.
| Station | Location | Sampling date | Sampling depth (m) | Temperature (°C) | Salinity | Chl a (μg l−1) | Water depth (m)a |
|---|---|---|---|---|---|---|---|
| North Atlantic Ocean | |||||||
| UTK202 | 76°06′W; 29°17′N | 23 May 2005 | 1 | 24.3 | 36.5 | 0.063 | 5019 |
| UTK211 | 56°47′W; 35°18′N | 27 May 2005 | 1 | 20.3 | 36.6 | 0.085 | 5171 |
| UTK220 | 29°W; 37°52′N | 1 June 2005 | 1 | 18.9 | 36.2 | 0.115 | 842 |
| UTK229 | 16°04′W; 55°05′N | 12 June 2005 | 1 | 13.6 | 35.4 | 0.843 | 747 |
| UTK240 | 24°10′W; 63°12′N | 28 June 2005 | 1 | 9.0 | 35.2 | 1.863 | 914 |
| Pacific Ocean | |||||||
| UTK250 | 174°32′E; 5°39′N | 10 January 2007 | 1 | 29.1 | 34.5 | 0.860 | 5216 |
| UTK255 | 172°18′W; 7°04′S | 15 January 2007 | 1 | 30.5 | 34.3 | 0.450 | 5390 |
| UTK262 | 162°33′W; 34°09′S | 28 January 2007 | 1 | 22.5 | 35.7 | 0.680 | 5469 |
| South China Sea | |||||||
| C7 | 118°E; 20°N | 16 July 2007 | 5 | 30.2 | 33.8 | 0.090 | 3010 |
| 25 | 30.1 | 33.8 | 0.092 | ||||
| 50 | 27.9 | 34.0 | 0.110 | ||||
| 75 | 25.2 | 34.4 | 0.370 | ||||
| 100 | 23.4 | 34.5 | 0.713 | ||||
| 150 | 19.7 | 34.7 | 0.103 | ||||
| Bering Sea and Chukchi Sea | |||||||
| NP-15 | 56°03′W; 171°19′N | 15 July 2008 | 60 | 3.91 | 33.1 | 0.06 | 2755 |
| NP-14 | 56°17′W; 171°02′N | 18 July 2008 | 30 | 5.15 | 32.6 | 0.27 | 140 |
| MN-5 | 59°53′W; 170°23′N | 9 July 2008 | 40 | −0.87 | 31.3 | 0.57 | 62 |
| SL-8 | 62°12′W; 172°42′N | 12 July 2008 | 50 | −1.59 | 32.2 | 0.60 | 58 |
| COM-20 | 71°12′W; 168°18′N | 6 August 2009 | 40 | −0.97 | 32.5 | 2.80 | 51 |
| COM-37 | 72°02′W; 166°19′N | 7 August 2009 | 40 | 0.28 | 32.5 | 1.20 | 48 |
Water depths data for stations in North Atlantic and Pacific Oceans were from Google Earth (http://earth.google.com/); others were measured during cruises.
DNA extraction, PCR amplification, cloning and sequencing
The bacterial community genomic DNA was extracted using a phenol–chloroform method as previously described (Kan et al., 2006). Picocyanobacterial ITS sequences were amplified following the same protocol in our previous study (Cai et al., 2010). It is noteworthy that multiple sizes (∼600–900 bp) of PCR products were observed on the electrophoresis gels for a single subtropical/tropical sample. The PCR products were excised and purified using the QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany), and cloned using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA, USA) by following the manufacturer's instructions. Clones were sequenced using BigDye terminator chemistry and an ABI 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) at the Institute of Marine and Environmental Technology, UMCES. The sequences recovered in this study were deposited in GenBank (accession numbers: HQ722936-HQ723207, HQ723209-HQ723233, HQ723235, HQ723237-HQ723240, HQ723242, HQ723244-HQ723313, HQ723315-HQ723381, HQ723383-HQ723419, and HQ723421-HQ724283).
Phylogenetic and statistic analyses
The DNA sequences were aligned using the ClustalX2 program (Larkin et al., 2007), and the alignments were manually corrected by using MEGA 4.0 (http://www.megasoftware.net/). The PAUP* software version 4.0b10 (Sinauer Associates, Inc. Publishers, Sunderland, MA, USA) and the PHYLIP software package (http://evolution.genetics.washington.edu/phylip.html) were used to perform the distance analysis independently, and the RAxML web-server (http://phylobench.vital-it.ch/raxml-bb/) was used to carry out the maximum likelihood analysis, respectively. Canonical correspondence analysis, based on sequence distribution combined with environmental factors (temperature, salinity, depth, latitude and concentration of chlorophyll a) was performed by using the Canoco version 4.5 (Biometris, Wageningen, The Netherlands). Three sequences ungrouped into any clades (UTK255_33, UTK211_35, C7_5m_69) were not included in statistical analysis.
Search picocyanobacterial ITS sequences in the GOS data set
Eighty-three selected ITS sequences from all the picocyanobacterial clades described in the phylogenetic analysis were input to BLASTN search against the GOS expedition ‘All Metagenomic Sequence Reads' data set through the CAMERA interface (http://camera.calit2.net/; Rusch et al., 2007). An optimized E-value threshold of 10−25 was finally determined after a touchdown examination from 10−100 to 10−20. In total, 658 different GOS sequence reads were hit and retrieved, which was in the similar range of a previous GOS reads recruitment that harvested 532 sequences for cyanobacteria using 16S rRNA gene sequences (Biers et al., 2009). Then 17 non-picocyanobacterial sequences were discarded after an examination (BLASTN against the NCBI genome database), and 641 sequences were used to perform a local BLASTN search against those 83 representative sequences and then were assigned to potential picocyanobacterial clades through comparing E-values, identities and scores.
