Abstract
The Moorea Coral Reef Long Term Ecological Research (LTER) Site (17.50°S, 149.83°W) comprises the fringe of coral reefs and lagoons surrounding the volcanic island of Moorea in the Society Islands of French Polynesia. As part of our Microbial Inventory Research Across Diverse Aquatic LTERS biodiversity inventory project, we characterized microbial community composition across all three domains of life using amplicon pyrosequencing of the V6 (bacterial and archaeal) and V9 (eukaryotic) hypervariable regions of small-subunit ribosomal RNA genes. Our survey spanned eight locations along a 130-km transect from the reef lagoon to the open ocean to examine changes in communities along inshore to offshore gradients. Our results illustrate consistent community differentiation between inshore and offshore ecosystems across all three domains, with greater richness in all domains in the reef-associated habitats. Bacterial communities were more homogenous among open ocean sites spanning >100 km than among inshore sites separated by <1 km, whereas eukaryotic communities varied more offshore than inshore, and archaea showed more equal levels of dissimilarity among subhabitats. We identified signature communities representative of specific geographic and geochemical milieu, and characterized co-occurrence patterns of specific microbial taxa within the inshore ecosystem including several bacterial groups that persist in geographical niches across time. Bacterial and archaeal communities were dominated by few abundant taxa but spatial patterning was consistent through time and space in both rare and abundant communities. This is the first in-depth inventory analysis of biogeographic variation of all three microbial domains within a coral reef ecosystem.
Keywords: coral reef, MIRADA-LTERS, pyrosequencing, V6, V9
Introduction
Microorganisms from all three domains of life have a critical role in mediating biogeochemical cycles in marine ecosystems due in part to their high abundance, surface area and biomass (Pomeroy et al., 2007), as well as their vast diversity of metabolic activities (Whitman et al., 1998; Azam and Malfatti, 2007; Massana and Pedrós-Alió, 2008). Sequencing of small-subunit ribosomal RNA (rRNA) genes has described many major taxa of marine microbes (Sogin et al., 2006; Dinsdale et al., 2008; Huse et al., 2008; Massana and Pedrós-Alió, 2008; Galand et al., 2009a, 2009b; Stoeck et al., 2010), whereas recent technological advances such as high-throughput amplicon pyrosequencing have facilitated deeper sequence recovery including observations of a wide diversity of rare phylotypes (Curtis et al., 2002; Sogin et al., 2006; Huse et al., 2008; Galand et al., 2009a, 2009b). Recent reports shed new light on a microbial community pattern where few, abundant phylotypes numerically dominate marine bacterial and archaeal communities, whereas the majority of the diversity exists within less abundant phylotypes (Gilbert et al., 2009; Brazelton et al., 2010). Large-scale exploration of taxonomic diversity has also provided insights into landscape-scale patterns of population structure including differences in geographic distribution, temporal shifts, spatial differentiation and environmental forcing (Nelson, 2008; Andersson et al., 2009; Brown et al., 2009; Carlson et al., 2009; Gilbert et al., 2009).
Massively parallel sequencing has further improved descriptions of microbial eukaryotic diversity, suggesting dominance and diversity patterns similar to bacterial and archaeal communities (Massana and Pedrós-Alió, 2008; Edgcomb et al., 2009; Cheung et al., 2010; Stoeck et al., 2010; Amaral-Zettler et al., 2011). However, gaps remain in our understanding of comparative biogeography among microbial organisms, specifically with regard to patterns of covariation and community differentiation along environmental gradients. Therefore, a key challenge is to simultaneously inventory the diversity of bacterial, archaeal and microeukaryotic phylotypes in a well-characterized ecosystem to gain insight to the potential biotic and abiotic controls across spatial and temporal scales.
Here we report the first in-depth analysis of bacterial, archaeal and microbial eukaryotic community structure within a coral reef ecosystem, the Moorea Coral Reef Long Term Ecological Research (MCR LTER) site. The MCR LTER comprises a band of coral reefs and lagoons that surround the small volcanic island of Moorea in the Society Islands of French Polynesia. The eastern-most deep-water bay (Paopao Bay, also known as Cooks Bay) on the northern coast extends into a lagoon system bounded by three primary coral reef habitat types (fringing reef, back reef and fore reef); bay and reef habitats comprise the main study sites of ongoing LTER microbial research (Nelson et al., 2011). The Moorea tropical ecosystem is currently the subject of an intensive, island-wide biotic inventory to accumulate a database of, among other identifying information, genetic markers for every species of non-microbial marine and terrestrial form of life on the island (Moorea Biocode Project, http://biocode.berkeley.edu). Our study provides an important complement to the Biocode Project—initiating a much-needed inventory of microbial life present in the rich tropical environment of the Moorea coral reef ecosystem.
In this study our primary goals were to: (a) extensively inventory the bacterial, archaeal and microbial eukaryotic marine communities of Moorea along a transect extending from inshore (bay, fore reef, fringing and back reef) to 130 km offshore in oligotrophic open water and (b) quantify and describe landscape-scale spatial and temporal changes in the diversity, distribution and associations of microbial taxa. We collected a total of 17 duplicate discrete inshore and offshore water samples in January 2008 and May 2009 and described the biogeographic patterns in microbial communities within each of the three domains. We found consistent patterns of bacterial community spatial differentiation in both seasons, with clear differentiation between inshore and offshore communities persisting despite temporal offsets in both habitats across the two seasons. Communities of archaea and microbial eukaryotes also showed inshore–offshore differences, and all three domains of life examined in concert exhibited greater variation among inshore habitats and greater homogeneity among offshore samples likely influenced by the bacterial community. These large- and mid-scale spatial patterns were identified within both abundant and rare members of the microbial populations, suggesting biogeographical rather than random distributions.
