Abstract
An analysis of the microbial metabolism is fundamental to understanding globally important element transformations. One culture-independent approach to deduce those prokaryotic metabolic functions is to analyze metatranscriptomes. Unfortunately, since mRNA is extremely labile, it is unclear whether the abundance patterns detected in nature are vulnerable to considerable modification in situ simply due to sampling procedures. Exemplified on comparisons of metatranscriptomes retrieved from pelagic suboxic zones of the central Baltic Sea (70–120 m depth), earlier identified as areas of high aerobic ammonium oxidation activity, and quantification of specific transcripts in them, we show that different sampling techniques significantly influence the relative abundance of transcripts presumably diagnostic of the habitat. In situ fixation using our newly developed automatic flow injection sampler resulted in an abundance of thaumarchaeal ammonia monooxygenase transcripts that was up to 30-fold higher than that detected in samples obtained using standard oceanographic sampling systems. By contrast, the abundance of transcripts indicative of cellular stress was significantly greater in non-fixed samples. Thus, the importance of in situ fixation in the reliable evaluation of distinct microbial activities in the ecosystem based on metatranscriptomics is obvious. In consequence, our data indicate that the significance of thaumarchaeota to aerobic ammonium oxidation could yet have been considerably underestimated. Taken these results, this could in general also be the case in attempts aimed at an unbiased gene expression analysis of areas below the epipelagic zone, which cover 90% of the world's oceans.
Keywords: amoA, recA, suboxic zones, metatranscriptomics, in situ fixation, central Baltic Sea
Introduction
Microorganisms are the driving catalysts of virtually all biogeochemical cycles on this planet. Therefore, knowledge regarding microbial abundances, diversities and activities in the environment is of fundamental interest not only to microbiologists, but also for understanding globally important element cycles.
A challenge for investigating microbes and especially their activities is that most of all environmental prokaryotes have yet not been cultured. Fifteen years ago an amount of 99% of ‘non-cultivable' microorganisms was estimated (Amann et al., 1995; Pace, 1997); nowadays, it could be assumed that a fraction of 90% still resists cultivation. Consequently, the study of microbial assemblages largely depends on cultivation-independent methods. Concerning the linkage of microbial activity and identity, one of the most promising approaches is the investigation of principally all functional mRNA transcripts isolated from an environmental sample (metatranscriptomics). Transcripts are a useful tool because (a) they guide protein synthesis and provide insights into the metabolic pathways that are important to microbial assemblages at the time of sampling and (b) they offer a perspective on microbial gene expression in situ that is less biased than that of other approaches, which are often incubation dependent. Technically, the recent development of ultrahigh-throughput cDNA sequencing technology (pyrosequencing; Margulies et al., 2005) in combination with new and advanced bioinformatics tools has further advanced the field of metatranscriptomics by enabling the generation and management of huge transcriptional data sets (DeLong, 2009). This has already been demonstrated for environments ranging from sandy lawn soil (Urich et al., 2008) to North Pacific subtropical gyres (Frias-Lopez et al., 2008; Poretsky et al., 2009), coastal waters (Gifford et al., 2010; Hollibaugh et al., 2010) and mesocosms in a Norwegian fjord (Gilbert et al., 2008). Thus, it is very likely that for ecosystems as diverse as the world's oceans, understanding and predicting in situ gene expression patterns will overwhelmingly become the domain of metatranscriptomics (Moran, 2009).
Unfortunately, the half-life of some transcripts can be very short. For the marine cyanobacterium Prochlorococcus, a median half-life of 2.4 min and a median decay rate of 2.6 min for expressed genes were determined (Steglich et al., 2010), but half-lives of less than a minute have also been reported (Belasco, 1993; Steglich et al., 2010). These observations imply that the ability of prokaryotes to very rapidly adapt their expression patterns may cause strong alterations in mRNA profiles during longer sampling procedures or when environmental conditions are changed as a result of sample processing. Thus, the question arises whether gene expression analyses of more challenging oceanic habitats, such as marine oxygen minimum zones, the deep sea or areas below the epipelagic zone, describe the true in situ expression patterns of the community or are instead severely biased by the sampling procedure.
We hypothesized that different water-sampling procedures influenced the relative abundance of transcripts in microbial transcriptomes, potentially resulting in biased conclusions regarding the relevance of different microbially driven biogeochemical cycles in the studied habitat. To test this hypothesis, we sampled hypoxic basins of the central Baltic Sea, specifically the suboxic zone (with <10 μmol l−1 of oxygen; Coolen et al., 2007) at depths between 70 and 120 m (Figure 1). This zone has been reasonably well studied and is known to share several microbial characteristics with other suboxic zones worldwide (Labrenz et al., 2007). One of the important features is the high cellular abundance (25%) of the thaumarchaeal cluster GD2 and the occurrence of high aerobic ammonium oxidation rates accompanied by high concentrations of thaumarchaeal amoA transcripts in a distinct area of low ammonium and oxygen concentrations within this environment (Labrenz et al., 2010). Frias-Lopez et al. (2008) noted already that the expression patterns of environmentally diagnostic genes can provide significant insight into microbial processes active in the environment. Following this hypothesis, we quantified thaumarchaeal amoA, amoB, amoC and NH4+ transporter expression, as indicators for habitat-specific ammonium modification activity, and bacterial recA expression, given the recA protein's essential role in DNA repair and maintenance and thus as an indicator of cellular stress (Little et al., 1980; Anderson et al., 2006) (experimental pipeline, see Supplementary Figure 1).