Results and discussion
Geographic areas and strategy of constructing clone libraries
Specific PCR products were amplified from a total of 20 DNA extracts of samples collected from diverse marine ecosystems, including the tropical, subtropical and high-latitude North Atlantic Ocean, equatorial Pacific Ocean, subtropical South Pacific Ocean, South China Sea, Bering Sea and Chukchi Sea (Table 2, Figure 1). Also included were six samples from a stratified depth profile (surface water to the bottom of the euphotic zone, Supplementary Figure S1) collected from South China Sea. The PCR primers were designed based on more than 50 ITS sequences of picocyanobacteria including Prochlorococcus and marine Synechococcus from subclusters 5.1 and 5.2 (Cai et al. 2010, Supplementary Figure S2). Twenty libraries were constructed, and ca. 70 clones were randomly picked and sequenced for each sample. A total of 1339 sequences were retrieved.
Novel lineages of HL-adapted Prochlorococcus
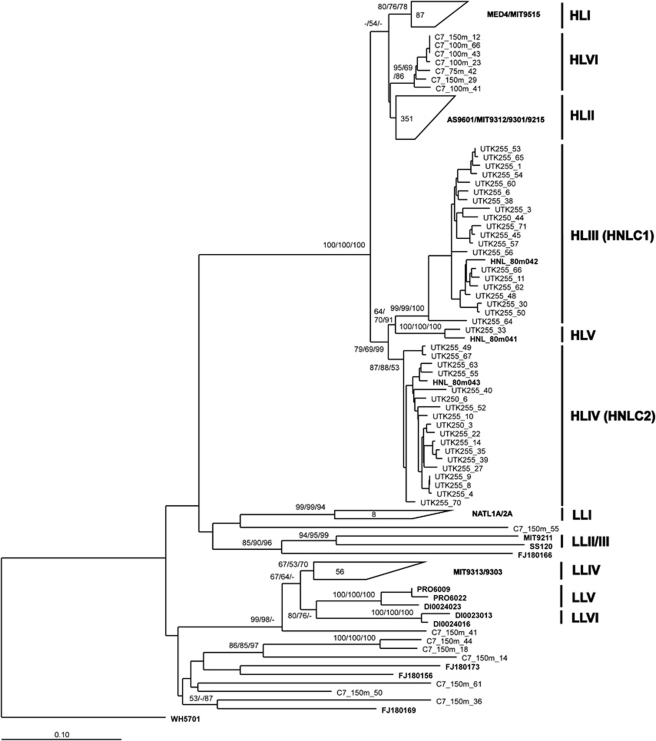
The majority of Prochlorococcus sequences in our study fell into the two well-characterized HL-adapted ecotypes, clades HLI (16% of all Prochlorococcus sequences), HLII (63%) and one LL-adapted ecotype, clade LLIV (10%). Four newly designated lineages of Prochlorococcus (HLIII, HLIV, HLV and HLVI) were phylogenetically more closely related to the existing HL-adapted clades (HLI and HLII) than the LL-adapted clades (Figure 2, Supplementary Figure S3). No previously reported sequences were clustered into clade HLVI, suggesting the presence of unknown HL-adapted Prochlorococcus in the ocean. Sequences in clades HLIII, HLIV and HLV had <89% identities to sequences in clades HLI and HLII, while sequences in clade HLVI were more closely related to HLI and HLII (<92% and <95% identities, respectively; Table 3). The lowest identities between sequences within clade HLI and HLII were 94% and 93%, respectively. Although sequences in clade HLVI may not be distinguishable to those in clade HLI or HLII, based on the identity range, seven sequences formed a monophyletic clade as HLVI, whose position and presence were supported by the bootstrap values (Figure 2, also see Supplementary Figure S3).
Figure 2.
Phylogenetic tree, based on 16S-23S rRNA ITS sequences, showing the relationships among Prochlorococcus genotypes. Sequence positions of 812 bp without tRNAs were used for tree constructions. The showing phylogenetic tree was inferred using distance method with HKY85 model and heuristic search by using the PAUP* software. Parallel distance bootstrap supporting were estimated by using PAUP* (Neighbor-joining with HKY85 model) and PHYLIP (Jukes–Cantor model). Maximum likelihood (ML; GTR–GAMMA model) inferences were also performed by using RAxML. Bootstrap values, with re-sampling for 1000 and 100 replicates for distance and ML analyses, respectively, were shown at the nodes in the order of NJ-PAUP/NJ-PHYLIP/ML. The numbers of environmental sequences retrieved in this study were shown in the trapezoids. Cultivated strains and referential environmental sequences were shown in bold. NJ, neighbor joining.
Table 3. Identities of 16S-23S rDNA ITS sequences between high-light-adapted Prochlorococcus clades.
|
Environmental sequences derived in this study |
||||||
|---|---|---|---|---|---|---|
| HLI | HLII | HLIII | HLIV | HLV | HLVI | |
| HLI strains | >94% | <92% | <89% | <89% | <89% | <92% |
| HLII strains | <92% | >93% | <88% | <89% | <89% | <95% |
Abbreviation: ITS, internal transcribed spacer.
Prochlorococcus strains used in this analysis were: MIT9302, MIT9201, MIT9312, MIT9311, MIT9401, MIT9321, MIT9322, AS9601, SB, MIT9314, MIT9301, MIT9107, MIT9116, MIT9123, RS810, MED4 and MIT9515.
