Abstract
Despite a long-suspected role in the development of human colorectal cancer (CRC), the composition of gut microbiota in CRC patients has not been adequately described. In this study, fecal bacterial diversity in CRC patients (n=46) and healthy volunteers (n=56) were profiled by 454 pyrosequencing of the V3 region of the 16S ribosomal RNA gene. Both principal component analysis and UniFrac analysis showed structural segregation between the two populations. Forty-eight operational taxonomic units (OTUs) were identified by redundancy analysis as key variables significantly associated with the structural difference. One OTU closely related to Bacteroides fragilis was enriched in the gut microbiota of CRC patients, whereas three OTUs related to Bacteroides vulgatus and Bacteroides uniformis were enriched in that of healthy volunteers. A total of 11 OTUs belonging to the genera Enterococcus, Escherichia/Shigella, Klebsiella, Streptococcus and Peptostreptococcus were significantly more abundant in the gut microbiota of CRC patients, and 5 OTUs belonging to the genus Roseburia and other butyrate-producing bacteria of the family Lachnospiraceae were less abundant. Real-time quantitative PCR further validated the significant reduction of butyrate-producing bacteria in the gut microbiota of CRC patients by measuring the copy numbers of butyryl-coenzyme A CoA transferase genes (Mann–Whitney test, P<0.01). Reduction of butyrate producers and increase of opportunistic pathogens may constitute a major structural imbalance of gut microbiota in CRC patients.
Keywords: colorectal cancer, gut microbiota, pyrosequencing
Introduction
Colorectal cancer (CRC), also known as large bowel cancer, is one of the most common forms of cancer and the third most common cause of cancer mortality in the world (Jemal et al., 2011). Although heredity is tightly associated with some forms of CRC (Lynch and de la Chapelle, 2003), diet is considered to be the most significant risk factor of this disease by many researchers. Incidence of up to 80% of CRC has been attributed to dietary factors in western populations by epidemiological surveys (Bingham, 2000). High rates of CRC are found in populations consuming diets high in animal fat, red meat and processed meat, and low in unrefined grains, dietary fiber and vegetables (Chan and Giovannucci, 2010; Dahm et al., 2010).
A highly diverse and dense microbiota exists in the lower part of the human gastrointestinal tract. Recent studies have shown that diet has a dominating role in shaping the structure of gut microbiota (De Filippo et al., 2010; Walker et al., 2010; Zhang et al., 2010), which anaerobically ferments dietary components that are not completely digested and absorbed in the upper gastrointestinal tract (van Hylckama Vlieg et al., 2011). Metabolites and antigens produced by gut microbiota may have significant roles in influencing CRC risk through their interactions with host metabolism and immunity (Davis and Milner, 2009; Saleh and Trinchieri, 2011).
The types of bacteria and their metabolites that may affect the progression of CRC have long been of interest. For example, acetaldehyde-producing bacteria, sulphate-reducing bacteria and 7α-dehydroxylating bacteria have been implicated as possible contributing factors to the risk of CRC, due to their metabolites possessing colonic inflammatory and tumor-inducing toxicities, including acetaldehyde (Homann et al., 2000), hydrogen sulfide (Huycke and Gaskins, 2004) and secondary bile acids (Tong et al., 2008; Bernstein et al., 2009), so on. On the other hand, bacterial metabolites that may reduce CRC risk include butyrate and other short-chain fatty acids (Scharlau et al., 2009), and conjugated linoleic acids (Palombo et al., 2002), so on. Producers of these compounds, identified by isolation and in vitro characterization, have thus been regarded as beneficial bacteria for human colon health. These bacteria are widely distributed in different phylogenetic groups, including the butyrate producers in the order Clostridiales (Louis and Flint, 2009) and conjugated linoleic acids producers in the orders Clostridiales, Lactobacillales, Bifidobacteriales, and Actinomycetales, so on (Devillard et al., 2007).
Researchers have also made efforts to search for gut bacterial types possibly related with CRC by comparing their abundance between CRC patients/high-risk populations and healthy populations. Moore and Moore (1995) found through cultivation of human fecal microbiota that Bacteroides vulgatus, Eubacterium spp., Ruminococcus spp., Streptococcus hansenii, Bifidobacterium spp. and Faecalibacterium prausnitzii were significantly higher in people with high CRC risk, while species Lactobacillus S06 and Eubacterium aerofaciens were significantly higher in people with low CRC risk. Using the same method, O'Keefe et al. (O'Keefe et al., 2007) associated 7-α dehydroxylating bacteria to high CRC risk populations and Lactobacillus plantarum to low CRC risk populations. Using group-specific quantitative PCR (qPCR) assays, fecal amounts of Desulfovibrio sp. (Scanlan et al., 2009) and Enterococcus faecalis (Balamurugan et al., 2008) were found to be significantly higher in CRC patients than in healthy controls, while Bacteroides/Prevotella levels were significantly lower (Sobhani et al., 2011). Differences were also observed in the overall structure of the gut microbiota between four healthy persons and four persons with colonic adenoma (Shen et al., 2010), as well as between six healthy persons and six CRC patients (Sobhani et al., 2011).
However, adequate knowledge has not yet been provided by previous studies due to the well-known limitations of culture-based methods and DNA fingerprinting techniques, or the small sample size. With the development of high-throughput DNA-sequencing technology and application of multivariate statistical tools, researchers are now able to do a ‘microbiome-wide association study' to reveal microbial variations that are possibly linked to environmental changes such as the host health status, with higher resolution and fewer biases (Andersson et al., 2008; Dethlefsen et al., 2008). In this study, we employed this type of strategy to reveal a significant segregation of the gut microbiota of CRC patients from healthy volunteers, and identified the bacterial phylotypes important for this segregation.