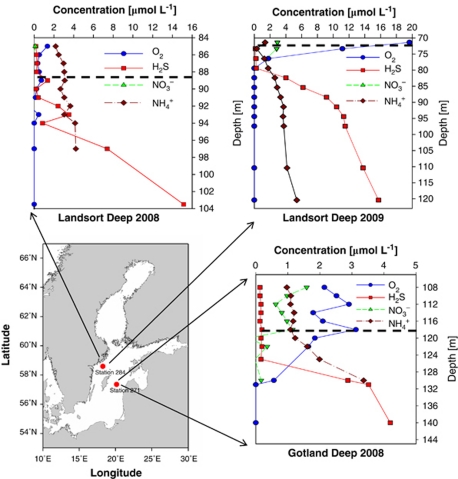
Figure 1.
Map of the central Baltic Sea including chemical profiles of the investigated pelagic redoxclines. Red dots indicate the positions of sampling stations 284 (Landsort Deep, 58°35.00′N, 18°14.06′E) and 271 (Gotland Deep, 57°19.89′N, 20°10.28′E). Embedded graphs present chemical data recorded during sampling in August 2008 and September 2009. The dashed line marks the sampling depths: 118 m (Gotland Deep 2008), 89 m (Landsort Deep 2008) and 72 m (Landsort Deep 2009). Data source for the map: Seifert T, Tauber F, Kayser B (2001). A High Resolution Spherical Grid Topography of the Baltic Sea (Revised Edition). Baltic Sea Science Conference, 25–29 November 2001; Stockholm. Poster #147 (http://www.io-warnemuende.de/iowtopo).
Traditional sampling from a ship using conventional water-sampling bottles usually takes 20–30 min (for depths down to 100–200 m), ultimately exposing the samples to conditions far from those in situ while the water is filtered in the ship's laboratory to collect the nucleic acids. We compared this standard sampling procedure with two alternative treatments of samples taken at the same time and place at 70–120 m depth (Figure 1). (a) The sampling retrieval time was reduced to ∼10 min by using a PUMP-CTD system (Strady et al., 2008). (b) The mRNA molecules present in the samples were stabilized and RNase activity inhibited by fixing and preserving the samples in situ within several seconds. For this purpose, we developed an automatic flow injection sampler (AFIS; Figure 2). In the laboratory, mRNA was (a) pyrosequenced, followed by metatranscriptomics or (b) subject to reverse transcriptase quantitative PCR (RT-qPCR) analyses.
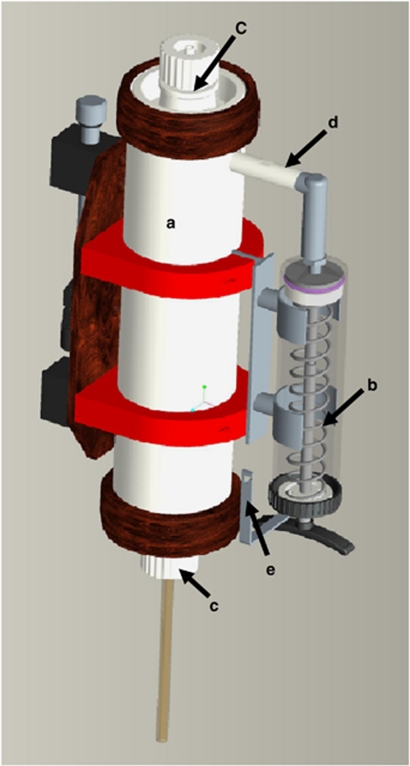
Figure 2.
Design of the AFIS. (a) Teflon water bottle (2.7 l volume); (b) syringe (hand syringe GENIUS 450 ml) with maximal volume of 300 ml; (c) bottle lids connected with closing devices on the CTD-bottle rosette; (d) adapter between syringe and bottle, coupled with a dispenser tube; (e) escapement for the syringe plunger, directly connected with the closing devices for the bottle lids. The operating mode is based on the following principles: (1) the syringe plunger is pushed by a spring; (2) the release of the spring is coupled with the closing devices for the bottle lids; (3) the solution passes through the dispenser tube into the bottle; (4) the dispenser tube extends from the top to the bottom of the bottle; (5) the solution is equally distributed through perforations of increasing diameters made in the dispenser tube.
In this study, analyses of samples prepared by the newly developed or more traditional methods revealed that autecologically important transcripts as thaumarchaeal amoA can virtually be lost without the application of in situ fixation procedures.
Materials and methods
The entire experimental pipeline of this study is provided in Supplementary Figure 1.
Sampling
Water samples were collected from suboxic zones of the central Baltic Sea stations 271 (Gotland Deep, 57°19.89′N, 20°10.28′E) and 284 (Landsort Deep, 58°35.00′N, 18°14.06′E) on board the RV Poseidon during cruise P370 in August 2008, and on board the RV Maria S Merian during cruise MSM12/4b in September 2009, using free-flow bottles (Hydrobios, Kiel, Germany) and the AFIS attached to a conductivity-temperature-depth (CTD) rosette, respectively. In addition, samples were also obtained using the IOW PUMP-CTD system (technical assembly described by Strady et al., 2008). This sampling system has been used repeatedly to access deeper waters for gene expression analyses from, for example, the Black Sea (Lam et al., 2007). For RNA extraction, in 2008, water was collected from depths of 89 m (station 284) and 118 m (station 271) (Figure 1). In 2009, water was sampled from a depth of 72 m (station 284). In all cases, 1000 ml of sample water was filtered through a 0.22-μm filter (47 mm diameter, polyvinylidene fluoride or polytetrafluorethylene, Millipore, Billerica, MA, USA), which took 15–20 min, and the filters were shock frozen. Further on, these filters were stored without interruption at −80 °C in the laboratory until used for total RNA extraction.