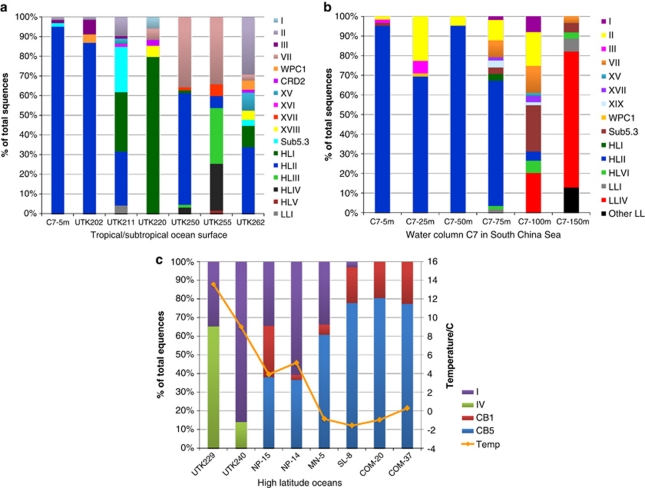
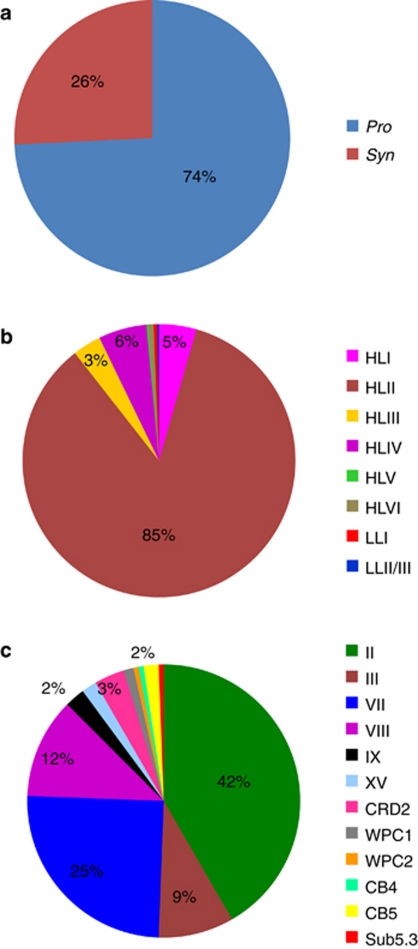
Among the 20 widely collected samples, clades HLIII, HLIV and HLV were only detected in the surface water samples UTK255 and UTK250 collected from the Equatorial Pacific Ocean (Figure 2, Supplementary Figure S4). At station UK255, clades HLIII and HLIV together contribute ca. 50% of the clone library (Figure 4a). When searched against the GOS database, nearly 9% of the GOS Prochlorococcus reads could be attributed to clades HLIII and HLIV (Figure 5b). Furthermore, the vast majority (95%) of clades HLIII and HLIV reads in GOS database occurred in the surface water between latitudes 10°N and 10°S of Pacific and Indian Oceans (Supplementary Figure S5). These results indicated that the HL-adapted Prochlorococcus clades HLIII–HLV may be confined to the equatorial ocean. A recent study based on the metagenomic exploration of GOS database identified two novel HL-adapted Prochlorococcus clades (HNLC1 and HNLC2), which are restricted to high-nitrate, high-temperature and low-iron HNLC waters in the Equatorial and South Pacific and the tropical Indian Oceans (Rusch et al., 2010). Quantitative PCR analysis revealed that HNLC clades occurred more frequently in upper euphotic zone (0–80 m) than in deeper waters (80–100 m), suggesting their HL preference (West et al., 2010). Our sequences were clustered with known HNLC Prochlorococcus ITS sequences (Figure 2), indicating that HLIII and HLIV clades refer to HNLC1 and HNLC2 clades, respectively (West et al., 2010, personal communication with N West). Another HNLC genotype, clade HLV, contained much less sequences than HLIII and HLIV clades (one sequence found in this study and two in West et al., 2010). This seeming rare Prochlorococcus clade suggests that HNLC ecotypes adapted to a relatively narrow biogeographic area have been diversified. Reconstruction of consensus genomes from HNLC clades suggested that these Prochlorococcus have adapted to iron-depleted environments by eliminating a number of iron-containing genes (Rusch et al., 2010). Cultivation of these novel genotypes will be an important step toward understanding their adaptation to this unique ecosystem.
High-light-adapted Prochlorococcus clade HLVI was discovered in the middle-to-lower euphotic zone (75–150 m) of the South China Sea station C7 (Figure 2, Supplementary Figure S4). Only seven ITS sequences of this novel lineage were recovered, which accounted for the 2% of Prochlorococcus sequences at station C7. Searching for clade HLVI ITS sequences in GOS database also provided very few hits (Figure 5b). Clade HLVI was slightly divergent from clades HLI and HLII, the two most dominant ecotypes in the world's oceans (Figure 2, Supplementary Figure S3). Interestingly, a recent metagenomic study revealed that HL-like Prochlorococcus dominated the deep chlorophyll maximum (125 m) water at the Hawaii Ocean Time-series Station ALOHA, and further suggested that an unknown HL-like population may be well adapted to the lower euphotic zone (Shi et al., 2011). It appears that the HL-adapted clade HLVI also displays favorite of LL condition. The niche adaptation of Prochlorococcus genotypes/ecotypes sometimes displays complexity, such as the fact that the abundance of the LL ecotype eNATL (that is, LLI) often peaks in upper euphotic zone (Zinser et al., 2007; Malmstrom et al., 2010) and it shows characteristics of both HL and LL ecotypes (Coleman and Chisholm, 2007; Kettler et al., 2007). Partensky and Garczarek (2011) suggested eNATL as an intermediate ecotype. Whether the genetically HL-like Prochlorococcus clade HLVI has adapted to LL environment or serves as another ‘intermediate ecotype' needs further investigation.
