Abstract
Two novel magnetotactic bacteria (MTB) were isolated from sediment and water collected from the Badwater Basin, Death Valley National Park and southeastern shore of the Salton Sea, respectively, and were designated as strains BW-2 and SS-5, respectively. Both organisms are rod-shaped, biomineralize magnetite, and are motile by means of flagella. The strains grow chemolithoautotrophically oxidizing thiosulfate and sulfide microaerobically as electron donors, with thiosulfate oxidized stoichiometrically to sulfate. They appear to utilize the Calvin–Benson–Bassham cycle for autotrophy based on ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) activity and the presence of partial sequences of RubisCO genes. Strains BW-2 and SS-5 biomineralize chains of octahedral magnetite crystals, although the crystals of SS-5 are elongated. Based on 16S rRNA gene sequences, both strains are phylogenetically affiliated with the Gammaproteobacteria class. Strain SS-5 belongs to the order Chromatiales; the cultured bacterium with the highest 16S rRNA gene sequence identity to SS-5 is Thiohalocapsa marina (93.0%). Strain BW-2 clearly belongs to the Thiotrichales; interestingly, the organism with the highest 16S rRNA gene sequence identity to this strain is Thiohalospira alkaliphila (90.2%), which belongs to the Chromatiales. Each strain represents a new genus. This is the first report of magnetite-producing MTB phylogenetically associated with the Gammaproteobacteria. This finding is important in that it significantly expands the phylogenetic diversity of the MTB. Physiology of these strains is similar to other MTB and continues to demonstrate their potential in nitrogen, iron, carbon and sulfur cycling in natural environments.
Keywords: biomineralization, chemolithoautotrophs, magnetotactic bacteria, magnetite, sulfur oxidation
Introduction
Magnetotactic bacteria (MTB) are a metabolically, morphologically and phylogenetically heterogeneous group of mainly aquatic prokaryotes that passively align and actively swim along magnetic-field lines (Bazylinski and Frankel, 2004). This behavior, called magnetotaxis, is due to the presence of intracellular single-magnetic-domain crystals of magnetite (Fe3O4) and/or greigite (Fe3S4) that are each surrounded by a lipid bilayer and called magnetosomes. Magnetosomes are generally organized in chain(s) within the cell and impart a permanent magnetic dipole to the cell (Bazylinski and Frankel, 2004). MTB are considered ubiquitous in aquatic habitats that are near neutral pH and have oxygen concentration and redox gradients in the water column or sediments (Bazylinski and Frankel, 2004).
Known cultured and uncultured magnetite-producing MTB are phylogenetically affiliated with the Alphaproteobacteria and Deltaproteobacteria classes, of the Proteobacteria and the Nitrospirae phylum (Bazylinski and Frankel, 2004). Greigite-producing MTB have not yet been cultured and include a group of morphologically similar multicellular magnetotactic prokaryotes affiliated with the Deltaproteobacteria (Abreu et al., 2007; Simmons and Edwards, 2007) and large rod-shaped bacteria (Pósfai et al., 1998), whose phylogeny, in general, has not been studied in detail.
The Gammaproteobacteria class of the phylum Proteobacteria is one of the most genera-rich taxa among the prokaryotes, containing about 250 genera (Garrity et al., 2005), while the phylogeny of this group is reportedly difficult to resolve (Williams et al., 2010). There is only a single report of a magnetotactic bacterium, a putative greigite-producing rod, in this group (Simmons et al., 2004). However, the mineral present in its magnetosomes was never unequivocally identified and a more thorough examination of the phylogenetic relationship of this organism raised doubts to its affiliation with the Gammaproteobacteria (Amann et al., 2007). Amann et al. (2007) point out that the probe used by Simmons et al. (2004) for fluorescent in situ hybridization of the putative greigite-producing rod, the group-specific 23S RNA-targeted probe GAM42a (Manz et al., 1992), does not allow for the unambiguous linkage of any gammaproteobacterial-specific sequences to this bacterium.
Saline lakes are globally distributed on the Earth and constitute 45% of total inland water bodies (Hahn, 2006). Saline and high-saline environments are relatively common in the southwestern United States and have not been thoroughly examined for the presence of MTB or for the presence of magnetofossils. Microbial research in saline environments is important for multiple reasons. Because of the presence of salt deposits and saline environments on Mars (Catling, 1999; Mancinelli et al., 2004; Squyres et al., 2004; Langevin et al., 2005), studies of microbial diversity in terrestrial saline environments may shed light on the forms of extinct and/or extant life on the Mars, if life ever existed on this planet. In addition, primordial life on the Earth might have started in hypersaline environments (Dundas, 1998; Knauth, 1998); thus, research on microbes in saline environments bears relevance to our understanding of the early evolution of the biosphere on Earth.