Materials and methods
Sampling stations and seawater collection
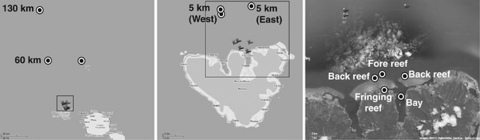
We collected a total of 17 seawater samples in January 2008 and May 2009 (Figure 1) in duplicate sterile 1 l polyethylene terephthalate bottles at five core LTER sampling stations: Paopao Bay (17.490°S, 149.823°W), fore reef (17.475°S, 149.837°W), back reef (17.477°S, 149.820°W), fringing reef (17.485°S, 149.834°W) and 5 km offshore (17.430°S, 149.863°W). We also collected offshore samples from the SSV Robert C. Seamans (Sea Education Association, Woods Hole, MA, USA) ranging from 5 km to 130 km off the North shore of Moorea using a Sea-Bird SBE 32SC rosette water sampler (Sea-Bird Electronics, Bellevue, WA, USA). We filtered replicate water samples through a 0.2-μm Sterivex filter (Millipore, Billerica, MA, USA) and added 2.0 ml of Puregene lysis buffer (Qiagen, Valencia, CA, USA) as described in Amaral-Zettler et al. (2009). We extracted DNA as described previously (Amaral-Zettler et al., 2009) and stored our samples at −20 °C until amplification. Geochemical and biological analyses were carried out on sample replicates. Ancillary biological and geochemical analytical methods are listed in the Supplementary Information (Supplementary Figure S1, Supplementary Table S1).
Figure 1.
A map of offshore and inshore sample locations at the Moorea Coral Reef LTER site.
Pyrosequencing and sequence analysis
We amplified bacterial and archaeal V6 16S rRNA gene hypervariable regions using a cocktail of primers designed to amplify a wide range of microbial groups as described previously (Huber et al., 2007). We amplified eukaryotic V9 hypervariable regions as described in Amaral-Zettler et al. (2009). Briefly, we amplified 10 ng of genomic DNA from each sample in triplicate 50 μl polymerase chain reactions, and pooled and purified amplicons using the Qiagen QIAquick column-based purification kit (Qiagen). We sequenced amplicons on a 454 Genome Sequencer FLX (Roche, Basel, Switzerland) using the manufacturer's suggested amplicon protocol for the GS-FLX platform and the LR70 kit. We trimmed sequences of adapter and primer sequences and removed low-quality reads as described previously (Huse et al., 2007) and assigned sequences to operational taxonomic units (OTUs) using the 2% single-linkage preclustering and pair-wise alignment with average linkage at a final 3% sequence identity clustering level (97% similarity; SLP/PW-AL; Huse et al., 2010) for bacteria and archaea. Although the V9 region is among the most hypervariable in eukaryotic small-subunit rRNA genes, because of the microheterogeneity associated with multiple copies of eukaryotic rRNA, we chose to use a 6% cluster level (94% similarity) for eukaryotic data. OTU identifiers used in this study are represented by the domain followed by a numerical code corresponding to a specific cluster. Representative sequences for each OTU can be accessed through the Visualization and Analysis of Microbial Population Structures database at http://vamps.mbl.edu/.
Richness and evenness estimates
We used bacterial and archaeal abundance matrices to generate OTU frequency count data to estimate richness using parametric modeling via the CatchAll program (Bunge, 2011). We implemented CatchAll to choose the best of multiple parametric models based on the largest τ value (where τ is the upper frequency cutoff and, therefore, inclusive of the greatest range of data) for which a goodness of fit exceeds 0.01. Comparative bacterial and archaeal richness estimates for OTUs clustered at the 94% and 90% sequence similarity threshold are provided in Supplementary Table S2. We calculated eukaryotic richness estimates using the non-parametric Chao2 estimator as described previously (Amaral-Zettler et al., 2009) and implemented in SPADE (Chao and Shen, 2010) based on presence/absence matrices. We compared richness estimates of sample replicates for similarity as biological replicates using a two-sample z-test; sample replicates did not differ in richness (P>0.05) with the exception of the bacterial and archaeal samples from MCR_5 (back reef; January) and bacterial samples from MCR_11 (fringing reef; May). Pooled sample replicate data were used for OTU richness calculations. Replicates were analyzed separately for analyses comparing community similarity between sample stations where indicated. We calculated evenness for bacteria and archaea sampled at the January timepoint using Simpson's (1−λ′) index in the PRIMER-E package (v6; Plymouth, UK; Clarke and Gorley, 2006). All sequences conform to the Minimum Information about a MARKer gene Sequence standard (Yilmaz et al., 2011) and were deposited in the National Center for Biotechnology Information Sequence Read Archive under the accession numbers SRA030397, SRP007814, and SRP007926.
Multivariate statistical analyses
We performed distance-based multivariate analyses using the non-metric Multidimensional Scaling, Analysis of Similarity and Similarity Percentage routines within the PRIMER-E package (v6; Plymouth, UK; Clarke and Gorley, 2006) using pooled sample replicates where indicated in the text. Community similarity matrices for bacterial and archaeal data sets were calculated from square-root transformed OTU relative abundance matrices using the Bray-Curtis metric; for eukaryotic data sets Jaccard similarity matrices were generated from OTU presence–absence matrices to eliminate potential bias from multiple rRNA gene copy number. Archaeal OTU network analysis used significant linear Pearson's correlations between the relative abundance of archaeal OTUs and bacterial OTUs (P<0.01) as input for network maps generated with Cytoscape (http://www.cytoscape.org).
Results
Microbial biogeographic patterns
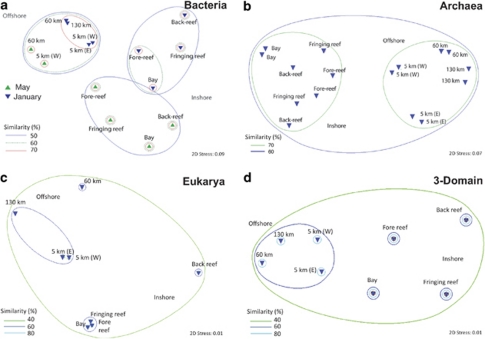
All three microbial domains showed clear community differentiation between offshore and inshore regions (Analysis of Similarity P<0.01 for each domain), with different domains showing different degrees of differentiation among local subhabitats (Figures 2a–c). We observed a significant seasonal shift in the inshore and offshore bacterial communities (2-way Analysis of Similarity, P=0.001). Similarity among all offshore stations across both seasons was >60%, whereas inshore stations clustered separately in January and May (similarity <50%, Analysis of Similarity P=0.029; Figure 2a) largely because of elevated levels of Actinobacteria and Deferribacteres in May (Figure 3). Moreover, the orientation of inshore–offshore differentiation was consistent across both seasons (Figure 2a). To determine whether patterns of bacterial community differentiation were only due to very common or very rare taxa, we compared hierarchical clustering patterns of sample similarity matrices derived only from abundant (>1% of total sequences) and rare (<0.01%) OTUs. We found nearly identical patterns of differentiation at all levels, with offshore, inshore and deep communities clearly distinct, and subhabitat relationships maintained in the inshore environment (Supplementary Figure S2).