Physicochemical structure of the water column
Data regarding the physical structure of the water column were recorded by a CTD unit (Seabird 911 plus) equipped with a SBE oxygen sensor (Seabird Electronics, Bellevue, WA, USA). Chemical profiles of oxygen, hydrogen sulfide, ammonia and nitrate were determined from the water samples according to standard methods (Grasshoff et al., 1983).
In situ fixation
Our self-developed AFIS enabled the direct fixation of water samples, that is, at the time of sampling and at the original habitat. The system consists of a 2.7-l Teflon water bottle and a 300-ml syringe for the fixation solution. The construction and operating mode are described in detail in Figure 2. Two different fixation solutions were tested: (a) stop solution (5% water-saturated phenol in absolute ethanol), which is often used for preparing microarray samples (Khodursky et al., 2003) and (b) Carnoy fixative (30% chloroform and 10% acetic acid in absolute ethanol), which is predominantly used for preparing microscopy tissue slices (Wartman, 1960). In situ, ∼2 s after the AFIS bottles were closed at the depth of interest, 300 ml of fixative was injected into the closed bottle (taking ∼5 s) and equally distributed, resulting in an ∼1:10 dilution of fixation solution in the water sample. Samples fixed in this manner were then filtered as described above.
Total cell numbers
Total cell numbers were determined by 4′,6-diamidino-2-phenylindole (DAPI) staining as described previously (Masín et al., 2006).
Cell activity measurements
To determine the fixation capacities of the stop solution and Carnoy fixative, 10 n 3H-methyl-thymidine (3H-TdR) (60.1 Ci mmol−1, Moravek Biochemicals) and 50 n 14C-leucine (261 mCi mmol−1, Moravek Biochemicals, Brea, CA, USA) were added to triplicate 5 ml samples of brackish surface water of the Warnow River estuary (July 2009), with dual-label incorporation determined after incubation at the in situ temperature. Incubations were stopped after 0 min (directly after addition of stop solution and Carnoy fixative, respectively), 5, 10, 20, 40 and 60 min by additionally fixing the cells with formaldehyde (10% v/w) for at least 1 h in the dark at 5 °C. Following fixation, samples were filtered on 0.22 μm polycarbonate filters (Millipore) and extracted with cold trichloroacetic acid according to Chin-Leo and Kirchman (1988). Four microliters of scintillation cocktail was added to the dry filters and the incorporated substrates then counted in a scintillation counter Tri-Crab 2800TR (Perkin Elmer, Shelton, CT, USA).
The in situ fixation capacity using AFIS was analyzed for August 2008 samples by incorporation of 20 nmol l−1 3H-TdR (25 Ci mmol−1, Moravek Biochemicals) and 3H-acetate (20 Ci mmol−1, Moravek Biochemicals). Radioactively labeled substrates were added to non-fixed and AFIS samples on board ∼25 min after the bottles were closed in the suboxic zone. Incubation time was at least 1.5 h for 3H-TdR samples and 2 h for 3H-acetate samples at room temperature, with the reactions finally stopped by formaldehyde fixation (10% v/w) in the dark at 5 °C for 3 h (3H-TdR) or 8 h (3H-acetate).
RNA preparation
Unless stated otherwise, all kits employed in this study were used according to the manufacturer's instructions. RNA was extracted from filters using the RNeasy kit (Qiagen, Hilden, Germany) and the RNA extract treated with DNase I (DNA-Free kit, Ambion, Austin, TX, USA) to remove any contaminating DNA. The mRNA fraction was enriched by the enzymatic digestion of rRNA molecules (mRNA-ONLY Prokaryotic mRNA Isolation kit, Epicentre Biotechnologies, Madison, WI, USA) followed by subtractive hybridization of rRNA with capture oligonucleotides (MICROBExpress kit, Ambion). In order to obtain sufficient transcripts to analyze rarely expressed specific genes, the mRNA isolates were amplified using the MessageAmp II-Bacteria kit (Ambion). Briefly, the remaining RNA molecules were polyadenylated and reverse transcribed into cDNA. Multiple copies of antisense RNA (aRNA) were then obtained by linear in vitro transcription. The RNA quality and quantity were assessed spectrophotometrically using the 2100 Electrophoresis Bioanalyzer (Agilent Technologies, Böblingen, Germany).
Random hexamer primers were then used to convert aRNA into double-stranded cDNA (Universal RiboClone cDNA Synthesis System, Promega, Madison, WI, USA). The entire procedure, ranging from total RNA extraction to cDNA generation, is described in detail elsewhere (Poretsky et al., 2009).
Metatranscriptomic analyses
Five metatranscriptomic libraries representing each of the different sampling procedures (AFIS: stop solution, AFIS: Carnoy fixative, free-flow bottle reference for AFIS; PUMP-CTD, free-flow bottle reference for PUMP-CTD) were generated from the Gotland Deep (2008) sample of 118 m depth (Figure 1). The metatranscriptomic pipeline is shown in Supplementary Figure 2. Double-stranded DNA with flush ends was pyrosequenced by LGC Genomics GmbH (Berlin, Germany) using the Roche/454 GS FLX titanium technology. A BLASTN search against the RDP-II, SILVA SSURef+LSURef database (Urich et al., 2008) identified hits with an e-value ⩽0.001 and a bit score of ⩾40 as rRNA reads and sequences, but no hits as possible protein-encoding mRNA reads. These potential mRNA reads were phylogenetically assigned using the PhymmBL annotation program (Brady and Salzberg, 2009); cutoffs were as described in the section ‘Parameters settings for weighted voting in PhymmBL'. Functional analysis was done using the rapid annotation subsystem technology (MG-RAST; Meyer et al., 2008) and investigated with cutoffs of e-value ⩽0.01, P-value ⩾30, percent identity ⩾40 and alignment length ⩾50.