During an ecological study of inland saline aquatic environments in the deserts of the southwestern United States, we isolated two novel strains of MTB that are unequivocally, phylogenetically affiliated with the Gammaproteobacteria class of the phylum Proteobacteria. In this report, we describe these organisms, their significance to the study and evolution of magnetotaxis.
Materials and methods
Study sites and sample collection
Water and sediment samples were taken from various parts of Badwater Basin in Death Valley National Park and the Salton Sea, both sites are located in the California, USA. Badwater Basin (36.05°N, 116.63°W) is an endorheic basin (a terminal or closed watershed with no outflow of water), located in the southern part of Death Valley and noted as the lowest point in North America (86 m below sea level). It consists of a small spring-fed pool; groundwater discharge comes from the porous carbonates of the Panamint Range to the west or from fractured metamorphic rocks of the Black Mountains to the east (Hunt, 1975). The accumulated salts of the surrounding basin make the water extremely saline in certain areas. The Salton Sea is a saline endorheic rift lake located directly on the San Andreas Fault. Like Badwater Basin, it is located below sea level (∼69 m). The lake is fed by the New, Whitewater and Alamo Rivers. Principally because of agricultural runoff, the lake has become increasingly saline with a current average salinity of 47–48 ppt (Reese et al., 2008).
Samples were collected from the shore. One liter plastic bottles were filled to about 0.2–0.3 of their volume with sediment, then filled to their capacity with water that overlaid the sediment. Air bubbles were excluded. Once in the laboratory, samples were stored under dim light at room temperature (∼25 °C).
Isolation and growth of magnetotactic Gammaproteobacteria
Cells of MTB were magnetically concentrated by placing the south pole of a magnetic stirring bar next to sample bottles at the sediment–water interface for ∼30 min, and then further purified using the capillary magnetic racetrack technique (Wolfe et al., 1987). Cells were used as inocula in a modified semisolid, oxygen-gradient, enrichment medium similar to that described by Bazylinski et al. (2004). However, the salinity was adjusted to that of the samples by using a basal medium that contained a salt mixture similar to that of artificial seawaters. For the Badwater Basin samples, the formula used was (per liter): 19.45 g NaCl, 5.9 g MgCl2•6H2O, 3.24 g Na2SO4, 1.8 g CaCl2•2H2O and 0.55 g KCl. For the Salton Sea samples (per liter): 37.8 g NaCl, 5.4 g MgCl2•6H2O, 5.4 g Na2SO4, 0.5 g CaCl2•2H2O and 0.9 g KCl. To a liter of these mixtures, 5 ml modified Wolfe's mineral elixir (Wolin et al., 1963; Bazylinski et al., 2000), 0.2 ml 1% aqueous resazurin and 0.3 g NH4Cl were added and the pH was adjusted to 7.0. Two grams of Bacto-Agar (Difco Laboratories, Detroit, MI, USA) was then added after which the medium was autoclaved. After autoclaving, the following was added as sterile stock solutions (per liter): 0.5 ml of vitamin solution (Frankel et al., 1997); 2.8 ml of 0.5 KHPO4 buffer (pH 7.0); 3 ml of 40% sodium thiosulfate (Na2S2O3•5H2O) or 5 ml of neutralized 100 m sodium sulfide (Na2S•9H2O); 3 ml of 10 m FeSO4•7H2O (in 0.02 HCl); 10 ml of a filter-sterilized, freshly made 5.0% sodium bicarbonate (NaHCO3) solution; and 10 ml of a filter-sterilized, freshly made, neutralized cysteine•HCl•2H2O solution. Screw-capped glass culture tubes were filled to ∼80% of their volume with medium. The medium was allowed to sit at room temperature for several hours to solidify and to allow the O2 concentration gradient to form as evidenced by the presence of a pink (oxidized) zone near the surface and a colorless (reduced) zone in the lower part of the tubes.
Axenic cultures were obtained by dilution to extinction three times in succession in the semisolid growth medium containing thiosulfate as the electron donor. Purity of the cultures was determined using light microscopy.