Figure 2.
Non-metric Multidimensional Scaling ordination of Bray-Curtis ((a) bacterial and (b) archaeal) or Jaccard ((c) eukaryotic and (d) three domain) similarity matrices. Bacterial and archaeal analyses were based on 97% clustering; eukaryotic analyses were based on 94% similarity clustering. The three domain similarity matrix was based on the presence/absence transformed abundance data from all three domains. Bacterial ordination included surface water samples collected in January 2008 and May 2009 (200-m communities were very different and were excluded from analysis for clarity). The archaeal, eukaryotic (minus metazoan OTUs) and three-domain ordinations were based on samples collected in January 2008 only. Archaeal ordination included biological replicates to illustrate the degree of similarity between replicates collected from a single site (excluded from other ordinations for clarity).
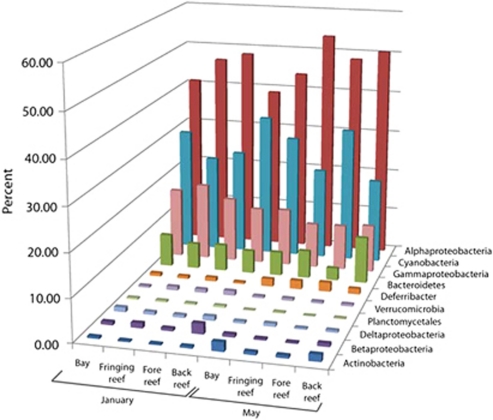
Figure 3.
Relative abundance of the most frequently identified bacterial taxa within inshore sample stations across two seasons.
Archaeal communities also formed distinct inshore and offshore groups that shared <60% similarity, but exhibited equal levels of dispersion within the two regions, unlike bacterial communities where inshore differentiation was higher among subhabitats (Figure 2b). Although the microbial eukaryotic community displayed a similar inshore–offshore differentiation pattern analogous to that of the bacterial and archaeal communities, there was a striking dissimilarity between back-reef and other inshore stations, where the community shared <60% similarity with the adjacent inshore sample sites (Figure 2c). With the exception of this backreef outlier, inshore samples were remarkably homogenous and offshore stations heterogeneous, in direct contrast to the patterns observed in bacterioplankton. To assess biogeographic patterning across all three domains among sites in January 2008, we combined presence–absence matrices for bacteria, archaeal and non-metazoan eukaryotes and calculated Jaccard similarities among samples. Overall microbial communities showed relative homogeneity among offshore sampling locations (similarity >60%) and clear differentiation from inshore samples, with each inshore sampling location being distinct (similarities <60% Figure 2d).
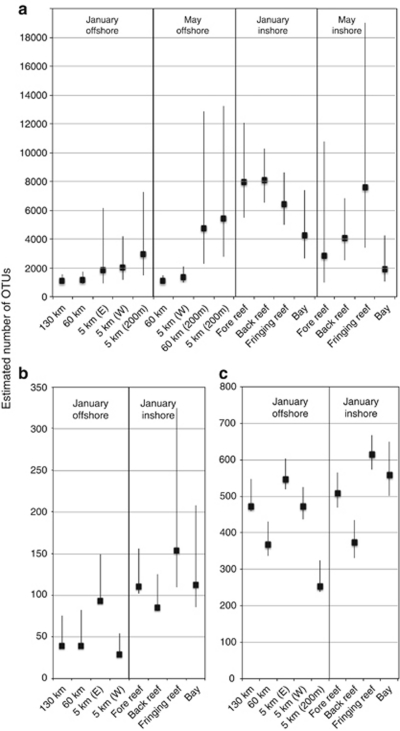
Microbial richness and evenness
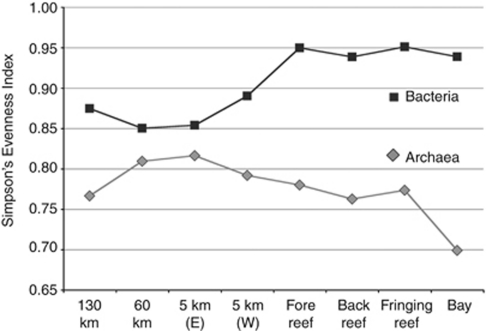
We sequenced a total of 442 565 bacterial V6 amplicons from eight locations within the MCR LTER across the two major seasons, which clustered into 7151 observed OTUs (Table 1). Offshore surface waters (5 km, 60 km and 130 km from shore) generally had fewer observed OTUs in both seasons than inshore communities (Table 1). CatchAll estimates of actual OTU richness indicated that both inshore and offshore sites were more diverse than this study has uncovered but showed similar patterns of elevated inshore diversity, particularly in the January rainy season (Figure 4). One notable outlier from this trend was the inshore Paopao Bay site, which demonstrated lower OTU richness estimates in both January and May compared with the inshore average (Table 1; Figure 4). Simpson's evenness index (1−λ′) revealed a more even distribution of bacterial sequences among OTUs at inshore surface stations compared with offshore surface sites (Figure 5). Archaeal communities contained far lower OTU richness and evenness than bacterial communities at all sites along the transect, but showed a similar spatial trend of greater richness estimates inshore relative to offshore (Figure 4b). Bacterial and archaeal evenness estimates illustrated similar trends when sequences were clustered at the 94% and 90% sequence similarity cutoffs (data not shown). A total of 64 073 archaeal V6 tags sequenced from the eight locations sampled in January 2008 clustered into 185 OTUs (3% threshold). In contrast with spatial patterning of richness, Simpson's evenness index revealed greater evenness for offshore archaeal communities compared with inshore archaeal communities (Figure 5). Eukaryotic sequences analyzed in this study included 31 883 sequences from the eight locations sampled yielding a total of 1218 OTUs with sequence similarity ⩾94% (exclusive of metazoa). The 94% similarity threshold was chosen primarily to accommodate the highly variable small-subunit rRNA copy numbers in the eukaryotic domain, and as such may represent a conservative estimate of OTU richness. Figure 4c demonstrates that microbial eukaryotic richness estimates were only slightly greater inshore compared with offshore. Richness was more pronounced in surface samples than at depth. Eukaryotic evenness was not assessed because of the biases associated with assessing relative abundances using eukaryotic rRNA gene data.