Materials and methods
Sampling
All the 46 CRC patients, aged 42–77 years, were from Shanghai Cancer Hospital, Shanghai Medical College, Fudan University (Shanghai, China). All patients were categorized according to histopathological features by TNM classification of malignant tumors after surgery. None of the patients were on any medication before sample collection. The 56 healthy volunteers, aged 40–54 years, were selected as controls during a routine physical examination and none of them had had gastrointestinal tract disorders or any antibiotics in the past 3 months before sample collection (Table 1). All participants were local residents of Shanghai city for more than 10 years. Informed consent was given by all individuals before participation. One fecal sample was collected from each control, and from each patient before surgery. Fecal samples were frozen immediately after sampling and stored at −80 °C.
Table 1. Clinical characteristics of all individuals.
| Healthy volunteers | CRC patients | |
|---|---|---|
| n | 56 | 46 |
| Age (median, range) | 49, 40–54 | 60, 42–77 |
| Male/female | 27/29 | 24/22 |
| BMI (median, range) | 21.07, 20–22.68 | ND |
| Stage I | 8 | |
| Stage II | 18 | |
| Stage III | 17 | |
| Stage IV | 3 |
Abbreviations: BMI, body mass index; CRC, colorectal cancer; ND, no data.
DNA extraction
Genomic DNA was extracted from each fecal sample by bead-beating extraction and phenol–chloroform purification, as described previously (Li et al., 2008). The amount of DNA was determined by Fluorescent and Radioisotope Science Imaging Systems FLA-5100 (Fujifilm, Tokyo, Japan). Integrity and size of DNA were checked by 0.8% (w/v) agarose gel electrophoresis in 0.5 mg ml−1 ethidium bromide. All DNA samples were stored at −20 °C until further processing.
Pyrosequencing
The V3 region of the 16S ribosomal RNA (rRNA) gene from each DNA sample was amplified using the bacterial universal forward primer 5′-NNNNNNNNCCTACGGGAGGCAGCAG-3′ and the reverse primer 5′-NNNNNNNNATTACCGCGGCTGCT-3′. The NNNNNNNN was the sample-unique 8-base barcode for sorting of PCR amplicons into different samples, and the underlined text indicates universal bacterial primers for the V3 region of the 16S rRNA gene (Muyzer et al., 1993). PCR amplification, pyrosequencing of the PCR amplicons, and quality control of raw data were performed, as described previously (Zhang et al., 2010).
Bioinformatics and multivariate statistics
All reads were sorted into different samples according to barcodes. After removal of barcodes, the sequences were clustered using the program CD-HIT with 99.8% similarity (Li and Godzik, 2006). The most abundant sequence of each cluster was selected as a representative and aligned by NAST multi-aligner with template length ⩾100 bases and percent identity ⩾75% (DeSantis et al., 2006). RDP Classifier was used for taxonomical assignments of all sequences at 80% confidence level (Cole et al., 2009). Relative abundances of different phyla and genera in each sample were calculated and compared between healthy volunteers and CRC patients using the Student's t-test (data of normalized distribution) or Mann–Whitney test (data of non-normalized distribution) via software SPSS 17.0 (SPSS Inc, Chicago, IL, USA). All aligned sequences were imported into ARB (Ludwig et al., 2004) to construct a neighbor-joining tree. The tree, together with sequence abundance data, was then used for online Fast UniFrac analysis based on weighted metric (Hamady et al., 2010). Operational taxonomic unit (OTU) was classified with DOTUR (Schloss and Handelsman, 2005). For each library, OTUs defined at 98% similarity level were used for coverage analysis using Good's coverage by [1− (n/N)] × 100 (Good, 1953), and richness and diversity estimations using Rarefaction analysis (aRarefact-Win software, S Holland, Stratigraphy Laboratory, University of Georgia, Athens, GA, USA, http://www.uga.edu/strata/software), Shannon diversity index (H') (R package 2.7.1), as well as Simpson's diversity index (1-D), and Buzas and Gibson's evenness index (exp(H')/S) (PAST software, Ø Hammer, University of Oslo, Oslo, Norway, http://www.nhm.uio.no/norlex/past/download.html). The most abundant sequence of each OTU (98% similarity) was BLAST searched against the RDP database (Release 10) to determine the phylogeny of the OTU. Relative abundances of OTUs were used for principal component analysis, multivariate analysis of variance, and redundancy analysis via Matlab R2010a (The MathWorks, Natick, MA, USA) and Canoco for Windows 4.5 (Microcomputer Power, NY, USA).
