Over the last two decades, many new groups of deeply branching uncultivated archaea have been unveiled by molecular screening of 16S rRNA genes. Among these, Thaumarchaeota (Brochier-Armanet et al., 2008) are now known to represent a highly diversified and ancient phylum present in a wide variety of ecosystems, including marine and fresh waters, soils and also hot environments (Pester et al., 2011). The Thaumarchaeota have rapidly gained much attention after the discovery that some of them are able to oxidize ammonia aerobically, providing the first example of nitrification in the Archaea and therefore extending the range of microorganisms capable of this important metabolism, which was previously thought to be restricted to a few proteobacterial lineages (Konneke et al., 2005). The interest raised by Thaumarchaeota is witnessed by the growing availability over the last 4 years of isolated representatives (or enrichment cultures) and of genomic data. This opens up a whole new perspective on the diversity of Archaea and on ancient evolution.
A diverse and ancient phylum
Isolates or enrichment cultures from groups I.1a (Cenarchaeum symbiosium and Nitrosopumilus maritimus), I.1b (‘Ca. Nitrososphaera viennensis') and ThAOA/HWCG III (‘Ca. Nitrosocaldus yellowstonii') have now been reported. In addition, complete genome sequences are now available from three Thaumarchaeota belonging to the marine group I.1a (C. symbiosium, N. maritimus and ‘Candidatus (Ca.) Nitrosoarchaeum limnia'), and partial genome data has been released from a moderately thermophilic strain from soil, ‘Ca. Nitrososphaera gargensis' (group I.1b) (see Brochier-Armanet et al. (2011) and references therein). Importantly, these sequence data have confirmed the genomic features and phylogenetic distinctiveness of the Thaumarchaeota (Spang et al., 2010). Currently available genomic data are, however, far from covering the whole diversity of this phylum. In fact, many lineages of uncultivated archaea have been reported based on 16S rRNA sequences (Prosser and Nicol, 2008), some of them belonging to the Thaumarchaeota (for example, group I.1c, 1A/pSL12, ThAOA/HWCG III, SAGMCG-I, SCG and FSCG, etc.) (Pester et al., 2011). However, the situation is less clear for a few other uncultivated lineages (for example, HWCG I, Miscellaneous Crenarchaeotic Group, Marine Benthic Group B) whose position is unresolved in rRNA phylogenies. Regarding HWGC I, Nunoura and coworkers have recently reconstituted the genome of one of its members, ‘Ca. Caldiarchaeum subterraneum', using a metagenomic library prepared from a geothermal water stream collected in a subsurface gold mine (Nunoura et al., 2011). Based on the analysis of the genome sequence, they proposed that ‘Ca. C. subterraneum' represents a novel archaeal phylum, which was tentatively called ‘Aigarchaeota'. However, in contrast to rRNA trees, ‘Ca. C. subterraneum' appears robustly grouped with Thaumarchaeota in protein-based phylogenies (Brochier-Armanet et al., 2011, Nunoura et al., 2011). This suggests that HWCG I might represent a basal thaumarchaeal lineage, although more data from additional representatives is surely necessary to clarify the issue.
Further exploration of Thaumarchaeota—including uncultivated subgroups—will be essential to delineate more precisely this phylum from a biological, ecological and evolutionary point of view. For instance, it will be interesting to know if crenarchaeol, a specific lipid identified in the membranes of various Thaumarchaeota and recently proposed to be renamed thaumarchaeol (Pester et al., 2011), is a specific feature of the whole phylum. Similarly, it will be important to explore all thaumarchaeal lineages for the presence of members capable of ammonia oxidation. Indeed, it is possible that not all Thaumarchaeota are ammonia oxidizers, and a key question will be to understand whether this important metabolism is ancestral in the phylum, or rather appeared later during its diversification. For example, the ‘Ca. C. subterraneum' composite genome seems to lack the genes coding for ammonia-oxidation enzymes. This might suggest that ammonia oxidation appeared in Thaumarchaeota after the divergence of HWCG I, but further data from additional HWCG I representatives and from other deeply branching uncultivated thaumarchaeal lineages are required to conclude about this important issue. Moreover, the exploration of the biology and ecological diversity of Thaumarchaeota is expected to continue revealing a high diversity of cellular forms and lifestyles. For example, marine giant Thaumarchaeota forming multicellular filaments possibly associated with bacterial ectosymbionts have been recently reported from sulfide rich environments of a mangrove tropical swamp (Muller et al., 2010). Finally, the availability of genomic data from a wide variety of thaumarchaeal lineages will provide material for the establishment of a detailed classification of Thaumarchaeota, and will help clarifying their phylogenetic relationships with the other major archaeal phyla.
A hot-loving ancestor?