Table 1. Observed and estimated richness for Moorea samples using 97% bacterial and archaeal and 94% microbial eukaryotic similarity values.
| Sample name | Transect location | Depth (m) | Month |
Bacteria |
Archaea |
Eukaryaa |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of reads | Observed OTUs | Estimated OTUs | No. of reads | Observed OTUs | Estimated OTUs | No. of reads | Observed OTUs | Esimated OTUs | ||||
| MCR_1 | 5 km offshore (W) | 2 | Jan | 31 148 | 630 | 2045 | 6887 | 23 | 29 | 2884 | 380 | 471 |
| MCR_2 | 5 km offshore (W) | 200 | Jan | 8544 | 790 | 2943 | — | — | — | 2474 | 234 | 253 |
| MCR_3 | 60 km offshore | 2 | Jan | 34 677 | 553 | 1179 | 8788 | 24 | 39 | 2454 | 309 | 366 |
| MCR_4 | 130 km offshore | 2 | Jan | 37 737 | 557 | 1109 | 6911 | 25 | 39 | 5732 | 455 | 472 |
| MCR_5 | Back reef | 1 | Jan | 35 193 | 2071 | 8080 | 5896 | 57 | 85 | 774 | 235 | 373 |
| MCR_6 | Fringing reef | 5 | Jan | 30 701 | 1759 | 6419 | 8493 | 82 | 153 | 3898 | 477 | 613 |
| MCR_7 | Paopao Bay | 5 | Jan | 42 836 | 910 | 4235 | 9988 | 48 | 113 | 6150 | 414 | 557 |
| MCR_8 | 5 km offshore (E) | 5 | Jan | 37 230 | 566 | 1859 | 7786 | 58 | 93 | 4912 | 501 | 545 |
| MCR_9 | Fore reef | 5 | Jan | 46 137 | 1495 | 7930 | 9324 | 71 | 110 | 2605 | 389 | 508 |
| MCR_10 | Paopao Bay | 2 | May | 12 391 | 391 | 1923 | — | — | — | — | — | — |
| MCR_11 | Fringing reef | 2 | May | 11 816 | 934 | 7555 | — | — | — | — | — | — |
| MCR_12 | Back reef | 1 | May | 11 081 | 760 | 4052 | — | — | — | — | — | — |
| MCR_13 | Fore reef | 5 | May | 13 773 | 467 | 2817 | — | — | — | — | — | — |
| MCR_14 | 5 km offshore (W) | 2 | May | 17 684 | 490 | 1374 | — | — | — | — | — | — |
| MCR_15 | 5 km offshore (W) | 200 | May | 22 280 | 1435 | 5426 | — | — | — | — | — | — |
| MCR_18 | 60 km offshore | 200 | May | 17 321 | 1214 | 4730 | — | — | — | — | — | — |
| MCR_19 | 60 km offshore | 2 | May | 32 016 | 500 | 1095 | — | — | — | — | — | — |
Abbreviations: MCR, Moorea Coral Reef; OTU, operational taxonomic units.
Parametric richness estimates were calculated from two-pooled replicates (Bacteria and Archaea) for each sample or as separate replicates (Eukarya) in non-parametric estimation. See text for details.
Metazoan reads removed.
Figure 4.
Species richness estimates of: (a) bacterial, (b) archaeal and (c) eukaryotic OTUs with Bonferroni corrected 95% confidence intervals for all samples across a given domain. (W) and (E) indicate samples collected West or East of Avaroa Pass. Bacterial and archaeal estimates were calculated using CatchAll, whereas eukaryotic estimates were calculated using Chao2 as implemented in SPADE.
Figure 5.
Simpson's evenness index (1–λ′) of bacterial and archaeal communities sampled at the January timepoint using 97% similarity OTU clusters. Calculations based on log-transformed abundance data.
Microbial community composition among surface habitats
Of the nearly 204 000 inshore bacterial amplicons sequenced, 81–92% were classified within the Alphaproteobacteria, Cyanobacteria and Gammaproteobacteria (Figure 3) for both sampling periods. The alphaproteobacterial OTUs fell predominantly within the SAR11 cluster and Rhodobacteraceae groups inshore, whereas offshore stations were primarily dominated by sequences clustering within two different SAR11 groups. Bacterial communities were dominated by a few highly abundant OTUs and OTUs containing rare, low-frequency sequences comprising the majority of the observed diversity: the 15 most abundant bacterial OTUs (those containing >1.0% of the total sequences in a given sample) encompassed 66% of the sequences within inshore communities, whereas the remaining sequences were distributed among 4715 OTUs. By the same definition of abundant, offshore stations contained only 10 abundant OTUs, also within the Alphaproteobacteria, Cyanobacteria and Gammaproteobacteria groups, with a single cyanobacterial OTU containing >32% of the total offshore surface water bacterial sequences.