Real-time qPCR
Fecal DNA from each sample was subjected to real-time qPCR assays to determine the amounts of total bacteria and Bacteroides spp. through detection of 16S rRNA genes, and the amount of butyrate-producing bacteria through detection of butyryl-coenzyme A (CoA) CoA transferase genes. PCR reactions were performed using iQ SYBR Green SuperMix (Bio-Rad, Richmond, CA, USA) on a DNA Engine Opticon 3 system (MJ Research, Waltham, MA, USA). Each 25-μl reaction system contained 12.5 pmol of each primer for amplification of 16S rRNA genes of total bacteria, or 7 pmol of each primer for amplification of Bacteroides spp. 16S rRNA genes, or 25 pmol of each primer for amplification of butyryl-CoA CoA transferase genes. Primers and amplification conditions are listed in Supplementary Table S1. For each reaction, fluorescence detection was performed at the end of each cycle at 80 °C for 5 s. After all cycles, amplification specificity was monitored via dissociation curve analysis of PCR end products by increasing the temperature at a rate of 0.5 °C per 10 s from 60 to 95 °C with continuous fluorescence collection. Amplification efficiency was determined using Opticon Monitor Software (Version 3.1, MJ Research) from the slope of the line during the linear phase of the PCR reaction. Reactions with efficiency of 1.8–2.2 were selected for further analysis. A plasmid containing a Bacteroides spp. full-length 16S rRNA gene from a previous study (Li et al., 2008) was prepared using the QIAGEN Plasmid Mini Kit (Qiagen, Hilden, Germany) and diluted from 1 × 103 to 1 × 109 (copies μl−1) to construct standard curves for the detection of total bacteria and Bacteroides spp. A plasmid containing a single butyryl-CoA CoA transferase gene amplicon was prepared and diluted from 1 × 103 to 1 × 109 (copies μl−1) to construct standard curves for the detection of butyrate-producing bacteria. Reactions for standard samples were performed in triplicate and others in duplicate. Copy number of the targeting gene in each reaction was determined by comparison to the standard curve via Opticon Monitor Software. The Bacteroides spp. specific 16S rRNA gene copy number or butyryl-CoA CoA transferase gene copy number in relation to total 16S rRNA gene copy number was calculated for each sample, and compared between healthy volunteers and CRC patients by Mann–Whitney test as above. The amounts of Bacteroides spp. measured by qPCR and pyrosequencing were compared using Spearman's rank correlation coefficient via SPSS 17.0 (SPSS Inc).
Accession number
The sequence information in this paper has been deposited in the GenBank Sequence Read Archive with accession number SRP005150.
Results
Richness and diversity analysis
In total, 278 877 usable raw reads were obtained from all 102 samples, with an average of 2734±460 reads per sample. After CD-HIT clustering and NAST alignment, 21 043 unique representative sequences were generated. OTUs were delineated at 98% similarity level because higher thresholds generated a dramatic increase of OTU numbers, which might represent the microdiversity at subspecies level (Supplementary Figure S3) (Zhang et al., 2009). The total number of OTUs at 98% similarity level was 4249 (Supplementary Table S2). The values of Good's coverage of all libraries were above 94%. While no rarefaction curve plateaued with the current sequencing, the Shannon diversity estimates of all samples already reached stable values at this sequencing depth, suggesting that although new phylotypes would be expected with additional sequencing, most diversity had already been captured (Supplementary Figure S1). There were no statistically significant differences with Shannon diversity index, Simpson's diversity index or Buzas or Gibson's evenness index between the gut microbiota of healthy volunteers and CRC patients (Student's t-test, P>0.05; Supplementary Table S3).
Taxonomy-based comparisons of gut microbiota at the phylum and genus levels between healthy volunteers and CRC patients
The amounts of different phyla and genera were assessed by taxonomic assignment of all sequences using RDP Classifier. Among all bacterial groups revealed by the interpretable sequences, Firmicutes was the most predominant phylum, contributing 57.2% and 63.1% of the gut microbiota in healthy volunteers and CRC patients, respectively, followed by Bacteroidetes, which contributed 32.0% and 22.7%, respectively. Proteobacteria, Actinobacteria and Fusobacteria constituted the next most dominant phyla, contributing 2.81%, 2.22% and 2.20% of healthy volunteers, and 4.68%, 4.55% and 1.59% of CRC patients, respectively (Supplementary Figure S2). Microbial compositions showed high inter-individual variability. For example, Firmicutes accounted for 16.6–94.5%, and Bacteroidetes 0.13–75.0% among all individuals (Supplementary Table S4). Bacteroidetes was statistically significantly more abundant in the gut microbiota of healthy volunteers than that of CRC patients, and Proteobacteria significantly less abundant in the gut microbiota of healthy volunteers (P=0.002 and 0.045, respectively).
At the genus level, Firmicutes in the gut microbiota of CRC patients mainly consisted of Faecalibacterium, Blautia, Enterococcus, Subdoligranulum, Dorea, Roseburia, Megamonas, and Streptococcus, each constituting more than 1% of total bacteria in this population. Relatively abundant genera (>1%) in other phyla included Bacteroides, Prevotella, Parabacteroides and Porphyromonas of Bacteroidetes; Bifidobacterium and Collinsella of Actinobacteria; and Escherichia/Shigella of Proteobacteria. Statistically, the relative abundance of five genera: Bacteroides, Roseburia, Alistipes, Eubacterium and Parasutterella were found to be significantly higher in healthy volunteers than CRC patients, whereas another five genera: Porphyromonas, Escherichia/Shigella, Enterococcus, Streptococcus and Peptostreptococcus were significantly lower in healthy volunteers than CRC patients (P<0.05; Table 2).
Table 2. List of genera that were significantly different between H and C.
| Phylum | Genus |
Relative contribution (%)a |
Median, range (%) |
P-value | Tendency in CRC patients compared with healthy volunteersb | ||
|---|---|---|---|---|---|---|---|
| H | C | H | C | ||||
| Bacteroidetes | Bacteroides | 21.49 | 14.04 | 20.9, 0.08–49.8 | 10.7, 0–59.9 | 0.005 | ↓ |
| Firmicutes | Roseburia | 3.59 | 1.56 | 2.39, 0–26.0 | 0.57, 0–9.53 | 0.003 | ↓ |
| Bacteroidetes | Alistipes | 1.11 | 0.67 | 0.50, 0–5.7 | 0.15, 0–11.0 | 0.039 | ↓ |
| Firmicutes | Eubacterium | 0.52 | 0.13 | 0, 0–4.68 | 0, 0–2.51 | 0.028 | ↓ |
| Proteobacteria | Parasutterella | 0.44 | 0.13 | 0, 0–4.10 | 0, 0–1.80 | 0.032 | ↓ |
| Bacteroidetes | Porphyromonas | 4.1e–3 | 1.74 | 0, 0–0.08 | 0, 0–47.4 | 0.020 | ↑ |
| Proteobacteria | Escherichia/Shigella | 0.87 | 2.45 | 0.07, 0–18.2 | 0.39, 0–15.1 | 2e–4 | ↑ |
| Firmicutes | Enterococcus | 0.09 | 2.40 | 0, 0–5.16 | 0.04, 0–59.8 | 5e–4 | ↑ |
| Firmicutes | Streptococcus | 0.66 | 1.19 | 0.27, 0–6.52 | 0.49, 0–8.58 | 0.018 | ↑ |
| Firmicutes | Peptostreptococcus | 1.3e–3 | 0.59 | 0, 0–0.04 | 0.04, 0–7.85 | 5.5e–6 | ↑ |
Abbreviations: C, CRC (colorectal cancer) patients; H, healthy volunteers.