Diverse LL-adapted Prochlorococcus
Among the Prochlorococcus sequences (555) examined in this study, 13% were affiliated with LL-adapted ecotypes with 1.5% falling into clade LLI, 10% into clade LLIV and 1.5% into other undesignated LL clades. Sequences in clade LLI were detected at 75 m and 150 m of station C7 and in the surface water at station UTK211, whereas sequences in clade LLIV were only found at 100 m and 150 m of station C7 and dominated at 150 m. (Figure 4b, Supplementary Figure S4). Our study confirms the previous observation that clade LLI Prochlorococcus may be able to tolerate short-term exposure to high-intensity light and that their cell abundance peaked at shallower layers than other LL ecotypes (Table 1; Coleman and Chisholm, 2007; Zinser et al., 2007; Malmstrom et al., 2010). Several LL sequences recovered from the 150-m layer of station C7 were deeply branched (Figure 2), implying the presence of other potential novel lineages of Prochlorococcus in nature. The deeply branching LL Prochlorococcus have been observed in other studies (Zinser et al., 2006; Garczarek et al., 2007; Martiny et al., 2009; Lavin et al., 2010). None of our sequences were clustered with the clades LLV or LLVI, two recently defined phyletic groups (Lavin et al., 2010). Although some of our LL-adapted sequences appeared to cluster with the sequences in the previously described clade NC1 (Martiny et al., 2009), they did not form a monophyletic group. It has been suggested that clade NC1 might constitute multiple independent lineages (Martiny et al., 2009). Our study suggests that LL-adapted genotypes could be more diverse than HL-adapted ones. The higher genetic diversity of LL Prochlorococcus is also reflected by the wider genome size range of LL strains (1.69–2.68 Mbp) than HL strains (1.64–1.74 Mbp; Kettler et al., 2007). Specifically, members of clade LLIV, represented by the strains MIT9313 and MIT9303, have relatively large genome sizes, and this ecotype (eMIT9313) occupies the lower euphotic zone (Zinser et al., 2007). These two genomes show many features that distinguish them from other Prochlorococcus and enable them to adapt to LL environments (Rocap et al., 2003; Kettler et al., 2007). However, other four known LL strains have similar genome sizes to HL strains (Kettler et al., 2007). More LL Prochlorococcus genomes will help us understand more about how much genomic flexibility, and what potential adaptability can be obtained by the quite diverse LL genotypes.
Novel Synechococcus lineages
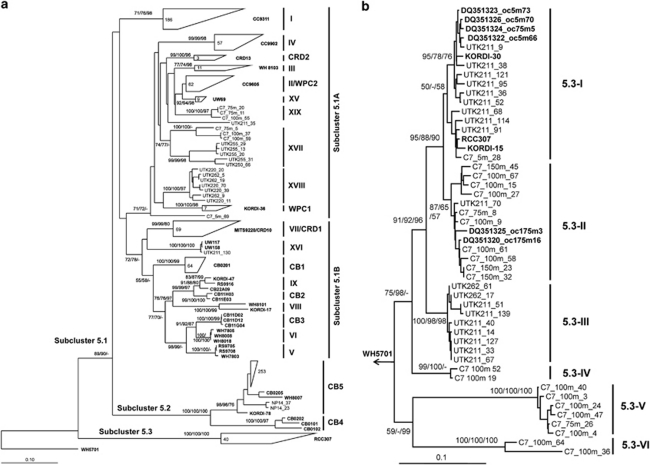
Three novel Synechococcus clades (XVII, XVIII and XIX) were identified in our study in addition to previously described clades I–X (Rocap et al., 2002; Fuller et al., 2003), clades XI–XIV (Penno et al., 2006), clades XV and XVI (Ahlgren and Rocap, 2006) and clades CB1–CB5 (Chen et al., 2006; Figure 3a). All of these new clades contained relatively few sequences and had limited distributions in the sea. Eight sequences in clade XVII were only found in the middle euphotic zone (75 m and 100 m) of the South China Sea station C7, and in the equatorial Pacific surface waters (UTK255 and UTK250); seven sequences in clade XVIII were exclusively detected in the surface water at station UTK220 and UTK262; three sequences in clade XIX were restricted to the middle euphotic zone at station C7 (75 m and 100 m; Figures 3a and 4). No GOS reads were recruited using the sequences from these three clades (Figure 5c). Two possible reasons may contribute to the lack of detection of Synechococcus clades XVII, XVIII and XIX in the GOS survey: (1) these genotypes may represent rare populations; (2) deep waters were not covered by the GOS expedition. In a most recent study (Mazard et al., 2011), three novel Synechococcus lineages (EnvA, EnvB and EnvC,) in the subcluster 5.1 were found from ‘intermediate' stations (that is, located in the transition zone between temperate mesotrophic and subtropical oligotrophic provinces), based on environmental sequences of gene petB (encoding the cytochrome b6 subunit of the cytochrome b6f complex). Although clade XVIII was also found at such transition zone-like stations (UTK220 and UTK262), owing to using different gene markers, it is not possible to compare or link clade XVIII with those three lineages.
Figure 3.
Phylogentic tree based on 16S-23S rRNA ITS sequences (866 bp, without tRNAs), showing the relationships among Synechococcus genotypes (a; see the legend of Figure 2 for more detail). Clade CRD1, represented by strain MITS9220, was incorporated into clade VII in this study, consistent with the 16S rDNA phylogeny (Scanlan et al., 2009). Clade WPC2 was resolved into clade II, which was also supported by the ITS phylogeny in a recent study (Mazard et al., 2011). An insert tree showing the phylogentic clustering of sequences in subcluster 5.3 (b). PAUP* software was used to perform the tree construction and bootstrap tests. The showing tree was constructed using distance method with HKY85 model and heuristic search. Neighbor-joining (NJ, 1000 replicates), maximum parsimony (MP, 100 replicates) and maximum likelihood (ML, 100 replicates) methods were used to estimate the bootstrap values (shown in the order of NJ/MP/ML). Cultivated strains and referential environmental sequences were shown in bold.
Figure 4.
Prochlorococcus and Synechococcus community composition calculated from environmental sequences recovered in this study including: community structure for individual samples from subtropical/tropical ocean surface (a), vertical profile of South China Sea station C7 (b) and high-latitude area (c).