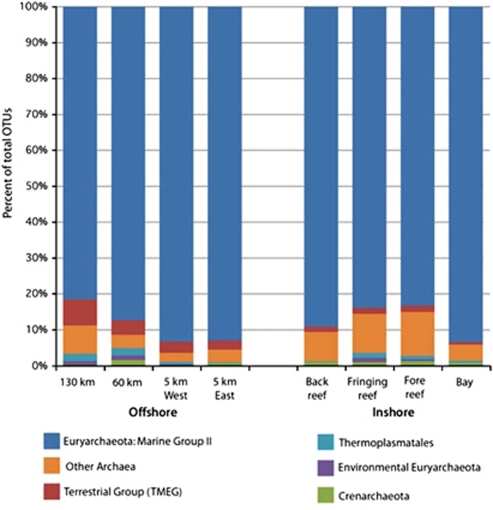
As with bacterial communities, rare archaeal sequences were responsible for the majority of observed diversity; 9 OTUs contained up to 97% of the total tags sequenced, whereas 130 OTUs were represented by rare sequences found with a frequency of <0.01%. Sequence tags corresponding to the Marine Group II (MG II) Euryarchaeaota division dominated each of the inshore and offshore stations, comprising >85% of all OTUs assigned in the data set (Figure 6), but different OTUs within this division were dominant in each habitat. For instance, one MG II OTU (Archaea_14) comprised 8–9% of the total tags sequenced from the back reef and fringing reef stations, whereas the same OTU represented <2% of Paopao Bay archaea, where a different MG II OTU (Archaea_4) comprised >42% of the archaeal sequences (resulting in the lowest evenness score in the data set (0.69); Figure 5). Another abundant MG II OTU (Archaea_47) common inshore (averaging 20% of total sequences), was far less abundant in offshore sites (<2% total sequences). A second abundant archaeal group, Halococcus, was well represented primarily in the back reef, fore reef and fringing reef sites, whereas less abundant or absent in Paopao Bay and offshore samples.
Figure 6.
Relative abundance of archaeal taxa along the offshore–inshore transect. Samples collected in January 2008.
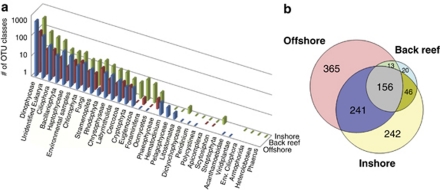
Dinophyceae were the most well-represented microbial eukaryotic group across all stations sampled (Figure 7) and most of the OTUs common to all samples, as well as those unique to specific samples, belonged in this class. Inshore, stramenopiles, rhodophytes and ciliate OTUs were well represented, whereas haptophyte OTUs were found predominantly in the offshore surface waters. Approximately 12% of the OTUs could not be classified beyond domain, and ∼20% of these unclassified OTUs were common to all samples. The Venn diagram in Figure 7 illustrates the classes of OTUs unique to inshore versus surface offshore versus back reef samples (see Figure 1 for locations), and clearly emphasizes that offshore samples have a larger pool of microbial eukaryotes not found inshore (378 OTUs) than the number found specifically inshore (242 OTUs) or distributed throughout (397 OTUs).
Figure 7.
(a) Relative abundance of eukaryotic OTU types that are unique to specific sample regions. (b) Venn diagram of eukaryotic OTUs clustered with 94% sequence identity illustrating the number of OTUs shared between inshore (fore-reef, fringing-reef and bay), surface water offshore (5 km E, 5 km W, 60 km and 130 km) and back-reef samples.
Archaeal network analysis
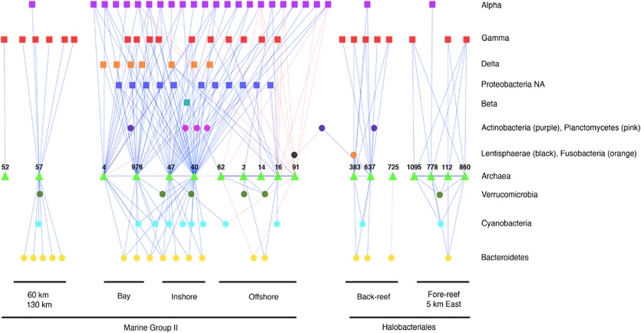
Most archaeal OTU taxonomic designations are indistinguishable because they have not been well-annotated in terms of phylogenetic classification. Therefore, it is worthwhile to parse out the archaeal community structure through ecological interactions such as examining patterns of co-occurrence. Using the correlations in relative abundance between archaeal and bacterial OTUs among samples, we found several well-supported co-occurring assemblages. Figure 8 illustrates a network diagram in which all archaeal OTUs with significant (P<0.01; R>0.9) positive and negative correlations to other archaeal and bacterial OTUs are shown as principal or central nodes in the network, along with their nearest phylogenetic neighbors. We found a total of 18 archaeal OTUs that had significant positive and negative correlations; all classified within the Euryarchaeota, specifically within the MG II or Halobacteriales subgroups. We found the greatest number of correlations between the archaeal OTUs and Alphaproteobacteria, Gammaproteobacteria and Bacteroidetes-affiliated OTUs, though significant correlations existed with members of the Deltaproteobacteria and Cyanobacteria. We found four strongly covarying archaeal OTU assemblages, in which each of the OTUs within the assemblage was found more frequently within a specific subhabitat than in other samples. Among the MG II-affiliated OTUs, one assemblage of two OTUs found predominantly with the Paopao Bay station (4 and 976) was positively correlated with two archaeal OTUs dominant in inshore stations (47 and 40), whereas collectively these two sets of OTUs were negatively correlated with an offshore archaeal assemblage comprising five co-occurring OTUs, emphasizing the specificity of archaeal taxa to certain habitats. One particular archaeal OTU found primarily offshore (Archaea_91) had a strong negative correlation to several Alphaproteobacterial OTUs, which in turn had equally strong positive correlations with inshore-specific archaeal assemblages.
Figure 8.
Cytoscape network diagram of Pearson's correlations (lines) between archaeal OTUs and all other bacterial and archaeal OTUs (symbols) across sample habitats in January 2008. Archaeal OTUs represented were those with significant positive (solid lines) or negative (dashed lines) correlation (P<0.01) to other bacterial or archaeal OTUs. Numbers represent identification numbers of archaeal OTUs. Alpha, Beta, Gamma and Delta-Proteobacterial groups. All OTUs were classified by phylum/domain and horizontally aligned with group identification.