As mentioned above, earlier studies showed that ∼25% of the microbial assemblages in aerobic ammonium oxidation zones of the central Baltic Sea were closely related to the Thaumarchaeum Candidatus Nitrosopumilus maritimus, and potentially catalyzing ammonium oxidation exclusively (Labrenz et al., 2010). Therefore, thaumarchaeal transcripts involved in ammonium modification were analyzed specifically. Abundances of amoB, amoC and NH4+ transporter transcripts were identified based on BLAST similarities (e-value ⩽0.01) against the genome of Candidatus N. maritimus, analyzed using MG-RAST (Meyer et al., 2008) as described above (and in Supplementary Figure 2), and calculated as the fraction of specific transcripts compared with total mRNA transcripts. Interestingly, concerning ammonium transporter transcripts only one (NCBI locus tag Nmar_1698, protein #ABX13594) of the two genes described for Candidatus N. maritimus (Walker et al., 2010) could exclusively be detected in the metatranscriptomes.
The transcript abundances of amoA and recA were determined by BLASTN against self-generated consensus sequence databases (Supplementary Table 1) because specific data sets existed for central Baltic Sea redoxclines already (Grote, 2009; Labrenz et al., 2010) enabling more distinct identifications of accordant transcripts. First potential mRNA reads were BLASTX queried (e-value ⩽0.01) against NCBI curated, non-redundant RefSeq (reference sequence database) to determine putative functions. A BLASTX search (cutoffs: sequence length >23 amino acids, percent identity >40) identified amoA/recA transcripts within the annotated sequences by a selection of restricted criteria within the sequence names (for example, ‘ammonia monooxygenase' or ‘recombinase A'). The alignment of all identified amoA and recA transcript sequences within each of the five metatranscriptomic libraries provided consensus sequences. A BLASTN search (cutoffs: e-value ⩽0.01, bit score ⩾30) of the five metatranscriptomic libraries against the amoA/recA consensus sequences database was even more specific than a BLASTX search based on gene notations, identifying, besides RefSeq annotated reads, possible fragments of sequences as specific transcripts. The thaumarchaeal amoA consensus database contained 16 and the bacterial recA consensus database contained 31 different sequences. Quantifications were based on the number of sequences within the mRNA reads of the five different samples exhibiting hits (cutoffs: e-value ⩽0.01, bit score ⩾30) against the databases and then presented as the ratio between reference and treatments.
Quantitative PCR analysis
Transcripts were quantified using the iCycler iQ RT PCR detection system (Bio-Rad, Hercules, CA, USA) in a 15-μl reaction volume. Thaumarchaeal amoA RT-qPCR was done as described earlier using the AOA-amoA-r/AOA-amoA-f primer system (Coolen et al., 2007). For the quantification of bacterial 16S rRNA, bacterial 519f/907r primer system amplification was done with a 35-cycle PCR (30 s 95 °C, 30 s 52 °C, 30 s 68 °C), and with a 40-cycle PCR (60 s 94 °C, 60 s 53 °C, 60 s 72 °C) for the GD1-recA-r822/GD1-recA-f614 primer system (Supplementary Table 2), followed by a terminal elongation step of 5 min. GD1 primers were established on 16S rRNA analyses of key organism ‘Sulfurimonas sp. GD 1', which was isolated from a central Baltic Sea redoxcline (Grote, 2009), and represents the in situ highly abundant ‘Sulfurimonas GD17' subgroup (Grote et al., 2007). A PCR control without an initial RT step was included with every set of reactions.
To analyze relative gene expression differences, reference vs treatment ratios of each gene transcript were determined by RT-qPCR amplification of a serial dilution of aRNA (in duplicate) and calculation of the differences in cycle threshold values (Ct) with respect to PCR efficiency (E). By comparing these ratios, that is, for the reference (R) vs the alternative treatment (T) approach, the relative gene expression ratio based on the expression factor (EF) was calculated using the formula EF=(1+ER)CtR/(1+ET)CtT (modified 2−ΔΔCt method by Frias-Lopez et al., 2008). The requirements to perform this calculation are equal initial amounts of RNA and a determination of the PCR efficiency and the threshold cycle at identical fluorescence signals.
Data deposition
The sequences reported in this paper have been submitted to GenBank, under accession number SRA026103.1.
Results and discussion
During cruises to the Baltic Proper in 2008 and 2009, suboxic zones of the Landsort Deep and the Gotland Deep were sampled. These deeps are characterized by pronounced redox gradients at depths ranging from 70 to 130 m, with decreases in oxygen and increases in both ammonium and sulfide with increasing depth and a peak in nitrate in the suboxic zone (Figure 1). Those water layers, exhibiting oxygen and ammonium concentrations between 0.3 and 11 μmol l−1 (Figure 1), were chosen for sampling because similar water masses have already been described as characteristic for pronounced aerobic ammonium oxidation activities of the Black Sea (Lam et al., 2007) as well as central Baltic Sea (Labrenz et al., 2010) and were most likely to contain habitat-diagnostic thaumarchaeal amoA transcripts in amounts quantifiable by molecular methods.