Figure 5.
Classification of Prochlorococcus and Synechococcus 16S-23S rRNA ITS sequences retrieved from the GOS database showing Prochlorococcus and Synechococcus ratio (a), Prochlorococcus (b) and Synechococcus (c) genotypes.
Diverse Synechococcus of subcluster 5.3
Synechococcus subcluster 5.3 was only re-established recently (Dufresne et al., 2008; Scanlan et al., 2009), and contains three Synechococcus strains, RCC307 (Ahlgren and Rocap, 2006), KORDI-15 and KORDI-30 (Choi and Noh, 2009). Many of our sequences fell within subcluster 5.3 and formed six independent lineages that were not fully recognized previously, indicating a need for understanding the ecological relevance of this group of picocyanobacteria.
A total of 40 sequences from three stations, C7, UTK211 (Atlantic Ocean) and UTK262 (Pacific Ocean), belong to subcluster 5.3 (Figure 3b). At station UTK211 and at 100 m of water column C7, members of subcluster 5.3 contributed ca. 20% of each clone library (Figures 4a and b). Nevertheless, few sequences in subcluster 5.3 were detected in the GOS database (Figure 5c), suggesting that this group of Synechococcus may be present in specific locations. Our data suggest that there are at least six different clades (5.3-I to 5.3-VI) in subcluster 5.3. Sequences in clades 5.3-I and 5.3-III were only present in surface waters of the three stations, whereas clades 5.3-II, 5.3-IV, 5.3-V and 5.3-VI prevailed in the medium to LL zones at station C7. The three Synechococcus strains (RCC307, KORDI-15 and KORDI-30) in clade 5.3-III were all isolated from surface or upper euphotic zone (Dufresne et al., 2008; Choi and Noh, 2009). Six environmental sequences recovered from a water column in the western Sargasso Sea (Ahlgren and Rocap, 2006) were also grouped corresponding to depth partitioning (Figure 3b). It appears that members of subcluster 5.3 are present in different oceans but some clades may be restricted to certain depths, suggesting a partitioning of this group of Synechococcus along the vertical profile. Whether these clades represent specific niche adaptation such as HL- and LL-adapted Prochlorococcus warrants future study.
Synechococcus lineages in high-latitude oceans
Four Synechococcus lineages (clades I, IV, CB1 and CB5) were found in high-latitude oceans (Figure 4c, Supplementary Figure S4). All sequences from the two North Atlantic stations UTK229 (55°N) and UTK240 (62°N) fell within the Synechococcus clades I and IV (Figure 4c). About 34% sequences from UTK229 belonged to Synechococcus clade I, whereas 87% of sequences from UTK240 clustered with clade I. Clade I also constituted a significant portion (ca. 40–60%) of the Synechococcus community in the southern Bering Sea stations NP-15, NP-14 and MN-5 (56–60°N), but was rarely or not detected at stations SL-8, COM-20 and COM-37 located in the northern Bering Sea and Chukchi Sea (62–72°N; Figure 4c). Instead, clades CB1 and CB5 Synechococcus made up nearly 20% and 80%, respectively, of the sequenced clones from stations SL-8, COM-20 and COM-37 (Figure 4c). Clade CB5 Synechococcus that dominated the highest latitude subzero waters, sampled in this study, were affiliated with subcluster 5.2, whereas clade CB1 fell into subcluster 5.1 (Figure 3a). It has been known that the abundance of picocyanobacteria decreases with increasing latitude and decreasing water temperature (Murphy and Haugen 1985; Olson et al., 1990a, 1990b). In this study, we also observed a clear shift of picocyanobacterial community structure with increasing latitude or decreasing water temperature in the Bering Sea and Chukchi Sea (Figure 4c). Synechococcus in clades CB1 (strain CB0201) and CB5 (strain CB0205) were originally isolated from the Chesapeake Bay, an estuarine ecosystem (Chen et al., 2006) and were found dominating the picocyanobacterial communities in the bay in summer (Cai et al., 2010). Clade CB5 Synechococcus were also isolated from the East China Sea (strain KORDI-78, Choi and Noh, 2009) and Gulf of Mexico (strain WH8007, Chen et al., 2004). It appears that estuarine or coastal Synechococcus dominate the picocyanobacterial community in the northern Bering Sea and the Chukchi Sea. The prevalence of these picocyanobacterial genotypes in such a large region suggests that they might be autochthonous to the Arctic Ocean rather than allochthonous inputs from freshwater or the open oceans. This further suggests that Synechococcus that inhabit polar and subpolar waters might be adapted to cold even subzero environments.
Picocyanobacterial cell abundance in the Arctic Ocean is typically in the range of 0–103 cells per ml (Gradinger and Lenz, 1995; Cottrell and Kirchman, 2009). Coastal waters in the Arctic Ocean can be greatly influenced by river discharges from North America, and allochthonous inputs from riverine picocyanobacteria (in contrast to typical marine picocyanobacteria) to the coastal Arctic waters have been reported (Waleron et al., 2006). However, those riverine picocyanobacteria were not detected in this study, although the primers used could amplify the ITS sequences from freshwater environmental samples and most freshwater Synechococcus strains tested (data not shown). Synechococcus clade I or IV has been detected in two Arctic stations near the Norwegian coastline (Zwirglmaier et al., 2008), but these typical oceanic Synechococcus were not detected in the Chukchi Sea (our study) and the Beaufort Sea (Waleron et al., 2006). As expected, clades I, IV, CB1 and CB5 Synechococcus were not or rarely detected in the GOS database (Figure 5c). It appears that picocyanobacterial genotypes present in high-latitude area may vary with specific environments. Picocyanobacteria can survive in the dark season in Arctic or are even more abundant than in spring/summer (Gradinger and Lenz, 1995; Cottrell and Kirchman, 2009). It is of particular interest to explore whether the picocyanobacterial communities are different in dark winter, as their known taxonomic information is mostly confined in summer/autumn. Although present in low abundance, picocyanobacteria in the Arctic Ocean may have the ability to respond quickly to the global climate anomaly. Therefore, it is important to understand how polar cyanobacteria survive, grow and interact with the surrounding biota and environments, as they may become more significant factors in the future.