Bacterial diversity and community structure in deep waters
We calculated bacterial diversity estimates within samples taken from 200 m depth at three of the offshore sites (5 km January, 5 km May and 60 km May). Approximately 48 000 sequenced tags defined 2173 observed OTUs. However, richness estimates predicted >6380 OTUs for samples at 200 m, a value greater than surface water samples collected at the same stations (Figure 4). Bacterial taxonomic distributions from these deeper waters included the alpha- and gammaproteobacterial OTUs found in surface waters, but with an increase in Deferribacteres, Actinobacteria and several unclassified proteobacterial taxa (data not shown). As expected, we found few cyanobacterial taxa represented in the 200 m samples. As in the surface inshore waters, actinobacterial taxa comprised >1% of the total sequences at 200 m in May, but were absent or rare (<0.01% of the total sequences) in January.
Discussion
This first microbial biodiversity inventory in Moorean waters showcases that biogeographic patterns span all three domains of microbial life in the Moorea coral reef ecosystem. The description of three-domain spatial patterns within marine microbial communities in the context of the oceanic waters surrounding the coral reef ecosystem of Moorea is a key step in understanding the diversity and stability of planktonic microbial populations between coastal and open water oligotrophic ecosystems. The barrier reef system surrounding Moorea's Paopao Bay and adjacent lagoons has been the subject of several long-term biological (Adjeroud and Salvat, 1996; Adjeroud, 2000; Adjeroud et al., 2002; Penin et al., 2007; Alldredge and King, 2009), biogeochemical and oceanographic (Schrimm et al., 2004; Hench et al., 2008) research studies, though this study and a parallel report (Nelson et al., 2011) are the first to describe this ecosystem's microbial populations, this report being the first one to do so across all three domains of microbial life.
A companion study recently demonstrated strong spatial patterns in dissolved organic carbon, bacterioplankton densities and bacterial community structure between and among the inshore and offshore habitats (Nelson et al. 2011). Both dissolved organic carbon and bacterioplankton concentrations were depleted inshore of the reef crest relative to offshore waters and Paopao Bay with each habitat maintaining distinct microbial assemblages despite the apparent high flushing rate of the inshore environment. The data presented here further resolve a transition of increasing microbial richness from the offshore sites into the reef and lagoon system across all three domains of life (Figures 4a and b), an observation consistent with the high macroorganismal diversity of coral reefs relative to the surrounding oceans.
Consistent with previous observations, our community analysis identified three bacterial community types: an offshore community with relatively high similarity across seasons and an inshore community with high subhabitat community heterogeneity and greater seasonal differentiation (Figure 2a). However, this study further demonstrates that although offshore–inshore community differences are consistent across all three domains, the observation of relatively homogenous offshore bacterial communities is not born out in archaeal or eukaryotic microbial communities (Figures 2b and c). Instead, archaeal OTUs show equal degrees of dissimilarity among oceanic samples and among inshore samples, and eukaryotic microbial communities appear to be somewhat more variable offshore than inshore. The mechanisms driving these variable degrees of subhabitat community differentiation remain unknown.
Our data generally confirm global analyses of microbial communities that have described a community model consisting of a few dominant microbial taxa and a large number of less abundant OTUs that comprise the majority of the total community diversity (Sogin et al., 2006; Galand et al., 2009a, 2009b). By analyzing community patterns along the offshore–inshore transect, we were able to clarify that these patterns are maintained across domains even when the dominant members of the assemblage shift in relative abundance over relatively short distances (for example, Figure 8). In addition, by showing that subhabitat differentiation is maintained whether analyses are carried out on abundant or rare members of the community in isolation, we show that even members of this ‘rare biosphere' exhibit biogeographic patterning. Such patterns are also echoed in the microbial eukaryotic data when OTUs are restricted to those OTUs only occurring ⩽10 times across the entire matrix (data not shown).
Habitat specificity and temporal shifts in specific bacterial classes
Inspection of individual distribution patterns of the most frequently recovered bacterial taxa (that is, Alphaproteobacteria, Cyanobacteria and Gammaproteobacteria) demonstrates the significance of abundant groups in defining distinct communities, reflecting the influence of ‘keystone' groups described in earlier seasonal community ordination descriptions (Nelson, 2008; Treusch et al., 2009; Supplementary Figure S3). Alphaproteobacteria, Gammaproteobacteria and Cyanobacteria, the most abundant bacterial classes in this system, exhibit clear differentiation between inshore and offshore communities in both May (winter) and January (summer), but also show a synchronous shift in structure in both regions between the two seasons (Supplementary Figures S3b, S3e, S3h). Although less abundant taxa also followed this general pattern (for example, Verrucomicrobia, Supplementary Figure S3j) many other groups show different spatial and temporal dynamics.
In this study we found that the Paopao Bay ecosystem harbored communities of bacterial taxa that were distinct from those found in the adjacent lagoon and near-reef sites, and that this differentiation was maintained across seasons. This pattern was driven by cyanobacterial and gammaproteobacterial communities, which form distinct bay-specific groups (as much as 55% dissimilarity to adjacent communities; Supplementary Figures S3e, S3h), especially well defined in the dry winter (May; Supplementary Figure S3e). The bay is essentially a semi-contained system with relatively low flow and intermittent mixing into Avaroa Pass and adjacent channels, and with unique zooplankton populations including holoplankton, larvacaea and abundant diatoms (Alldredge and King, 2009 and Alldredge, personal communication). However, we also noted that the overall inshore bacterial community showed signs of increased fore reef/bay mixing in the austral summer (rainy) season (Figure 2a), reflecting seasonally increased wave-driven bay to fore reef current flow (Hench et al., 2008). Increased summer wave energy flux on the North shore of the island drives offshore water across the barrier reef crest, which then circulates within the shallow back-reef area and mixes with the deep-water bay before flushing out through the narrow Avaroa Pass. This circulation pattern potentially enhances mixing between subhabitats, homogenizing inshore community structure relative to austral winter periods (May). Collectively, these results demonstrate that the bacterial communities exhibit temporal and spatial patterns, and that inshore spatial patterns, in part, reflect the region's hydrographic regime. Our results and those of Nelson et al. (2011) emphasize that current flow and mixing in contiguous marine coastal environments does not obscure community-level signatures within microbial populations.