The presence of Thaumarchaeota, not only in mesophilic but also in thermophilic environments (de la Torre et al., 2008), confirms the wide range of phenotypes present in this phylum. It is worth noting that the deepest thaumarchaeal branches are composed of uncultivated groups from various hot environments (de la Torre et al., 2008). This observation, together with the presence of a reverse gyrase in the genome of Ca. C. subterraneum (Nunoura et al. 2011), suggests that the ancestor of Thaumarchaeota and ‘Aigarchaeota' was at least thermophile, and that mesophily in some thaumarchaeal lineages is a derived character. The analysis of environmental sequences from Thaumarchaeota indicates that adaptation to mesophily may have been helped through horizontal gene transfer from bacteria or from mesophilic archaea (Lopez-Garcia et al., 2004). Because secondary adaptations to mesophily are also observed in Euryarchaeota, this would be consistent with the hypothesis of multiple independent adaptations to mesophily in the Archaea from thermophilic or hyperthermophilic ancestors. This scenario is also supported by recent inferences of ancestral archaeal rRNA and protein sequences (Groussin and Gouy, 2011). The growing availability of genomic data from Thaumarchaeota will now allow studying in a more precise way such adaptations from thermophily to mesophily, for example, whether they have followed similar or different evolutionary paths with respect to the Euryarchaeota.
A new perspective on archaeal biology and ancient evolution
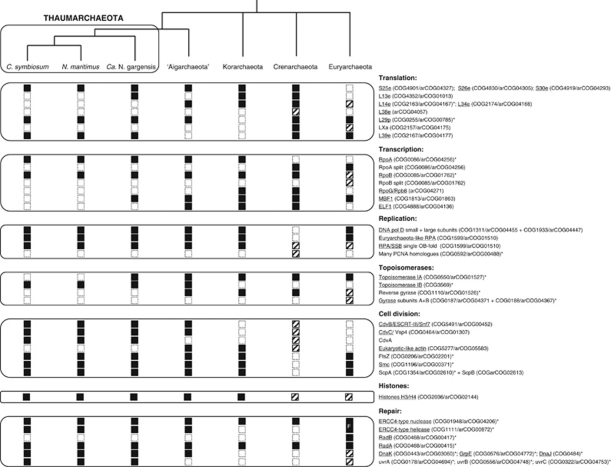
The discovery of Thaumarchaeota opens up new perspectives on the biological diversity and evolution of the Archaea, along with the relationships of this domain of life with Bacteria and Eucarya. An uneven distribution among archaeal lineages of components involved in important cellular systems, sometimes shared either with Bacteria or Eucarya, appears to be a common feature in Archaea (Brochier-Armanet et al., 2011). The availability of genomic data from the Thaumarchaeota has allowed making this phenomenon all the more evident. For instance, the analysis of thaumarchaeal genomes has revealed a unique combination of features shared either with Crenarchaeota, Euryarchaeota or Korarchaeota (Figure 1). A striking example is the presence of components of two very different cell division systems: Cdv, previously thought to be present only in some Crenarchaeota, and FtsZ, so far thought to be specific of Euryarchaeota and Korarchaeota (see Makarova et al. (2010) and references therein). A major question to be addressed is how these two cell division systems coexist and function in Thaumarchaeota. Interestingly, the major partner of FtsZ in Bacteria, MinD, is present in Euryarchaeota (Gerard et al., 1998) but not in Thaumarchaeota (unpublished observations), suggesting a different role for FtsZ in these two archaeal phyla. The forthcoming availability of genomic sequence data from basal uncultured groups will likely confirm the cellular systems in the Archaea. As an example, a eukaryotic-like ubiquitin modifier system has been found in the genome of ‘Ca. C. subterraneum' that is very different from those recently identified in prokaryotes (Nunoura et al., 2011), and a eukaryotic-like actin has been discovered in Korarchaeota, some Crenarchaeota (Makarova et al., 2010) and more recently in ‘Ca. C. subterraneum' (Brochier-Armanet et al., 2011) (Figure 1). The distribution of these components may result from horizontal gene transfers between Archaea and Eucarya, Archaea and Bacteria or among Archaea, but may also represent ancient traits that were present in the common ancestor of Archaea and were subsequently lost in different archaeal lineages. Investigating further the great plasticity of archaeal cellular systems, their origin and function, will surely provide important information on the evolutionary paths and adaptive strategies followed by Archaea along their diversification.
Figure 1.
Distribution of information-processing and cell division components in Archaea. Data were extracted from Spang et al. (2010) and Brochier-Armanet et al. (2011). Note that this list is susceptible to evolve with the availability of new archaeal genome sequences. The relationships among main archaeal lineages are still unclear apart from the grouping of Thaumarchaeota and ‘Aigarchaeota' in agreement with Brochier-Armanet et al. (2011) and Nunoura et al. (2011). Black and empty squares indicate the presence or absence of the corresponding gene in a given lineage, whereas hashed squares indicate the presence in only a few members. ‘F' indicates that the corresponding genes are fused. The genes that are also present in Eucarya are underlined, whereas those present also in Bacteria are indicated by asterisks.