Heterotrophic growth of the strains was determined using the growth medium described above for each strain except that thiosulfate was replaced by an organic carbon source at a concentration of 5 m. Carbon sources tested included acetate, succinate, pyruvate, lactate and glucose.
Thiosulfate oxidation to sulfate
Thiosulfate and sulfate concentrations in growth media were determined using liquid chromatography on a high-performance liquid chromatography system consisting of a Shimadzu (Kyoto, Japan) model CBM-20A system controller, model LC-20AD dual pumps, a CTO-20AC column oven and a CDD-10A VP detector. The system was linked to a computer containing the Shimadzu LCsolution system program, which was used for analysis. A SeQuant Anion Supressor column (Shodex IC SI-90G (PEEK) 4,6x10, Phenomenex, Torrance, CA, USA) coupled with a Continuous Anion Regeneration System (CARS) was used to separate the thiosulfate and sulfate. The mobile phase was composed of NaHCO3 (1 m) and Na2CO3 (3.2 m) (25:75, v/v) and was pumped at a flow rate of 1.5 ml min−1. Standard concentration curves were made for sulfate and thiosulfate for every experiment with r2 values >0.99.
For both strains BW-2 and SS-5, 600 μl of a microaerophilic band of cells formed 6 days after inoculation in a semisolid culture was removed and centrifuged for 3 min at 13 000 g to remove agar and cells. The supernate was filtered (0.2 μm), diluted with distilled deionized H2O (∼1:10) and injected into the injection loop (20 μl) of the high-performance liquid chromatography. All analyses were performed on triplicate cultures. Uninoculated growth medium was used as a negative control.
Ribulose-1,5-bisphosphate carboxylase/oxygenase activity
Ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) activity of cell-free extracts was performed as described by Beudeker et al. (1980), except that the dithiothreitol concentration was changed to 6.8 m and the pH was adjusted to 7.2 (Bazylinski et al., 2004). Radioactive 14C counts were determined using a Beckman model LS 6500 liquid scintillation counter (Beckman Instruments, Fullerton, CA, USA).
Nitrogenase activity
Nitrogenase activity of the strains grown in semisolid O2 concentration gradient medium was determined as described by Bazylinski et al. (2000) with some minor modifications. Ethylene concentrations were measured using Shimadzu model GC-2014 gas chromatograph equipped with dual-flame ionization detectors, and the stationary phase was a 3-mm × 1.8-m stainless steel column of Porapak N (80/100 mesh).
Generally, to test for nitrogenase activity, cells are grown in a medium lacking a fixed nitrogen source. In this case, we required cysteine as a reducing agent, so a fixed nitrogen source was present. However, the normal nitrogen sources used, NH4Cl and NaNO3, were omitted from the growth medium for the tubes used for nitrogenase testing. These compounds were included at 4 m as a negative control, as they should repress nitrogenase synthesis if they can be used as nitrogen sources for growth. Nitrogenase assays were performed with triplicate cultures for each condition.
Protein concentration determinations
For RubisCO assays, protein in cell-free extracts was measured using technique of Bradford (1976) with a Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, CA, USA). For nitrogenase assays, total cell protein was determined as described by Bazylinski et al. (2000) except that the cells and agar were not washed before digestion with trichloroacetic acid. Bovine gamma globulin was used as the standard for all protein assays.
Light and electron microscopy
The presence and behavior of magnetotactic microorganisms was determined using a Zeiss AxioImager M1 light microscope (Carl Zeiss MicroImaging, Inc., Thornwood, NY, USA) equipped with phase-contrast, fluorescence and differential interference contrast capabilities. The hanging-drop technique (Schüler, 2002) was used routinely in the examination of samples and for quantifying MTB. The local magnetic field used to determine magnetotaxis was reversed by rotating a large stirring bar magnet by 180° on the microscope stage. Swimming speeds and patterns of cells of both strains were calculated from images of swimming cells taken using dark-field light microscopy and a long exposure time of 200 ms.
The presence of magnetosomes, and the composition of magnetosome crystals and other intracellular inclusions were determined using combinations of transmission electron microscopy (TEM), energy-dispersive X-ray analysis (EDX) and selected-area electron diffraction (SAED) with a Tecnai (FEI Company, Hillsboro, OR, USA) Model G2 F30 Super-Twin TEM. The morphologies and structural details of the magnetosome crystals were studied using electron micrographs obtained with a Philips CM20 TEM (with an attached Noran Voyager EDS detector) and high-resolution images obtained using a JEOL 3010 TEM (JEOL USA, Inc., Peabody, MA, USA).