Biogeographic co-occurrence patterns between Archaea and Bacteria
Archaeal communities were also strongly separated into inshore–offshore communities, though no temporal data were collected for the archaeal domain. Both inshore and offshore archaeal communities were dominated by euryarchaeaotal taxa, particularly those affiliated with MG II archaea found previously dominant within coral reef ecosystems (Siboni et al., 2008). The variation in the inshore archaeal communities was not as pronounced as that of the bacteria over short horizontal distances. This could in part be due to lower overall diversity of archaea in the study site, the shallower sequencing depth (Supplementary Figure S4) or lesser biogeographic specificity in this region. However, network analysis identifying co-occurrence patterns between archaeal and bacterial OTUs does suggest the occurrence of potential niche partitioning within archaeal communities in close geographic proximity to each other (Figure 8). For instance there was a strong positive correlation between two halobacterial OTUs that were found primarily in the back reef and were rare or absent in the other stations sampled, including the adjacent bay station. These two back-reef-associated archaeal OTUs are part of a back-reef-specific assemblage that includes 12 bacterial OTUs (predominantly Gammaproteobacteria), 11 of which share no close correlations with archaeal OTUs found in other sample stations and the remaining Actinobacterial OTU with a strong negative correlation to offshore-specific archaea. Likewise, we found archaeal OTU assemblages that were specific to offshore stations, inshore stations (including Paopao Bay), and an assemblage found in both the back-reef and the eastern 5 km offshore site.
Microbial eukaryotic richness and community structure
Microbial eukaryotic richness patterns across the inshore and offshore locals were less distinct than observed for bacteria and archaea. It is unclear why this was true, but as with archaeal sampling, we cannot exclude the possibility that reduced sequencing effort may have contributed to this in part (Supplementary Figure S4). However, because our data did not exhibit a trend toward increased richness recovered with increased sampling effort (data not shown), we conclude that other factors likely explain these observations.
Our comparison of OTU composition between offshore, inshore and back reef sites indicated that more than half of the inshore microbial eukaryotic OTUs are shared with those from the offshore and that over two thirds of the back reef OTUs are shared with offshore and inshore sites. The majority of the OTUs unique to the offshore were rare (one or two occurrences) and encompassed a range of taxonomic groups from rhodophytes, diatoms, cercozoa, choanoflagellates, dinoflagellates, ciliates, apicomplexans, fungi and unicellular plants. Not surprisingly, dinoflagellate tags were the most abundant in our data set across locations and we attribute this to the large copy number of rRNA genes possessed by most members of this group and not necessarily their absolute abundance. Despite this fact, the dinoflagellates contained the largest numbers of different OTUs in our data set in all three locations. Certain protistan OTUs appeared to be restricted to offshore sites, these included ones related to Streptophyta, Acanthamoebidae and Phaeophyceae; whereas others were only detected inshore. Among the most ubiquitous OTUs that occurred in all replicates and all samples, were a gymnodinoid dinoflagellate, Ceratium, OTUs related to picoeukaryotic Syndiniales, Stichotrich ciliates and an unidentified ciliate. On the opposite end of the abundance spectrum, several of our ‘single-singleton'-OTUs that occurred one time in the entire data set had a 100% match to a relative in GenBank and included cercozoans, stichotrich ciliates, cryptophytes, dinoflagellates, fungi, haptophytes, rhodophytes, diatoms, oomycetes, pelagophytes and chlorophytes. Collectively, these protistan groups encompass photosynthetic, parasitic and heterotrophic lifestyles that extended from abundant to rare community members.
Conclusion
Our study provides the first simultaneous examination of alpha and beta diversity in all three microbial domains within a coral reef ecosystem. Our findings suggest that biogeographic patterning existing in the MCR ecosystem spans microbial communities across all three domains of life. These patterns are strong enough to extend to the rare, as well as abundant community members that characterize the coral reef. Elevated taxonomic richness across all domains within the reef environment relative to the surrounding oceans extends the macroorganismal concept of coral reefs as oases of biodiversity to encompass the microbial world. Finally, our records provide important baseline inventory diversity data against which we can compare future changes in this diverse and fragile marine habitat.
Acknowledgments
This work was supported by grants from the United States National Science Foundation (NSF) MIRADA LTERS grant DEB-0717390 (LAZ) and the Moorea Coral Reef Long Term Ecological Research project (NSF OCE-0417412) through minigrants to CAC and ALA and NSF OCE-0927411 to CAC. We thank John Bunge and Katelyn Giangregorio for their helpful discussions and assistance regarding statistical analyses.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adjeroud M. Zonation of macrobenthic communities along two bays in an insular coral reef ecosystem (Moorea, French Polynesia) Comp Rend Acad Sci. 2000;323:305–313. doi: 10.1016/s0764-4469(00)00134-7. [DOI] [PubMed] [Google Scholar]
- Adjeroud M, Salvat B. Spatial patterns in biodiversity of a fringing reef community along Opunohu Bay, Moorea, French Polynesia. Bull Mar Sci. 1996;59:175–187. [Google Scholar]
- Adjeroud M, Augustin D, Galzin R, Salvat B. Natural disturbances and interannual variability of coral reef communities on the outer slope of Tiahura (Moorea, French Polynesia): 1991 to 1997. Mar Ecol Prog Ser. 2002;237:121–131. [Google Scholar]
- Alldredge A, King J. Near-surface enrichment of zooplankton over a shallow back reef: implications for coral reef food webs. Coral Reefs. 2009;28:895–908. [Google Scholar]
- Amaral-Zettler LA, McCliment EA, Ducklow HW, Huse SM. A method for studying protistan diversity using massively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS One. 2009;4:e6372. doi: 10.1371/journal.pone.0006372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral-Zettler LA, Zettler ER, Theroux SM, Palacios C, Aguilera A, Amils R. Microbial community structure across the tree of life in the extreme Rio Tinto. ISME J. 2011;5:42–50. doi: 10.1038/ismej.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson AF, Riemann L, Bertilsson S. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 2009;4:171–181. doi: 10.1038/ismej.2009.108. [DOI] [PubMed] [Google Scholar]
- Azam F, Malfatti F. Microbial structuring of marine ecosystems. Nat Rev Micro. 2007;5:782–791. doi: 10.1038/nrmicro1747. [DOI] [PubMed] [Google Scholar]
- Brazelton WJ, Ludwig KA, Sogin ML, Andreishcheva EN, Kelley DS, Shen C-C, et al. Archaea and bacteria with surprising microdiversity show shifts in dominance over 1,000-year time scales in hydrothermal chimneys. Proc Natl Acad Sci. 2010;107:1612–1617. doi: 10.1073/pnas.0905369107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MV, Philip GK, Bunge JA, Smith MC, Bissett A, Lauro FM, et al. Microbial community structure in the North Pacific ocean. ISME J. 2009;3:1374–1386. doi: 10.1038/ismej.2009.86. [DOI] [PubMed] [Google Scholar]
- Bunge JA. Estimating the number of species with CatchAll. Proceedings of the 2011 Pacific Symposium on Biocomputing. 2011;16:121–130. doi: 10.1142/9789814335058_0014. [DOI] [PubMed] [Google Scholar]
- Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 2009;3:283–295. doi: 10.1038/ismej.2008.117. [DOI] [PubMed] [Google Scholar]
- Chao AN, Shen TJ. Program SPADE (Species Prediction And Diversity Estimates) 2010.