Relative contribution of a genus in healthy volunteers or CRC patients was calculated as percentage of the sequences of this genus to all sequences in this population.
‘↓' means decrease and ‘↑' means increase.
Structural comparison of gut microbiota between healthy volunteers and CRC patients with multivariate statistics
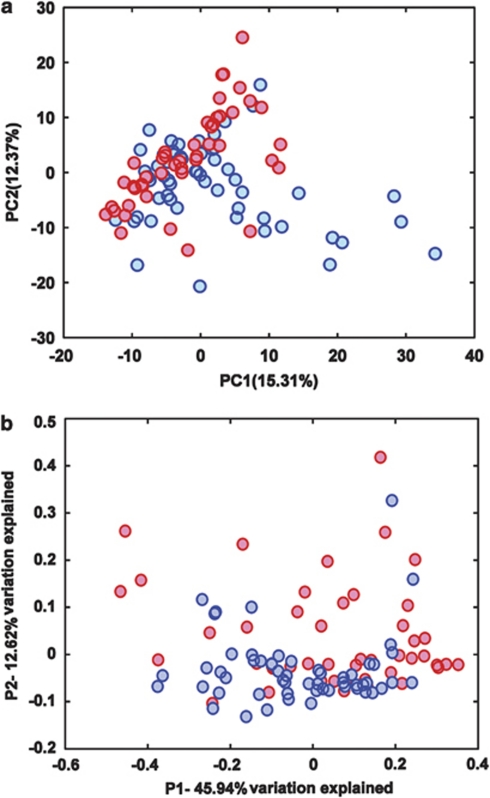
Multivariate statistical analyses were performed to compare the overall structure of gut microbiota of all samples. Principal component analysis based on the relative abundance of OTUs revealed a separation of the healthy and CRC individuals on the basis of the first two principal component (PC) scores, which accounted for 15.31% and 12.37% of the total variations (Figure 1a). A multivariate analysis of variance test further indicated that the variation of all individuals exhibited by the first two PC axes was probably explained by disease status (P=0.002) rather than gender (P=0.653) or age (age groups of 40–49, 50–59, 60–69, 70–79 years, P=0.165). Principal coordinate analysis based on weighted UniFrac metric confirmed the above findings, showing a significant separation of healthy volunteers and CRC patients (Figure 1b). At other similarity level of OTUs (95%, 97%, 99% and 100%), consistent results of principal component analysis and multivariate analysis of variance were obtained (Supplementary Figure S4).
Figure 1.
(a) Principal component analysis (PCA) scores plot based on the relative abundance of OTUs (98% similarity level). (b) PCoA (principal coordinate analysis) plot based on the weighted UniFrac metric. Blue circles represent healthy volunteers; red circles represent CRC patients.
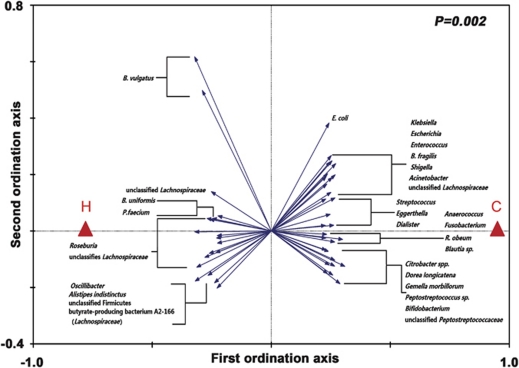
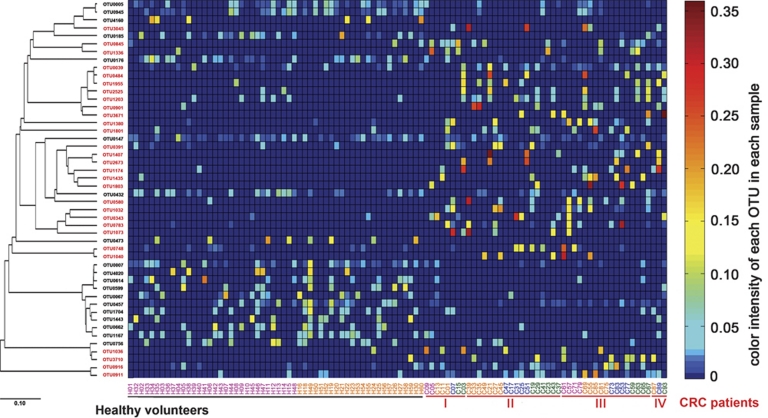
Redundancy analysis was performed using healthy or diseased as constrained explanatory variables and relative abundances of all OTUs as response variables. Monte Carlo Permutation Test showed that the constrained ordination model by healthy or diseased was significant (P=0.002) and 5.3% of the variance in OTU abundance data can be explained by the canonical axis. We identified 48 OTUs as key variables, which were well fitted by the sample scores on the canonical axis; each had at least 5% of the variability in its values explained by this axis (Figure 2). On the redundancy analysis ordination plot, 19 OTUs were enriched in the gut microbiota of healthy volunteers, among which 10 OTUs belonged to the genera Alistipes, Phascolarctobacterium, Oscillibacter, or unclassified genera of the order Clostridiales; 4 OTUs belonged to the butyrate-producing genus Roseburia, and 1 OTU was closely related to the butyrate-producing bacterium A2-166 (100% similarity) of the family Lachnospiraceae. On the contrary, 29 OTUs were enriched in the gut microbiota of CRC patients, which mainly belonged to genera Escherichia/Shigella, Klebsiella, or Citrobacter of the family Enterobacteriaceae, and genera Streptococcus, Enterococcus, Peptostreptococcus, Eggerthella, Fusobacterium, Gemella, or some other bacteria of the order Clostridiales. Interestingly, two OTUs closely related to B. vulgatus (100% and 98.7% similarity, respectively) and one OTU related to Bacteroides uniformis (97.5% similarity) were enriched in the gut microbiota of healthy volunteers, while one OTU closely related to Bacteroides fragilis (98.1% similarity) was enriched in that of CRC patients. Two OTUs of the order Bifidobacteriales were also enriched in the gut microbiota of CRC patients (Figure 3). Mann–Whitney tests showed that the relative abundances of 46 OTUs out of the 48 were significantly different between healthy and CRC individuals (P<0.05, Supplementary Table S5).