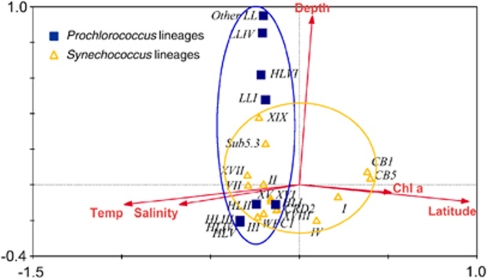
Biogeography of Prochlorococcus and Synechococcus lineages: global insights
Factor analysis showed that the distribution pattern of Prochlorococcus was predominantly influenced by the depth, whereas Synechococcus by both depth and latitude, and therefore other correlated parameters such as temperature and salinity (Figure 6). The global biogeography of picocyanobacteria in our study supports the known distribution patterns of marine Prochlorococcus and Synechococcus lineages in the ocean (Table 1). We observed that the vertical distributions of HL versus LL Prochlorococcus in the South China Sea water column are correlated with depth (Figures 4b and 6), and that the shift of clade HLII to clade HLI Prochlorococcus from the warm to cool oceanic waters has co-variation with latitude increasing (Figure 4a). These distribution patterns are consistent with previous studies (Ahlgren et al., 2006; Johnson et al., 2006; Zinser et al., 2007; Zwirglmaier et al., 2007, 2008). No Prochlorococcus were detected among the high-latitude sites (above 55°N) examined in our study. The biogeographical patterns of Synechococcus are also consistent with those reported previously (Zwirglmaier et al., 2008). For examples, clades I and IV Synechococcus are mostly confined to high-latitude, temperate waters (Figures 4 and 6), and clades II Synechococcus is the most abundant lineage, and widely distributed in the tropical and subtropical oceans (Figure 5c). In addition, our results showed that clade VII Synechococcus may occur more frequently in some equatorial regions or in the deeper euphotic zone, and clade VIII can inhabit hypersaline environment (Supplementary Figures S4 and S5). It is also suggested that Synechococcus in clades CB1 and CB5 may be more adaptive to high-latitude polar environments than other cold-adapted genotypes such as clades I and IV (Figure 6).
Figure 6.
Canonical correspondence analysis (CCA) showing the relationships between environmental factors and the distribution pattern of Prochlorococcus and Synechococcus lineages. Prochlorococcus and Syenchococcus lineages were, respectively, enclosed by blue and orange circles.
Suitability of PCR primers
Picocyanobacteria are a minor component of microbial community in polar seas, which makes an examination of their genetic diversity based on the PCR-clone library method difficult. When 16S rRNA-based cyanobacterial PCR primers (Nubel et al., 1997) were applied to polar samples, significant interference of 16S rRNA gene sequences from the plastids of eukaryotic algae were seen (data not shown). The plastid 16S rRNA sequences also contributed to the clone libraries of ‘cyanobacteria' communities in the Arctic Sea (Waleron et al., 2006). The PCR primers, based on picocyanobacterial ITS sequences, seem to avoid this problem, perhaps owing to the lack or difference of ITS in algal plastids. Only picocyanobacterial sequences were identified in our clone libraries from the Bering and Chukchi Seas. The design of the primers (Picocya16S-F and Picocya23-R) was based on the conserved regions of the ITS sequences from more than 50 marine and estuarine picocyanobacteria (Supplementary Figure S2). This first application of these primers to diverse marine environments shows that they are suitable for studying dynamic changes of complex picocyanobacterial communities in the global oceans.
Concluding remarks
Our study provides new insight into the diversity and distribution of picocyanobacteria in the global oceans. These findings include the presence of HL-adapted Prochlorococcus clades in the tropical oceans, that is, the occurrence of clades HLIII, HLIV and HLV in the iron-depleted equatorial areas and the detection of a HL Prochlorococcus lineage (clade HLVI) seemingly preferentially present in the lower euphotic zone. Further research on ‘intermediate-like' genotypes such as clade HLVI investigated here, and eNATL investigated previously by Coleman and Chisholm (2007) and Kettler et al., (2007) may help to better understand the niche adaptation and evolution of Prochlorococcus. Furthermore, we discovered the presence of estuarine or coastal Synechococcus (for example, clade CB5 in subcluster 5.2) in the Arctic and subarctic Oceans, and demonstrated the spatial partitioning of marine Synechococcus subcluster 5.3 along the vertical profile of euphotic zone. Potential niche adaptation (for example, cold- or light condition- adaptation) of these rarely described Synechococcus subclusters needs future studies. The 16S-23S rRNA ITS gene-based primers used in this study are able to detect the variation of picocyanobacterial communities in the oceans, and the ITS sequence-based phylogeny provides a high resolving power for analyzing the microdiversity among the closely related cyanobacterial lineages.