- Cheung MK, Au CH, Chu KH, Kwan HS, Wong CK. Composition and genetic diversity of picoeukaryotes in subtropical coastal waters as revealed by 454 pyrosequencing. ISME J. 2010;4:1053–1059. doi: 10.1038/ismej.2010.26. [DOI] [PubMed] [Google Scholar]
- Clarke KR, Gorley RN. PRIMERv6: user manual/tutorial. Primer-e: Plymouth routines in multivariate ecological research 2006.
- Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, Wegley L, et al. Microbial ecology of four coral atolls in the Northern Line Islands. PLoS One. 2008;3:e1584. doi: 10.1371/journal.pone.0001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgcomb V, Orsi W, Leslin C, Epstein S, Bunge J, Jeon S, et al. Protistan community patterns within the brine and halocline of deep hypersaline anoxic basins in the eastern Mediterranean Sea. Extremophiles. 2009;13:151–167. doi: 10.1007/s00792-008-0206-2. [DOI] [PubMed] [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Lovejoy C. Ecology of the rare microbial biosphere of the Arctic Ocean. Proc Natl Acad Sci. 2009a;106:22427–22432. doi: 10.1073/pnas.0908284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galand PE, Casamayor EO, Kirchman DL, Potvin M, Lovejoy C. Unique archaeal assemblages in the Arctic Ocean unveiled by massively parallel tag sequencing. ISME J. 2009b;3:860–869. doi: 10.1038/ismej.2009.23. [DOI] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T, et al. The seasonal structure of microbial communities in the Western English Channel. Environ Microbiol. 2009;11:3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x. [DOI] [PubMed] [Google Scholar]
- Hench JL, Leichter JJ, Monismith SG. Episodic circulation and exchange in a wave-driven coral reef and lagoon system. Limnol Oceanogr. 2008;53:2681–2694. [Google Scholar]
- Huber JA, Mark Welch DB, Morrison HG, Huse SM, Neal PR, Butterfield DA, et al. Microbial population structures in the deep marine biosphere. Science. 2007;318:97–100. doi: 10.1126/science.1146689. [DOI] [PubMed] [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Huber JA, Morrison HG, Sogin ML, Mark Welch D. Accuracy and quality of massively parallel DNA pyrosequencing. Gen Biol. 2007;8:R143. doi: 10.1186/gb-2007-8-7-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Mark Welch D, Morrison HG, Sogin ML. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol. 2010;12:1889–1898. doi: 10.1111/j.1462-2920.2010.02193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massana R, Pedrós-Alió C. Unveiling new microbial eukaryotes in the surface ocean. Curr Opin Microbiol. 2008;11:213–218. doi: 10.1016/j.mib.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Nelson CE. Phenology of high-elevation pelagic bacteria: the roles of meteorologic variability, catchment inputs and thermal stratification in structuring communities. ISME J. 2008;3:13–30. doi: 10.1038/ismej.2008.81. [DOI] [PubMed] [Google Scholar]
- Nelson CE, Alldredge AL, McCliment EA, Amaral-Zettler LA, Carlson CA. Depleted dissolved organic carbon and distinct bacterial communities in the water column of a rapidly flushing coral reef ecosystem. ISME J. 2011;5:1374–1387. doi: 10.1038/ismej.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penin L, Adjeroud M, Schrimm M, Lenihan HS. High spatial variability in coral bleaching around Moorea (French Polynesia): patterns across locations and water depths. Comptes Rendus Biologies. 2007;330:171–181. doi: 10.1016/j.crvi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Pomeroy LR, Williams PJI, Azam F, Hobbie JE. The microbial loop. Oceanography. 2007;20:28–33. [Google Scholar]
- Schrimm M, Buscail R, Adjeroud M. Spatial variability of the biogeochemical composition of surface sediments in an insular coral reef ecosystem: Moorea, French Polynesia. Est Coast Shelf Sci. 2004;60:515–528. [Google Scholar]
- Siboni N, Ben-Dov E, Sivan A, Kushmaro A. Global distribution and diversity of coral-associated Archaea and their possible role in the coral holobiont nitrogen cycle. Environ Microbiol. 2008;10:2979–2990. doi: 10.1111/j.1462-2920.2008.01718.x. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Morrison HG, Huber JA, Mark Welch D, Huse SM, Neal PR, et al. Microbial diversity in the deep sea and the underexplored ‘rare biosphere'. Proc Natl Acad Sci. 2006;103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeck T, David B, Markus N, Richard C, Jones MDM, Hans-Werner B, et al. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol Ecol. 2010;19:21–31. doi: 10.1111/j.1365-294X.2009.04480.x. [DOI] [PubMed] [Google Scholar]
- Treusch AH, Vergin KL, Finlay LA, Donatz MG, Burton RM, Carlson CA, et al. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 2009;3:1148–1163. doi: 10.1038/ismej.2009.60. [DOI] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: The unseen majority. Proc Natl Acad Sci. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz P, Kottmann R, Field D, Knight R, Cole JA, Amaral-Zettler LA, et al. The ‘Minimum Information about a MARKer gene Sequence' (MIMARKS) specification. Nat Biotechnol. 2011;29:415–420. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.