Evaluation of the AFIS approach in vitro and in situ
Before the AFIS could be applied, the fixation capacities of the stop solution as well as Carnoy fixative had to be confirmed. First, the abilities of the fixatives to stop microbial activity were tested by separately adding them to samples obtained from brackish surface waters of the Warnow River estuary and supplemented with radiolabeled thymidine or leucine tracer (Chin-Leo and Kirchman, 1988) as markers for prokaryotic activity. Both fixatives stopped tracer incorporation into cells immediately (Supplementary Figures 3a and b). Similarly, activity measurements for suboxic waters of the central Baltic Sea showed that cells incorporated radiolabeled thymidine tracer as well as acetate, but after in situ AFIS fixation incorporation was in the range of the normal blank values (Supplementary Figure 3c). Second, the influence of the fixatives on molecular analyses was examined in recA transcripts of our representative environmental Epsilonproteobacterium ‘Sulfurimonas sp. GD 1'. Only insignificantly different transcript abundances were obtained between unfixed and fixed cultures, as determined by specific RT-qPCR (Supplementary Figure 3d). Third, the influence on total cell numbers or 16S rRNA abundances was tested, revealing that the different sampling procedures did not cause significant differences between the treatments (Mann–Whitney test; Supplementary Figure 4). This result was expected because these parameters are considered to be more stable and do not change as quickly in time as, for example, mRNA levels (Deutscher, 2006). The median total cell numbers of 8.8 × 105 cells ml−1 (±1.8 × 105 cells ml−1) for all the different treatments were also consistent with the values determined in other studies, even those going 20 years back, of the central Baltic Sea (Brettar and Rheinheimer, 1991), indicating an overall comparability of the environmental samples investigated.
Taken together, the above-described tests revealed that the fixatives chosen stopped microbial activity but did not influence either the abundance of a specific transcript or more general ribosomal or cellular parameters. Thus, by stabilizing in situ gene expression profiles, the AFIS approach was, to the best of our knowledge, valid for further metatranscriptomic analyses.
Metatranscriptomic analyses
Five metatranscriptomic libraries were generated from suboxic waters of the Gotland Deep station 271, sampled in 15 August 2008 (Table 1). The CTD-based free-flow approach was used in the collection of samples AFIS fixed with stop solution or Carnoy fixative as well as an unfixed reference. Sampling by the PUMP-CTD and its attached free-flow bottle as reference was done 4.5 h later than the free-flow approach from the same body of water (ship regulations forbid parallel exposures of two sampling devices). Total mRNA reads for all metatranscriptomes generated ranged from 5400 to 8500 for the free-flow approach and 17 400 to 37 000 for the PUMP-CTD approach (Table 1). The average mRNA-read length was 304 bp. In order to confirm the adequacy of the expression patterns for the suboxic zone with respect to its phylogenetic composition, the entire metatranscriptomic data sets (only mRNA) were phylogenetically analyzed using the PhymmBL annotation program (Brady and Salzberg, 2009). Phylogenetically, the mRNA was primarily attributed to the phyla Proteobacteria, Thaumarchaeota and Firmicutes (Supplementary Figure 5a), groups previously shown to be important in Baltic Sea redoxclines (Labrenz et al., 2007, 2010) and comparable habitats of the Black Sea (Vetriani et al., 2003; Lam et al., 2007) and Cariaco Trench (Lin et al., 2006). Corresponding mRNA functional groupings were analyzed using the rapid annotation subsystem technology (MG-RAST; Meyer et al., 2008). The most abundant functional categories were related to protein and RNA metabolism, carbohydrates (as described in Urich et al., 2008 and Gilbert et al., 2008), respiration and nitrogen metabolism (Supplementary Figure 5b). This was not surprising because it is known that genes necessary to maintain the basic cellular machinery enabling growth and metabolism dominate microbial transcript pools (Moran, 2009); consequently, similar functional profiles have been identified earlier already (Gilbert et al., 2008; Urich et al., 2008). Thus, it can be assumed that the metatranscriptomes investigated were in the range of comparable analyses with respect to most abundant and especially to intracellularly important transcripts.
Table 1. Characteristics of the five metatranscriptomes generated from the suboxic zone of Gotland Deep station 271 (2008). Samples for the free-flow and the PUMP-CTD approach were taken 15 August, UTC 11.09 and UTC 15.46, respectively.
| Metatranscriptome parameter |
Free-flow approach |
PUMP-CTD approach |
|||
|---|---|---|---|---|---|
| Unfixed reference | In situ fixed by stop solution | In situ fixed by Carnoy fixative | Reference collected by free-flow | Sample collected by PUMP-CTD | |
| Total no. of readsa | 18 157 | 29 666 | 32 914 | 72 512 | 37 013 |
| rRNA reads (%)b | 70 | 76 | 74 | 49 | 53 |
| Total mRNA readsc | 5447 | 7120 | 8558 | 36981 | 17396 |
| Total size of potential mRNA reads (Mbp)a | 1.6 | 2.2 | 2.5 | 11.7 | 5.40 |
| Average length of potential mRNA reads (bp)a | 279 | 304 | 307 | 317 | 314 |
| NH4+ transporter transcripts (%)d (no. of reads) | 0.83 (45) | 2.32 (165) | 2.15 (184) | 2.89 (1067) | 1.28 (222) |
| amoA transcripts (%)e (no. of reads) | 0.18 (10) | 0.46 (33) | 0.23 (20) | 1.00 (371) | 0.20 (36) |
| amoB transcripts (%)d (no. of reads) | 0.02 (1) | 0.07 (5) | 0.02 (2) | 0.06 (24) | 0.07 (12) |
| amoC transcripts (%)d (no. of reads) | 0.04 (2) | 0.34 (24) | 0.13 (11) | 0.44 (161) | 0.33 (57) |
| recA transcripts (%)f (no. of reads) | 0.02 (1) | 0.01 (1) | 0.07 (6) | 0.30 (126) | 1.96 (341) |
Based on GS-FLX titanium pyrosequencing.
Identification of sequences using the Ribosomal Database Project II (RDP-II) and the SILVA project small subunit (SSU) data set.