Determination of 16S rRNA, RubisCO and mam gene sequences, and phylogenetic analysis
16S rRNA genes were amplified using bacteria-specific primers 27F 5′-AGAGTTTGATCMTGGCTCAG-3′ and 1492R 5′-TACGGHTACCTTGTTACGACTT-3′ (Lane, 1991). Partial sequences of the RubisCO form I ‘green' and form I ‘red' (both cbbL), and form II (cbbM) genes were amplified using the degenerate primer pairs RubIgF-RubIgR, RubIrF-RubIrR and RuIIF1-RuIIF3, respectively (Spiridonova et al., 2004). Primers for mam genes were designed from aligned mam gene sequences of known Magnetospirillum species and using the software AmplifX version 1.44 by Nicolas Jullien (http://ifrjr.nord.univ-mrs.fr/AmplifX-Home-page) (Supplementary Table 1). PCR products were cloned into pGEM-T Easy Vector (Promega Corporation, Madison WI, USA) and sequenced (Functional Biosciences, Inc., Madison WI, USA).
Alignment of 16S rRNA or RubisCO genes was performed using CLUSTAL W multiple alignment accessory application in the BioEdit sequence alignment editor (Hall, 1999). Phylogenetic trees were constructed using MEGA version 4 (Tamura et al., 2007) applying the neighbor-joining method (Saitou and Nei, 1987). Bootstrap values were calculated with 1000 replicates.
Nucleotide sequence accession numbers
16S rRNA gene sequences of the strains BW-2 and SS-5 are assigned to GenBank Accession numbers HQ595728 and HQ595729, respectively. The partial cbbM gene sequences of strains BW-2 and SS-5 carry GenBank Accession numbers HQ638677 and HQ638676, respectively and the partial cbbL gene sequence of strain SS-5, HQ638675.
Results
Description of sampling sites and samples
Sediment from Badwater Basin in Death Valley was black in color and reducing; the odor of hydrogen sulfide was readily apparent when sediments were disturbed. The salinity of Badwater Basin was 25 ppt at the underground opening of the spring and 67 ppt at the extremity of the pool formed by the spring. Where the salinity was at its maximum, 67 ppt, the sediment was mainly composed of salt. MTB were always present in samples collected from the area where the sediment was black and the salinities were between 25 and 31 ppt. Two different morphotypes of MTB were observed after magnetic enrichment: large rods, some containing numerous intracellular refractile globules, and a small helical bacterium. All the large rod-shaped MTB we examined contained greigite as the magnetic mineral phase as determined by a combination of EDX and SAED in the TEM (data not shown). In the lab, these MTB remained viable and motile in samples kept at 25 °C for several weeks.
Sediments in most areas sampled at the Salton Sea were similar to those of Badwater Basin, although in some beach areas the sediment consisted mainly of coarse sand. Salinities at the specific sampling sites at the Salton Sea varied widely from 2 to 52 ppt. The site from where the sample containing magnetotactic Gammaproteobacteria was taken was situated at the southeastern shore of the sea near the boat ramp at Red Hill Marina County Park (33.20°N, 115.61°W). The salinity here was 52 ppt and samples were collected from a reddish pool containing large numbers of halophilic prokaryotes as determined by microscopic observation. Immediately after collection and magnetic enrichment, the samples contained two morphotypes of MTB: magnetotactic multicellular prokaryotes and magnetotactic cocci. After several months of storage at 25 °C, the samples became devoid of multicellular prokaryotes and magnetotactic cocci, and only short, rod-shaped MTB, with north-seeking polarity (swam toward the south pole of the bar magnet), were observed using the hanging-drop technique.
Isolation of the magnetotactic Gammaproteobacteria
Magnetically purified cells from the Badwater Basin and the Salton Sea were used as inocula into semisolid, O2 concentration gradient growth medium. After several weeks of incubation at 25 °C, one tube inoculated with MTB from Badwater Basin and four tubes inoculated with MTB from the Salton Sea showed evidence of growth. Microaerophilic bands of cells were present in these tubes at the pink–colorless, oxic–anoxic interface (OAI). Microscopic examination of the cells in these cultures showed that the cells were magnetotactic and rod-shaped. The MTB from the Badwater Basin tube and from one of the Salton Sea tubes were diluted to extinction three successive times to obtain axenic cultures. The MTB strains from the Badwater Basin and the Salton Sea are designated as strains BW-2 and SS-5, respectively.