Acknowledgments
We thank Janet Rowe and Audrey Matteson for assistance in sample collection. We also thank Nyree West for exchanging information. This project was in part supported by the Xiamen University 111 program (to FC), the MOST project 2007CB815904, NSFC projects 41076063 and 40821063 and SOA project 201105021 (to NZJ), NSF Grants OCE0851113, OCE-0452409 and OCE-0825405 (to SWW) and Grant ARC-0732667 as part of the Arctic Natural Sciences Program of the National Science Foundation (to HRH).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ahlgren NA, Rocap G. Culture isolation and culture-independent clone libraries reveal new ecotypes of marine Synechococcus with distinctive light and N physiologies. Appl Environ Microbiol. 2006;72:7193–7204. doi: 10.1128/AEM.00358-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlgren NA, Rocap G, Chisholm SW. Measurement of Prochlorococcus ecotypes using real-time polymerase chain reaction reveals different abundances of genotypes with similar light physiologies. Environ Microbiol. 2006;8:441–454. doi: 10.1111/j.1462-2920.2005.00910.x. [DOI] [PubMed] [Google Scholar]
- Biers EJ, Sun S, Howard EC. Prokaryotic genomes and diversity in surface ocean water: interrogating the Global Ocean Sampling metagenome. Appl Environ Microbiol. 2009;75:2221–2229. doi: 10.1128/AEM.02118-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MV, Schwalbach MS, Hewson I, Fuhrman JA. Coupling 16S-ITS rDNA clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: development and application to a time series. Environ Microbiol. 2005;7:1466–1479. doi: 10.1111/j.1462-2920.2005.00835.x. [DOI] [PubMed] [Google Scholar]
- Cai H, Wang K, Huang S, Jiao N, Chen F. Distinct patterns of picocyanobacterial communities in winter and summer in the Chesapeake Bay. Appl Environ Microbiol. 2010;76:2955–2960. doi: 10.1128/AEM.02868-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell L, Nolla HA, Vaulot D. The importance of Prochlorococcus to community structure in the central north pacific-ocean. Limnol Oceanogr. 1994;39:954–961. [Google Scholar]
- Chen F, Wang K, Bachoon DS, Lu J, Lau S, Campbell L. Phylogenetic diversity of Synechococcus in the Chesapeake Bay revealed by ribulose-1,5-bisphosphate carboxylase-oxygenase (RuBisCO) large subunit gene (rbcL) sequences. Aquat Microb Ecol. 2004;36:153–164. [Google Scholar]
- Chen F, Wang K, Kan J, Suzuki MT, Wommack KE. Diverse and unique picocyanobacteria in Chesapeake Bay, revealed by 16S-23S rRNA internal transcribed spacer sequences. Appl Environ Microbiol. 2006;72:2239–2243. doi: 10.1128/AEM.72.3.2239-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DH, Noh JH. Phylogenetic diversity of Synechococcus strains isolated from the East China Sea and the East Sea. FEMS Microbiol Ecol. 2009;69:439–448. doi: 10.1111/j.1574-6941.2009.00729.x. [DOI] [PubMed] [Google Scholar]
- Coleman ML, Chisholm SW. Code and context: Prochlorococcus as a model for cross-scale biology. Trends Microbiol. 2007;15:398–407. doi: 10.1016/j.tim.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Cottrell MT, Kirchman DL. Photoheterotrophic microbes in the Arctic Ocean in summer and winter. Appl Environ Microbiol. 2009;75:4958–4966. doi: 10.1128/AEM.00117-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufresne A, Ostrowski M, Scanlan DJ, Garczarek L, Mazard S, Palenik BP, et al. Unraveling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 2008;9:R90. doi: 10.1186/gb-2008-9-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris MJ, Palenik B. Niche adaptation in ocean cyanobacteria. Nature. 1998;396:226–228. [Google Scholar]
- Fuller NJ, Marie D, Partensky F, Vaulot D, Post AF, Scanlan DJ. Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl Environ Microbiol. 2003;69:2430–2443. doi: 10.1128/AEM.69.5.2430-2443.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garczarek L, Dufresne A, Rousvoal S, West NJ, Mazard S, Marie D, et al. High vertical and low horizontal diversity of Prochlorococcus ecotypes in the Mediterranean Sea in summer. FEMS Microbiol Ecol. 2007;60:189–206. doi: 10.1111/j.1574-6941.2007.00297.x. [DOI] [PubMed] [Google Scholar]
- Gradinger R, Lenz J. Seasonal occurance of picocyanobacteria in the Greenland Sea and central Arctic Ocean. Polar Biol. 1995;15:447–452. [Google Scholar]
- Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science. 2006;311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- Kan J, Crump B, Wang K, Chen F. Bacterioplankton community in Chesapeake Bay: predictable or random assemblages. Limnol Oceanogr. 2006;51:2157–2169. [Google Scholar]
- Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S, et al. Patterns and Implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 2007;3:e231. doi: 10.1371/journal.pgen.0030231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. ClustalW2 and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lavin P, González B, Santibáñez JF, Scanlan DJ, Ulloa O. Novel lineages of Prochlorococcus thrive within the oxygen minimum zone of the eastern tropical South Pacific. Environ Microbiol Rep. 2010;2:728–738. doi: 10.1111/j.1758-2229.2010.00167.x. [DOI] [PubMed] [Google Scholar]
- Li WKW. Primary production of prochlorophytes, cyanobacteria, and eucaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol Oceanogr. 1994;39:169–175. [Google Scholar]
- Liu H, Nolla HA, Campbell L. Prochlorococcus growth rate and contribution to primary production in the equatorial and subtropical North Pacific Ocean. Aquat Microb Ecol. 1997;12:39–47. [Google Scholar]
- Liu H, Suzuki K, Minami C, Saino T, Watanabe M. Picoplankton community structure in the subarctic Pacific Ocean and the Bering Sea during summer 1999. Mar Ecol Progr Ser. 2002;237:1–14. [Google Scholar]