Sequences with no hits against the RDP-II and the SILVA project SSU and large subunit (LSU) data set.
Based on BLAST similarities (e-value ⩽0.01) against the genome of Candidatus Nitrosopumilus maritimus, analyzed using MG-RAST (Meyer et al., 2008) as described in Supplementary Figure 2, and calculated as the fraction of specific transcripts compared with total mRNA transcripts.
Based on hits against a self-generated thaumarchaeal amoA-consensus sequences database and calculated as the fraction of specific transcripts compared with total mRNA transcripts.
Based on hits against self-generated ‘Sulfurimonas sp. GD1'-based recA-consensus sequences database and calculated as the fraction of specific transcripts compared with total mRNA transcripts.
The specific abundances of thaumarchaeal NH4+ transporter, amoA, amoB, amoC and bacterial recA transcripts in the metatranscriptomes showed differences in the relative abundance of these five transcripts found among treatments. The relative number of thaumarchaeal transcripts was 1.2- to 9.3-fold higher in the AFIS-fixed samples than in the standard treatments, while in the PUMP-CTD sample it was up to 80% lower (Table 1; Figure 3a). Copy numbers of recA transcripts were at the limit of detection in the AFIS-fixed samples or their references but nearly sevenfold higher in the PUMP-CTD sample than in its reference (Table 1).
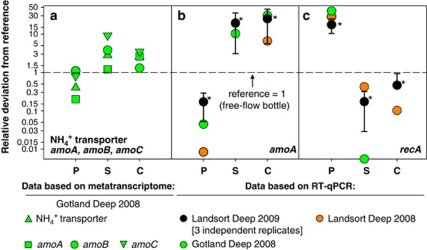
Figure 3.
Comparison of transcript abundances in PUMP-CTD and AFIS samples in relation to their references. Colors represent the investigated Deep: black, Landsort Deep (2009); orange, Landsort Deep (2008); green, Gotland Deep (2008). Deviations in gene expression were calculated by comparing PUMP-CTD or data of fixed samples with the accordant non-fixed free-flow bottle references. A factor of 1 denotes that the abundances were equal to those of the reference (marked as a dashed line). Investigated parameters were (a) abundances based on metatranscriptomics: thaumarchaeal NH4+ transporter, amoA, amoB and amoC transcripts. RT-qPCR-based abundances: (b) thaumarchaeal amoA transcripts; (c) ‘Sulfurimonas sp. GD1' recA transcripts. P, PUMP-CTD; S, stop solution; C, Carnoy fixative. *Based on mean±s.d. of three independent replicates, the non-parametric Mann–Whitney test showed a significant difference to the reference (P<0.01).
Thus, the analysis of these five diagnostic transcripts provided clear indications that their gene expression patterns differed between the different treatments. However, only recently Gifford et al. (2010) demonstrated that significant gains in comparative analyses of biogeochemically informative gene expression patterns potentially require greater sequencing investment than 105–106 sequence reads per metatranscriptome. Thus, with sequencing reads in the range from 103 to 104 (Table 1) our above-described metatranscriptomic results for diagnostic transcripts had to be confirmed by alternative methods, as described in the following.
Thaumarchaeal amoA and ‘Sulfurimonas sp. GD1'-specific recA abundances
In this study, specific RT-qPCR was used to again determine thaumarchaeal amoA and ‘Sulfurimonas sp. GD1' recA transcript abundances because RT-qPCR approaches can also provide absolute transcript numbers, and often with enhanced sensitivity (Church et al., 2010; Gifford et al., 2010). RT-qPCR analyses indicated for Gotland Deep and Landsort Deep samples taken in 2008 that the amoA expression could even be 6- to 30-fold higher in the AFIS-fixed samples and 96–99% lower in the PUMP-CTD samples (Figure 3b). Moreover, recA transcript abundance was 28- to 39-fold higher in the PUMP-CTD samples and 60–90% (or even undetectable) lower in the AFIS-fixed samples (Figure 3c).
These data were generated for single samples only. In order to stabilize the results statistically, RT-qPCR was used again to determine thaumarchaeal amoA and ‘Sulfurimonas sp. GD1' recA transcript abundances for a Landsort Deep (2009) sample (Figure 1) in three independent replicates. These statistically robust results confirmed the above-described data set:
RT-qPCR analyses revealed that the abundances of thaumarchaeal amoA transcripts were significantly higher (non-parametric Mann–Whitney test; P<0.001) for the AFIS-fixed samples than in those obtained by any of the other sampling procedures. Compared with the standard sampling procedure using free-flow bottles, mean amoA expression was 20- to 25-fold higher in the fixed samples and 83% lower in the PUMP-CTD samples (Figure 3b). Also statistically significant (non-parametric Mann–Whitney test; P<0.001), mean recA transcript abundance was 17-fold higher in the PUMP-CTD samples than in the free-flow bottle samples and 54–83% lower in the AFIS-fixed samples, indicating that handling of the samples and changing the environmental conditions stressed the cells significantly and led to partly aberrant expression profiles. These results support analyses of Gilbert et al. (2008), who found that the most abundant sequences with annotations for ocean mesocosms of a fjord close to Bergen were stress-induced chaperonin proteins. The authors concluded that these sequences could have been induced by the sampling procedure, in particular during filtration of the bacteria.