Figure 2.
Biplot of the RDA (redundancy analysis) based on the relative abundance of OTUs (98% similarity level). Constrained explanatory variables are indicated by red triangles: H represents healthy and C represents diseased. OTUs that have at least 5% of the variability in their values explained by the canonical axis are indicated by blue arrows. Upper right shows P-value of Monte Carlo Permutation Test. B, Bacteroides; P, Phascolarctobacterium; E, Escherichia; R, Ruminococcus.
Figure 3.
Relative abundances of the 48 OTUs identified as key variables for the differentiation of microbiota of healthy volunteers and CRC patients. OTUs shown in black are the ones enriched in healthy volunteers and those in red are the ones enriched in CRC patients. Phylogenetic tree of the OTUs on the left is constructed with neighbor-joining method in ARB. For each OTU, the color intensity in each sample is normalized to represent its relative ratio in all samples. Individuals at the bottom are arranged according to disease status (healthy; stage I, II, III and IV of CRC) and age inside each stage (from youngest to oldest, color code: pink for age 40–49 years; orange for age 50–59 years; blue for age 60–69 years; green for age 70–79 years).
Real-time qPCR
Group-specific qPCR was used to determine the relative amounts of Bacteroides spp. and butyrate-producing bacteria in all samples (Supplementary Table S6). Bacteroides spp. was found to be significantly more abundant in healthy volunteers than CRC patients (P=0.005). Quantification of Bacteroides spp. by qPCR and pyrosequencing showed a significantly high degree of consistency (Spearman correlation, r=0.932, P<0.001), indicating that differences at genus level between healthy volunteers and CRC patients found by pyrosequencing are reliable. The abundances of butyrate-producing bacteria, as represented by butyryl-CoA CoA transferase genes, were significantly higher in the gut microbiota of healthy volunteers than CRC patients (P=1.5e–4).
Discussion
Gut microbiota interact extensively with the host through metabolic exchange and co-metabolism of substrates to maintain the normal functions and health status of the intestinal tract and the whole body (Nicholson et al., 2005). In this study, we revealed differing gut microbiota structure patterns in CRC patients compared with healthy persons. Significance of the associated structural shift of gut microbiota in the progression of human diseases has been highlighted by several recent publications. In our earlier study, we observed significant differentiation of gut microbiota between healthy rats and carcinogen-treated rats that developed colonic precancerous lesions, but were still apparently healthy (Wei et al., 2010). Alterations of gut microbiota have also been suggested to occur in Crohn's disease patients, ulcerative colitis patients (Frank et al., 2007; Willing et al., 2009, 2010), children with celiac disease (Nadal et al., 2007), and infants suffering from allergic inflammation (Kalliomaki and Isolauri, 2003).
An important part of the structural imbalance is a significant depletion of butyrate-producing bacteria in the gut microbiota of CRC patients. Adequate amounts of butyrate has been implicated in the protection against colitis and CRC by reducing oxidative damage to DNA, inducing apoptosis in DNA-damaged cells, inhibiting tumor cell growth, and decreasing the activity of co-carcinogenic enzymes (Pryde et al., 2002; Rose et al., 2007). The in vivo supply of butyrate largely depends on the butyrate-producing bacteria, which are mainly classified into two groups: the Clostridium leptum cluster (or clostridial cluster IV, represented by F. prausnitzii) and the Clostridium coccoides cluster (or clostridial cluster XIVa, represented by Eubacterium rectale/Roseburia spp.) of the phylum Firmicutes (Louis and Flint, 2009). Here we showed reduced amounts of C. coccoides (Roseburia spp. and Eubacterium spp.) in the gut microbiota of CRC patients by pyrosequencing analysis. We further confirmed the depletion of butyrate producers in CRC patients by measuring the levels of partial butyryl-CoA CoA transferase gene, which is involved in the butyrate formation pathway in the majority of human colonic butyrate producers (Louis and Flint, 2007).