- Malmstrom RR, Coe A, Kettler GC, Martiny AC, Frias-Lopez J, Zinser ER, et al. Temporal dynamics of Prochlorococcus ecotypes in the Atlantic and Pacific oceans. ISME J. 2010;4:1252–1264. doi: 10.1038/ismej.2010.60. [DOI] [PubMed] [Google Scholar]
- Martiny AC, Tai AP, Veneziano D, Primeau F, Chisholm SW. Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Environ Microbiol. 2009;11:823–832. doi: 10.1111/j.1462-2920.2008.01803.x. [DOI] [PubMed] [Google Scholar]
- Mazard S, Ostrowski M, Partensky F, Scanlan DJ.2011Multi-locus sequence analysis, taxonomic resolution and biogeography of marine Synechococcuse Environ Microbiol; doi: 101111/j.1462-2920.2011.02514.x. [DOI] [PubMed] [Google Scholar]
- Moore LR, Chisholm SW. Photophysiology of the marine cyanobacterium Prochlorococcus: ecotypic differences among cultured isolates. Limnol Oceanogr. 1999;44:628–638. [Google Scholar]
- Moore LR, Rocap G, Chisholm SW. Physiology and molecular phylogeny of co-existing Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- Murphy LS, Haugen EM. The distribution and abundance of phototrophic ultraplankton in the North Atlantic. Limnol Oceanogr. 1985;30:47–58. [Google Scholar]
- Nubel U, Garcia-Pichel F, Muyzer G. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl Environ Microbiol. 1997;63:3327–3332. doi: 10.1128/aem.63.8.3327-3332.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson RJ, Zettler ER, Altabet MA, Dusenberry JA, Chisholm SW. Spatial and temporal distributions of prochlorophyte picoplankton in the North Atlantic Ocean. Deep-Sea Res I. 1990a;37:1033–1051. [Google Scholar]
- Olson RJ, Zettler ER, Armbrust EV, Chisholm SW. Pigment, size and distribution of Synechococcus in the North Atlantic and Pacific oceans. Limnol Oceanogr. 1990b;35:45–58. [Google Scholar]
- Partensky F, Hess WR, Vaulot D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol Mol Biol Rev. 1999;63:106–127. doi: 10.1128/mmbr.63.1.106-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partensky F, Garczarek L. Prochlorococcus: advantages and limits of minimalism. Annu Rev Mar Sci. 2011;2:305–331. doi: 10.1146/annurev-marine-120308-081034. [DOI] [PubMed] [Google Scholar]
- Penno S, Lindell D, Post AF. Diversity of Synechococcus and Prochlorococcus populations determined from DNA sequences of the N-regulatory gene ntcA. Environ Microbiol. 2006;8:1200–1211. doi: 10.1111/j.1462-2920.2006.01010.x. [DOI] [PubMed] [Google Scholar]
- Rocap G, Distel DL, Waterbury JB, Chisholm SW. Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed spacer sequences. Appl Environ Microbiol. 2002;68:1180–1191. doi: 10.1128/AEM.68.3.1180-1191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, et al. Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation. Nature. 2003;424:1042–1047. doi: 10.1038/nature01947. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusch DB, Martiny AC, Dupont CL, Halpern AL, Venter JC. Characterization of Prochlorococcus clades from iron-depleted oceanic regions. Proc Natl Acad Sci USA. 2010;107:16184–16189. doi: 10.1073/pnas.1009513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito MA, Rocap G, Moffett JW. Production of cobalt binding ligands in a Synechococcus feature at the Costa Rica upwelling dome. Limnol Oceanogr. 2005;50:279–290. [Google Scholar]
- Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, et al. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev. 2009;73:249–299. doi: 10.1128/MMBR.00035-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Tyson GW, Eppley JM, DeLong EF. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2011;5:999–1013. doi: 10.1038/ismej.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo G, Palenik BP. Synechococcus diversity in the California current as seen by RNA polymerase (rpoC1) gene sequences of isolated strains. Appl Environ Microbiol. 1997;63:4298–4303. doi: 10.1128/aem.63.11.4298-4303.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo G, Palenik B. A Synechococcus serotype is found preferentially in surface marine waters. Limnol Oceanogr. 2003;48:1744–1755. [Google Scholar]
- Veldhuis MJW, Kraay GW, van Bleijswijk JDL, Baars MA. Seasonal and spatial variability in phytoplankton biomass, productivity and growth in the northwestern Indian Ocean: the southwest and northeast monsoon, 1992–1993. Deep-Sea Res Part I: Oceanogr Res Pap. 1997;44:425–449. [Google Scholar]
- Waleron M, Waleron K, Vincent WF, Wilmotte A. Allochthonous inputs of riverine picocyanobacteria to coastal waters in the Arctic Ocean. FEMS Microbiol Ecol. 2006;59:356–365. doi: 10.1111/j.1574-6941.2006.00236.x. [DOI] [PubMed] [Google Scholar]
- West NJ, Lebaron P, Strutton PG, Suzuki MT. A novel clade of Prochlorococcus found in high nutrient low chlorophyll waters in the South and Equatorial Pacific Ocean. ISME J. 2010;5:933–944. doi: 10.1038/ismej.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser ER, Coe A, Johnson ZI, Martiny AC, Fuller NJ, Scanlan DJ, et al. Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl Environ Microbiol. 2006;72:723–732. doi: 10.1128/AEM.72.1.723-732.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser ER, Johnson ZI, Coe A, Karaca E, Veneziano D, Chisholm SW. Influence of light and temperature on Prochlorococcus ecotype distributions in the Atlantic Ocean. Limnol Oceanogr. 2007;52:2205–2220. [Google Scholar]
- Zwirglmaier K, Heywood JL, Chamberlain K, Woodward EMS, Zubkov MV, Scanlan DJ. Basin scale distribution patterns of picocyanobacterial lineages in the Atlantic Ocean. Environ Microbiol. 2007;9:1278–1290. doi: 10.1111/j.1462-2920.2007.01246.x. [DOI] [PubMed] [Google Scholar]
- Zwirglmaier K, Jardillier L, Ostrowski M, Mazard S, Garczarek L, Vaulot D, et al. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages amongst oceanic biomes. Environ Microbiol. 2008;10:147–161. doi: 10.1111/j.1462-2920.2007.01440.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.