Besides providing insights into the early evolution of cellular features, the discovery of Thaumarchaeota is also crucial for understanding the nature of ancient microbial communities. For example, bacterial nitrification appears to be a recent feature because it is present only in a few late-emerging subgroups of γ- and β-proteobacteria (Konneke et al., 2005). Therefore, it is tempting to speculate that the very ancient traces of nitrification recently suggested by isotopic records from the late Archaean (Garvin et al., 2009) may have been produced by the activity of ancient ammonia-oxidizing Thaumarchaeota. In order to test this hypothesis, it will be essential to establish whether nitrification is widespread in Thaumarchaeota and whether it is an ancestral ability in this phylum.
Perspectives
Microbes have dominated most of the life history and are a fundamental part of the biosphere. The study of microbial diversity and evolution is therefore an essential issue in Biology, and many unexpected findings are being unveiled from the investigation of poorly known microbial lineages. The example of Thaumarchaeota underlines the fact that the exploration of many uncultivated or unexplored microbial lineages is likely to provide key information on all aspects—biological, evolutionary and ecological—of the microbial world.
Note added in proof
While this article was in press, the group of Rolf Bernander reported experimental data showing that Cdv is the main system implicated in cell division in the thaumarchaeon N. maritimus, whereas FtsZ appears not to be and is suggested to have developed a new function in Thaumarchaeota (Pelve EA, Linds AC, Martens-Habbena W, de la Torre JR, Stahl DA, Bernander R. (2011). Cdv-based cell division and cell cycle organization in the thaumarchaeon Nitrosopumilus maritimus. Mol Microbiol 82: 555–566).
Acknowledgments
We wish to thank three anonymous referees for their useful comments, and apologize to colleagues whose work could not be directly cited due to constraints on the number of allowed references.
References
- Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- Brochier-Armanet C, Forterre P, Gribaldo S. Phylogeny and evolution of the Archaea: one hundred genomes later. Curr Opin Microbiol. 2011;14:274–281. doi: 10.1016/j.mib.2011.04.015. [DOI] [PubMed] [Google Scholar]
- de la Torre JR, Walker CB, Ingalls AE, Konneke M, Stahl DA. Cultivation of a thermophilic ammonia oxidizing archaeon synthesizing Crenarchaeol. Environ Microbiol. 2008;10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- Garvin J, Buick R, Anbar AD, Arnold GL, Kaufman AJ. Isotopic evidence for an aerobic nitrogen cycle in the latest Archean. Science. 2009;323:1045–1048. doi: 10.1126/science.1165675. [DOI] [PubMed] [Google Scholar]
- Gerard E, Labedan B, Forterre P. Isolation of a minD-like gene in the hyperthermophilic archaeon Pyrococcus AL585, and phylogenetic characterization of related proteins in the three domains of life. Gene. 1998;222:99–106. doi: 10.1016/s0378-1119(98)00471-5. [DOI] [PubMed] [Google Scholar]
- Groussin M, Gouy M. Adaptation to environmental temperature is a major determinant of molecular evolutionary rates in archaea. Mol Biol Evol. 2011;28:2661–2674. doi: 10.1093/molbev/msr098. [DOI] [PubMed] [Google Scholar]
- Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P, Brochier C, Moreira D, Rodriguez-Valera F. Comparative analysis of a genome fragment of an uncultivated mesopelagic Crenarchaeote reveals multiple horizontal gene transfers. Environ Microbiol. 2004;6:19–34. doi: 10.1046/j.1462-2920.2003.00533.x. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Yutin N, Bell SD, Koonin EV. Evolution of diverse cell division and vesicle formation systems in Archaea. Nat Rev Microbiol. 2010;8:731–741. doi: 10.1038/nrmicro2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F, Brissac T, Le Bris N, Felbeck H, Gros O. First description of giant Archaea (Thaumarchaeota) associated with putative bacterial ectosymbionts in a sulfidic marine habitat. Environ Microbiol. 2010;12:2371–2383. doi: 10.1111/j.1462-2920.2010.02309.x. [DOI] [PubMed] [Google Scholar]
- Nunoura T, Takaki Y, Kakuta J, Nishi S, Sugahara J, Kazama H, et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011;39:3204–3223. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pester M, Schleper C, Wagner M. The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol. 2011;14:300–306. doi: 10.1016/j.mib.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser JI, Nicol GW. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol. 2008;10:2931–2941. doi: 10.1111/j.1462-2920.2008.01775.x. [DOI] [PubMed] [Google Scholar]
- Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, Spieck E, et al. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]