Balamurugan et al.(Balamurugan et al., 2008) found by qPCR assays that F. prausnitzii decreased approximately fourfold in 20 CRC patients compared with 17 healthy volunteers. In our study, Faecalibacterium constituted more than 11% of microbiota of either healthy volunteers or CRC patients, with no difference in between. No OTU related to F. prausnitzii was found to contribute to the structural differentiation between CRC patients and healthy controls. Instead, our results show that a decrease in Roseburia spp. is more closely associated with the occurrence of CRC. It has been suggested by two dietary intervention studies that, population densities of Roseburia/E. rectale spp. have a strong correlation with fecal butyrate concentrations in response to altered carbohydrate supply while F. prausnitzii does not (Duncan et al., 2007; Ramirez-Farias et al., 2009). This indicates the importance of Roseburia spp. and E. rectale in the production of butyrate in vivo. A culture-based study has also suggested that Roseburia spp. are the most important active conjugated linoleic acids producers in the human gut (Devillard et al., 2007). Our findings here have suggested that Roseburia spp. may have important roles in protecting hosts from CRC.
Another feature of the structural imbalance of gut microbiota in CRC patients is the significant increase in a variety of opportunistic pathogens. Some researchers have proposed hypotheses for the oncogenic potential of bacteria during the progression to CRC, while the mechanisms were suggested to include induction of inflammation or production of mutagenic toxins (Collins et al., 2010). Inflammation is commonly associated with CRC. Colitis-associated cancer, a subtype of CRC, develops directly from inflammatory bowel disease. Other sporadic CRCs, which are not associated with clinically detectable inflammatory bowel disease, also display robust inflammatory infiltration and increased expression of proinflammatory cytokines (Terzic et al., 2010). Inflammation associated with CRC has been suggested to be related to bacterial infection. Through isolation and cultivation from biopsy samples, Martin et al. (Martin et al., 2004) have found increased proportion of hemagglutinin expressing Escherichia coli in Crohn's disease and colon cancer patients compared with healthy controls. It has also been suggested that enteropathogenic E. coli possesses the ability to downregulate DNA mismatch repair proteins and, therefore, promotes colonic tumorigenesis (Maddocks et al., 2009). Streptococcus bovis promotes the progression of preneoplastic lesions by increasing cell proliferation and interleukin-8 production in a rat model (Ellmerich et al., 2000), and its wall-extracted antigens promote carcinogenesis in Caco-2 (human colorectal adenocarinoma epithelium) cells by overexpression of cyclo-oxygenase-2 (Biarc et al., 2004). E. faecalis produces extracellular superoxide that damages colonic epithelial cell DNA both in vitro and in vivo (Huycke and Moore, 2002; Huycke et al., 2002), and its population was found to be significantly higher in CRC patients compared with healthy volunteers by qPCR assays (Balamurugan et al., 2008). Both these studies and our results suggest that Enterococcus, Streptococcus, and Escherichia/Shigella may contribute to the differentiation of the gut microbiota of CRC patients from healthy controls.
Previous studies have suggested that a significant reduction of the phylum Bacteroidetes occurs in Crohn's disease/ulcerative colitis patients compared with controls (Ott et al., 2004; Frank et al., 2007). However, in our study, we have found different changes among genera of this phylum that, Bacteroides decreased while Porphyromonas increased significantly in the gut microbiota of CRC patients compared with controls. Furthermore, key OTUs in the genus Bacteroides also showed different enrichments between healthy volunteers and CRC patients. Previous studies have suggested that different Bacteroides strains may influence the host health in different ways. A B. vulgatus strain was shown to protect against E. coli-induced colitis in interleukin-2-deficient mice (Waidmann et al., 2003), while interleukin-10 knockout (Il10−/−) mice monoassociated with pig isolates of B. vulgatus have significantly reduced colitis-associated colorectal tumor multiplicity compared with common Il10−/− mice (Uronis et al., 2009). In contrast, the common colonic commensal, enterotoxigenic B. fragilis was reported to trigger cellular proliferation, colonic colitis and strongly induce colonic tumors in multiple intestinal neoplasia (Min) mice (Wu et al., 2009). These studies are in agreement with our finding that the OTU related to B. fragilis was more abundant in CRC patients and the two related to B. vulgatus were more abundant in healthy persons. It is also reported that non-toxigenic human commensal bacterium B. fragilis, which expresses a single microbial molecule polysaccharide A, has the potential to protect animals from experimental colitis induced by Helicobacter hepaticus infection (Mazmanian et al., 2008). The possible strain-specific bioactivities of Bacteroides group members in human gut suggest that more work at the gene or strain level is still needed to identify the effects of gut microbes relevant to the pathogenesis of CRC.
With this design of cross-sectional cohort study, it is not possible to untangle the causal relationship between gut microbiota and CRC. As samples were collected from patients already diagnosed with CRC, the changes of gut microbiota identified in this work might be just a consequence of CRC. However, these imbalanced changes might contribute to the further progression of CRC through the possible mechanisms discussed above. One way to help clarify the possible roles of gut microbiota in CRC could be correcting this structural imbalance by using designed nutritional interventions or gut microbiota transplantation from healthy donors (Borody et al., 2003).
In summary, by comparing the gut microbial composition of CRC patients and healthy volunteers, we have identified a structural imbalance of the gut microbiota, represented by the reduction of butyrate producers and the increase of opportunistic pathogens, which may be a significant feature of human CRC. The structural imbalance of gut microbiota revealed in this work may provide insights for in-depth analysis of host–microbe interactions and determination of their roles in cancer development.