It is interesting to note that sampling by the PUMP-CTD method seems to have had the most pronounced effect on the bacterial assemblages. In this method, water is sampled through a tube of 18 mm diameter down to 400 m depth in a relatively short time period. As an example, a 355-m depth sampling resulted in a flow rate of 2.875±0.025 l min−1, corresponding to a transit time of 3:51–3:57 min from the area of sampling to onboard arrival (Strady et al., 2008), which is less than the transit time of water using a CTD rosette. In general, sampling time should be an important factor affecting gene expression patterns (Frias-Lopez et al., 2008), but it seems that a potential mechanical stress or pressure differences within the tube also influence gene expression patterns extraordinarily. Thus, although the PUMP-CTD seems to be well suited for chemical analyses of gradients due to its exceptional vertical resolution (Strady et al., 2008) it may be particularly problematic for gene expression analyses.
Taken together, the above data show that for suboxic Baltic waters characterized by high ammonium oxidation activities properties such as 16S rRNA abundance or total cell numbers are not significantly influenced by differences in sampling procedures. However, in the case of specific transcripts, in this case the geochemically important thaumarchaeal amoA, differences in the handling of samples can lead to large statistically significant differences in transcript abundance. Moreover, the loss of amoA transcripts from non-fixed samples was accompanied by an increase in recA transcript abundance throughout the microbial assemblages, indicating changes in transcriptome profiles due to enhanced stress of the bacterial cells.
Plausibility of higher thaumarchaeal amoA transcript abundances
Although up to 30-fold higher amoA transcript copy numbers were retrieved with the AFIS sampler (Figure 3b), there is currently no way to prove that this accurately represents the in situ gene expression profile. However, previous studies, in which thaumarchaeal amoA transcript abundance was quantified after conventional sampling, indicated that amoA copy numbers are greatly underestimated. For example, maximal—theoretical—amoA transcript numbers of only 0.04 copies per thaumarchaeal cell were determined for a Baltic Sea suboxic zone (Labrenz et al., 2010), for Black Sea suboxic zones even less numbers can be estimated based on data of Lam et al. (2007). In both studies, these samples covered the presumed nitrification zone, where amoA activities were maximal; however, based on these numbers at most 4% of the amoA-containing archaeal assemblages could have expressed this gene, based on a maximum of one amoA transcript copy per cell. In consequence, with a mean of 20-fold higher amoA abundances in the in situ fixed samples, as determined in this study, up to 80% of the amoA-containing archaeal assemblages described in the studies of Lam et al. (2007) and Labrenz et al. (2010) could have expressed this gene. This is comparable to numbers of active cells as determined by 5-cyano-2,3-ditolyl tetrazolium chloride measurements for different bacteria during their exponential and stationary growth, as well as for environmental samples (Créach et al., 2003), and seems to be a more realistic abundance for this microbially catalyzed function, and we assume that this number better represents environmental conditions.
Consequences of in situ fixation for mRNA analyses
Exemplified for suboxic waters of the central Baltic Sea characterized by higher ammonium oxidation activities and accordant thaumarchaeal amoA or specific bacterial recA abundances, our data indicate the importance of in situ fixation of transcript samples for a reliable sampling of microbial metatranscriptomes and, thereby, analyses of microbial functions, interactions, or even the modeling of microbial activities. Since most previous amoA transcript analyses were based on non-fixed samples and determined by RT-qPCR, our data suggest that the significance of thaumarchaeota to aerobic ammonium oxidation has been considerably underestimated. Thus, even though other studies have proposed a significant role for archaeal nitrification in the global nitrogen cycle (Francis et al., 2005; Wuchter et al., 2006; Erguder et al., 2009; Church et al., 2010), it may actually be more prominent. Based on the results presented here, it is probable that many low abundance or short half-life mRNAs, some of which are, no doubt, autecologically very important, will only be detectable using a proper in situ fixation technique. This could also be true for sampling areas below the epipelagic zone, covering 90% of the world's oceans, where sample retrieval times are long.
Acknowledgments
We thank the captains and the crews of the RV Poseidon and RV Maria S Merian. We are very grateful to Hanno Teeling (MPI Bremen) for providing fruitful discussion and Thomas Schott for support on metatranscriptomic analyses. This study was funded by the WGL-pact project ‘REAL' (Leibniz Association) to KJ.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KL, Roberts C, Disz T, Vonstein V, Hwang K, Overbeek R, et al. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J Bacteriol. 2006;188:6739–6756. doi: 10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. Control of Messenger RNA Stability. Academic Press: San Diego; 1993. pp. 3–11. [Google Scholar]
- Brady A, Salzberg SL. Phymm and PhymmBL: metagenomic phylogenetic classification with interpolated Markov models. Nat Methods. 2009;6:673–676. doi: 10.1038/nmeth.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brettar I, Rheinheimer G. Denitrification in the central Baltic: evidence for H2S-oxidation as motor of denitrification at the oxic-anoxic interface. Mar Ecol Prog Ser. 1991;77:157–169. [Google Scholar]