Acknowledgments
This work was supported by Project 2006BAI11B08-02 of the National Science & Technology Pillar Program, Key Project 30730005 of the National Nature Science Foundation of China (NSFC), Project 2009ZX10004-601 of National Science and Technology Major Project of China and Key Project 2007CB513002 of Chinese National Programs for Fundamental Research and Development (973 program).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Andersson AF, Lindberg M, Jakobsson H, Backhed F, Nyren P, Engstrand L. Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS One. 2008;3:e2836. doi: 10.1371/journal.pone.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. J Gastroenterol Hepatol. 2008;23 (Part 1:1298–1303. doi: 10.1111/j.1440-1746.2008.05490.x. [DOI] [PubMed] [Google Scholar]
- Bernstein H, Bernstein C, Payne CM, Dvorak K. Bile acids as endogenous etiologic agents in gastrointestinal cancer. World J Gastroenterol. 2009;15:3329–3340. doi: 10.3748/wjg.15.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biarc J, Nguyen IS, Pini A, Gosse F, Richert S, Thierse D, et al. Carcinogenic properties of proteins with pro-inflammatory activity from Streptococcus infantarius (formerly S.bovis) Carcinogenesis. 2004;25:1477–1484. doi: 10.1093/carcin/bgh091. [DOI] [PubMed] [Google Scholar]
- Bingham SA. Diet and colorectal cancer prevention. Biochem Soc Trans. 2000;28:12–16. doi: 10.1042/bst0280012. [DOI] [PubMed] [Google Scholar]
- Borody TJ, Warren EF, Leis S, Surace R, Ashman O. Treatment of ulcerative colitis using fecal bacteriotherapy. J Clin Gastroenterol. 2003;37:42–47. doi: 10.1097/00004836-200307000-00012. [DOI] [PubMed] [Google Scholar]
- Chan AT, Giovannucci EL.2010Primary prevention of colorectal cancer Gastroenterology 1382029–2043.e2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37 (Database issue:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins D, Hogan AM, Winter DC. Microbial and viral pathogens in colorectal cancer. Lancet Oncol. 2010;12:504–512. doi: 10.1016/S1470-2045(10)70186-8. [DOI] [PubMed] [Google Scholar]
- Dahm CC, Keogh RH, Spencer EA, Greenwood DC, Key TJ, Fentiman IS, et al. Dietary fiber and colorectal cancer risk: a nested case-control study using food diaries. J Natl Cancer Inst. 2010;102:614–626. doi: 10.1093/jnci/djq092. [DOI] [PubMed] [Google Scholar]
- Davis CD, Milner JA. Gastrointestinal microflora, food components and colon cancer prevention. J Nutr Biochem. 2009;20:743–752. doi: 10.1016/j.jnutbio.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Jr, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM, et al. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 2006;34 (Web Server issue:W394–W399. doi: 10.1093/nar/gkl244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16s rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devillard E, McIntosh FM, Duncan SH, Wallace RJ. Metabolism of linoleic acid by human gut bacteria: different routes for biosynthesis of conjugated linoleic acid. J Bacteriol. 2007;189:2566–2570. doi: 10.1128/JB.01359-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Belenguer A, Holtrop G, Johnstone AM, Flint HJ, Lobley GE. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol. 2007;73:1073–1078. doi: 10.1128/AEM.02340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellmerich S, Scholler M, Duranton B, Gosse F, Galluser M, Klein JP, et al. Promotion of intestinal carcinogenesis by Streptococcus bovis. Carcinogenesis. 2000;21:753–756. doi: 10.1093/carcin/21.4.753. [DOI] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good IJ. The population frequencies of species and the estimation of population parameters. Biometrika. 1953;40:237–264. [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME. 2010;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann N, Tillonen J, Salaspuro M. Microbially produced acetaldehyde from ethanol may increase the risk of colon cancer via folate deficiency. Int J Cancer. 2000;86:169–173. doi: 10.1002/(sici)1097-0215(20000415)86:2<169::aid-ijc4>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Huycke MM, Abrams V, Moore DR. Enterococcus faecalis produces extracellular superoxide and hydrogen peroxide that damages colonic epithelial cell DNA. Carcinogenesis. 2002;23:529–536. doi: 10.1093/carcin/23.3.529. [DOI] [PubMed] [Google Scholar]
- Huycke MM, Gaskins HR. Commensal bacteria, redox stress, and colorectal cancer: mechanisms and models. Exp Biol Med (Maywood) 2004;229:586–597. doi: 10.1177/153537020422900702. [DOI] [PubMed] [Google Scholar]
- Huycke MM, Moore DR. In vivo production of hydroxyl radical by Enterococcus faecalis colonizing the intestinal tract using aromatic hydroxylation. Free Radic Biol Med. 2002;33:818–826. doi: 10.1016/s0891-5849(02)00977-2. [DOI] [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Kalliomaki M, Isolauri E. Role of intestinal flora in the development of allergy. Curr Opin Allergy Clin Immunol. 2003;3:15–20. doi: 10.1097/00130832-200302000-00003. [DOI] [PubMed] [Google Scholar]
- Li M, Wang B, Zhang M, Rantalainen M, Wang S, Zhou H, et al. Symbiotic gut microbes modulate human metabolic phenotypes. Proc Natl Acad Sci USA. 2008;105:2117–2122. doi: 10.1073/pnas.0712038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Louis P, Flint HJ. Development of a semiquantitative degenerate real-time pcr-based assay for estimation of numbers of butyryl-coenzyme A (CoA) CoA transferase genes in complex bacterial samples. Appl Environ Microbiol. 2007;73:2009–2012. doi: 10.1128/AEM.02561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–932. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- Maddocks OD, Short AJ, Donnenberg MS, Bader S, Harrison DJ. Attaching and effacing Escherichia coli downregulate DNA mismatch repair protein in vitro and are associated with colorectal adenocarcinomas in humans. PLoS One. 2009;4:e5517. doi: 10.1371/journal.pone.0005517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, et al. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology. 2004;127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Moore WE, Moore LH. Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol. 1995;61:3202–3207. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal I, Donat E, Ribes-Koninckx C, Calabuig M, Sanz Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J Med Microbiol. 2007;56 (Part 12:1669–1674. doi: 10.1099/jmm.0.47410-0. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- O'Keefe SJD, Chung D, Mahmoud N, Sepulveda AR, Manafe M, Arch J, et al. Why do African Americans get more colon cancer than Native Africans. J Nutr. 2007;137:175S–182S. doi: 10.1093/jn/137.1.175S. [DOI] [PubMed] [Google Scholar]
- Ott SJ, Musfeldt M, Wenderoth DF, Hampe J, Brant O, Folsch UR, et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut. 2004;53:685–693. doi: 10.1136/gut.2003.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palombo JD, Ganguly A, Bistrian BR, Menard MP. The antiproliferative effects of biologically active isomers of conjugated linoleic acid on human colorectal and prostatic cancer cells. Cancer Lett. 2002;177:163–172. doi: 10.1016/s0304-3835(01)00796-0. [DOI] [PubMed] [Google Scholar]
- Pryde SE, Duncan SH, Hold GL, Stewart CS, Flint HJ. The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett. 2002;217:133–139. doi: 10.1111/j.1574-6968.2002.tb11467.x. [DOI] [PubMed] [Google Scholar]
- Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr. 2009;101:541–550. doi: 10.1017/S0007114508019880. [DOI] [PubMed] [Google Scholar]
- Rose DJ, DeMeo MT, Keshavarzian A, Hamaker BR. Influence of dietary fiber on inflammatory bowel disease and colon cancer: importance of fermentation pattern. Nutr Rev. 2007;65:51–62. doi: 10.1111/j.1753-4887.2007.tb00282.x. [DOI] [PubMed] [Google Scholar]
- Saleh M, Trinchieri G. Innate immune mechanisms of colitis and colitis-associated colorectal cancer. Nat Rev Immunol. 2011;11:9–20. doi: 10.1038/nri2891. [DOI] [PubMed] [Google Scholar]
- Scanlan PD, Shanahan F, Marchesi JR. Culture-independent analysis of desulfovibrios in the human distal colon of healthy, colorectal cancer and polypectomized individuals. FEMS Microbiol Ecol. 2009;69:213–221. doi: 10.1111/j.1574-6941.2009.00709.x. [DOI] [PubMed] [Google Scholar]
- Scharlau D, Borowicki A, Habermann N, Hofmann T, Klenow S, Miene C, et al. Mechanisms of primary cancer prevention by butyrate and other products formed during gut flora-mediated fermentation of dietary fibre. Mutat Res. 2009;682:39–53. doi: 10.1016/j.mrrev.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen XJ, Rawls JF, Randall T, Burcal L, Mpande CN, Jenkins N, et al. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes. 2010;1:138–147. doi: 10.4161/gmic.1.3.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhani I, Tap J, Roudot-Thoraval F, Roperch JP, Letulle S, Langella P, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS One. 2011;6:e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzic J, Grivennikov S, Karin E, Karin M.2010Inflammation and colon cancer Gastroenterology 1382101–2114.e2105. [DOI] [PubMed] [Google Scholar]
- Tong JL, Ran ZH, Shen J, Fan GQ, Xiao SD. Association between fecal bile acids and colorectal cancer: a meta-analysis of observational studies. Yonsei Med J. 2008;49:792–803. doi: 10.3349/ymj.2008.49.5.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uronis JM, Muhlbauer M, Herfarth HH, Rubinas TC, Jones GS, Jobin C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS One. 2009;4:e6026. doi: 10.1371/journal.pone.0006026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hylckama Vlieg JE, Veiga P, Zhang C, Derrien M, Zhao L. Impact of microbial transformation of food on health-from fermented foods to fermentation in the gastro-intestinal tract. Curr Opin Biotechnol. 2011;22:211–219. doi: 10.1016/j.copbio.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Waidmann M, Bechtold O, Frick JS, Lehr HA, Schubert S, Dobrindt U, et al. Bacteroides vulgatus protects against Escherichia coli-induced colitis in gnotobiotic interleukin-2-deficient mice. Gastroenterology. 2003;125:162–177. doi: 10.1016/s0016-5085(03)00672-3. [DOI] [PubMed] [Google Scholar]
- Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X, et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2010;5:220–230. doi: 10.1038/ismej.2010.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Dong L, Wang T, Zhang M, Hua W, Zhang C, et al. Structural shifts of gut microbiota as surrogate endpoints for monitoring host health changes induced by carcinogen exposure. FEMS Microbiol Ecol. 2010;73:577–586. doi: 10.1111/j.1574-6941.2010.00924.x. [DOI] [PubMed] [Google Scholar]
- Willing B, Dicksved J, Halfvarson J, Andersson A, Lucio M, Zheng Z, et al. 2010A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes Gastroenterology 1391844–1854.e1. [DOI] [PubMed] [Google Scholar]
- Willing B, Halfvarson J, Dicksved J, Rosenquist M, Jarnerot G, Engstrand L, et al. Twin studies reveal specific imbalances in the mucosa-associated microbiota of patients with ileal Crohn's disease. Inflamm Bowel Dis. 2009;15:653–660. doi: 10.1002/ibd.20783. [DOI] [PubMed] [Google Scholar]
- Wu S, Rhee KJ, Albesiano E, Rabizadeh S, Wu X, Yen HR, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17T cell responses. Nat Med. 2009;15:1016–1022. doi: 10.1038/nm.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CH, Zhang MH, Wang SY, Han RJ, Cao YF, Hua WY, et al. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J. 2010;4:232–241. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- Zhang ML, Zhang MH, Zhang CH, Du HM, Wei GF, Pang XY, et al. Pattern extraction of structural responses of gut microbiota to rotavirus infection via multivariate statistical analysis of clone library data. FEMS Microbiol Ecol. 2009;70:177–185. doi: 10.1111/j.1574-6941.2009.00694.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.