- Chin-Leo G, Kirchman DL. Estimating bacterial production in marine waters from the simultaneous incorporation of thymidine and leucine. Appl Environ Microbiol. 1988;54:1934–1939. doi: 10.1128/aem.54.8.1934-1939.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church MJ, Wai B, Karl DM, DeLong EF. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ Microbiol. 2010;12:679–688. doi: 10.1111/j.1462-2920.2009.02108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coolen MJL, Abbas B, van Bleijswijk J, Hopmans EC, Kuypers MMM, Wakeham SG, Sinninghe Damste JS. Putative ammonia-oxidizing Crenarchaeota in suboxic waters of the Black Sea: a basin-wide ecological study using 16S ribosomal and functional genes and membrane lipids. Environ Microbiol. 2007;9:1001–1016. doi: 10.1111/j.1462-2920.2006.01227.x. [DOI] [PubMed] [Google Scholar]
- Créach V, Baudoux A-C, Bertru G, Rouzic BL. Direct estimate of active bacteria: CTC use and limitations. J Microbiol Methods. 2003;52:19–28. doi: 10.1016/s0167-7012(02)00128-8. [DOI] [PubMed] [Google Scholar]
- DeLong EF. The microbial ocean from genomes to biomes. Nature. 2009;459:200–206. doi: 10.1038/nature08059. [DOI] [PubMed] [Google Scholar]
- Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erguder TH, Boon N, Wittebolle L, Marzorati M, Verstraete W. Environmental factors shaping the ecological niches of ammonia-oxidizing archaea. FEMS Microbiol Rev. 2009;33:855–869. doi: 10.1111/j.1574-6976.2009.00179.x. [DOI] [PubMed] [Google Scholar]
- Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford SM, Sharma S, Rinta-Kanto JM, Moran MA. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J. 2010;5:461–472. doi: 10.1038/ismej.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Field D, Huang Y, Edwards R, Li W, Gilna P, et al. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS ONE. 2008;3:e3042. doi: 10.1371/journal.pone.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasshoff K, Erhardt M, Kremling K.1983Methods of Seawater Analysis, vol. 2.Verlag Chemie Weinheim: New York [Google Scholar]
- Grote J.2009Physiology, ecology, and genomics of facultative chemoautotrophic Epsilonproteobacteria in marine pelagic redoxclines. PhD thesis,University of Rostock, Germany [Google Scholar]
- Grote J, Labrenz M, Pfeiffer B, Jost G, Jürgens K. Quantitative distribution of Epsilonproteobacteria and a specific Sulfurimonas subgroup in pelagic redoxclines of the central Baltic Sea. Appl Environ Microbiol. 2007;73:7155–7161. doi: 10.1128/AEM.00466-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollibaugh JT, Gifford SM, Sharma S, Bano N, Moran MA. Metatranscriptomic analysis of ammonia-oxidizing organisms in an estuarine bacterioplankton assemblage. ISME J. 2010;5:866–878. doi: 10.1038/ismej.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodursky AB, Bernstein JA, Peter BJ, Rhodius V, Wendisch VF, Zimmer DP.2003Escherichia coli spotted double-strand DNA microarraysBrownstein MJ, Khodursky AB (eds). Functional Genomics Humana Press Inc.: Totowa; 61–78. [DOI] [PubMed] [Google Scholar]
- Labrenz M, Jost G, Jürgens K. Distribution of abundant prokaryotic organisms in the water column of the central Baltic Sea with an oxic-anoxic interface. Aquat Microb Ecol. 2007;46:177–190. [Google Scholar]
- Labrenz M, Sintes E, Toetzke F, Zumsteg A, Herndl GJ, Seidler M, et al. Relevance of a crenarchaeotal subcluster related to Candidatus Nitrosopumilus maritimus to ammonia oxidation in the suboxic zone of the central Baltic Sea. ISME J. 2010;4:1496–1508. doi: 10.1038/ismej.2010.78. [DOI] [PubMed] [Google Scholar]
- Lam P, Jensen MM, Lavik G, McGinnis DF, Müller B, Schubert CJ, et al. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc Natl Acad Sci USA. 2007;104:7104–7109. doi: 10.1073/pnas.0611081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Wakeham SG, Putnam IF, Astor YM, Scranton MI, Chistoserdov AY, et al. Comparison of vertical distributions of prokaryotic assemblages in the anoxic Cariaco Basin and Black Sea by use of fluorescence in situ hybridization. Appl Environ Microbiol. 2006;72:2679–2690. doi: 10.1128/AEM.72.4.2679-2690.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little JW, Edmiston SH, Pacelli LZ, Mount DW. Cleavage of the Escherichia colilexA protein by the recA protease. Proc Natl Acad Sci USA. 1980;77:3225–3229. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masín M, Zdun A, Ston-Egiert J, Nausch M, Labrenz M, Moulisová V, et al. Seasonal changes and diversity of aerobic anoxygenic phototrophs in the Baltic Sea. Aquat Microb Ecol. 2006;45:247–254. [Google Scholar]
- Meyer F, Paarmann D, D′Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran MA. Metatranscriptomics: Eavesdropping on complex microbial communities. Microbe. 2009;4:329–335. [Google Scholar]
- Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- Poretsky RS, Hewson I, Sun S, Allen AE, Zehr JP, Moran MA. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol. 2009;11:1358–1375. doi: 10.1111/j.1462-2920.2008.01863.x. [DOI] [PubMed] [Google Scholar]
- Steglich C, Lindell D, Futschik M, Rector T, Steen R, Chisholm S. Short RNA half-lives in the slow-growing marine cyanobacterium Prochlorococcus. Genome Biol. 2010;11:R54. doi: 10.1186/gb-2010-11-5-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strady E, Pohl C, Yakushev EV, Krüger S, Hennings U. PUMP-CTD-System for trace metal sampling with a high vertical resolution. A test in the Gotland Basin, Baltic Sea. Chemosphere. 2008;70:1309–1319. doi: 10.1016/j.chemosphere.2007.07.051. [DOI] [PubMed] [Google Scholar]
- Urich T, Lanzén A, Qi J, Huson DH, Schleper C, Schuster SC. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS ONE. 2008;3:e2527. doi: 10.1371/journal.pone.0002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetriani C, Tran HV, Kerkhof LJ. Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the Black Sea. Appl Environ Microbiol. 2003;69:6481–6488. doi: 10.1128/AEM.69.11.6481-6488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wartman WB. Leitfaden der Mikroskopischen Technik. Arch Intern Med. 1960;106:739. [Google Scholar]
- Walker CB, De la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuchter C, Abbas B, Coolen MJL, Herfort L, van Bleijswijk J, Timmers P, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci USA. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.