Physical description of strains BW-2 and SS-5
Cells of strain BW-2 are relatively large rods with an average length of 4.4±0.6 μm and a width of 2.2±0.2 μm (n=62) that contain a number of inclusions observed using light microscopy (Figures 1a and b). Two types of inclusions, based on size, were identified in cells of BW-2 using TEM. The largest inclusions were sulfur-rich as shown by EDX analysis and severely damaged by the electron beam of the TEM (data not shown). EDX analysis and TEM revealed that the smaller inclusions were phosphorous-rich but devoid of sulfur and likely represent polyphosphate bodies (data not shown).
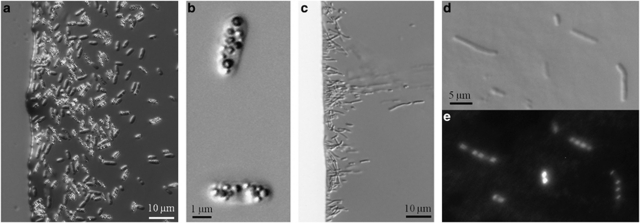
Figure 1.
Light microscope images of the novel MTB of the Gammaproteobacteria. (a, c) Differential interface contrast (DIC) light micrographs of north-seeking cells of strains BW-2 and SS-5, respectively, at the edge of a ‘hanging drop' in a magnetic field. (b) High-magnification DIC micrograph of cells of BW-2 showing the numerous, refractile, intracellular, sulfur-rich globules. (d) High-magnification DIC micrograph of very-long cells of strain SS-5 common in older cultures. (e) Fluorescence microscope image of the same SS-5 cells shown in (d) but stained with 4′,6-diamidino-2-phenylindole (DAPI) to show the numerous nucleoids present in very-long cells.
Cells of strain SS-5 are also rod-shaped but smaller than those of BW-2, having an average length of 2.5±0.5 μm and a width of 1.2±0.1 μm (n=64) (Figure 1). During stationary phase, very large cells (∼20–25 μm) became apparent suggesting that cells have trouble dividing at this growth stage. Long cells of strain SS-5 stained with 4′,6-diamidino-2-phenylindole show numerous nucleoids, which further suggests this to be the case (Figures 1d and e). The long cells were magnetotactic, although they appeared to have trouble reversing their direction of motility when the direction of the local magnetic field was reversed. Examination of strain SS-5 using TEM and EDX revealed that cells contain phosphate-rich inclusions. There was no evidence for the presence of sulfur-containing inclusions.
Chemolithoautotrophic and chemoorganoheterotrophic growth
Cells of the axenic cultures were grown routinely in semisolid O2 concentration gradient media containing thiosulfate or sulfide as electron donor and in liquid media with thiosulfate. Cells grew in this medium as a microaerophilic band of cells at the OAI evidenced by the presence of the band at the pink–colorless interface when the redox indicator resazurin was present in the growth medium. Sodium bicarbonate was the sole carbon source although a relatively small amount of organic carbon was present as vitamins and as the reducing agent cysteine. Because of the inclusion of this organic carbon, growth in this medium alone is not good evidence for chemolithoautotrophic growth. Thus, we tested the strains for the activity and presence of the gene for the CO2-fixing enzyme RubisCO of the Calvin–Benson–Bassham cycle of autotrophy as this pathway is the most widespread autotrophic pathway among the aerobic members of the Alpha-, Beta- and Gammaproteobacteria (Tourova et al., 2010).
Cell-free extracts of both strains BW-2 and SS-5, grown microaerobically under presumably autotrophic conditions with S2O32− as the electron donor and HCO3−/CO2 as the major carbon source, showed RubisCO activity (Table 1). Carbon dioxide fixation in the assays was always ribulose bisphosphate dependent and did not occur when ribulose bisphosphate was absent from the reaction mix (Table 1).
Table 1. RubisCO activities in cell-free extracts of the magnetotactic gammaproteobacterial strains BW-2 and SS-5 grown autotrophically in semisolid (O2)-gradient medium.
| Bacterial strain | Growth mode | Electron donor/ carbon source | Electron acceptor | RubisCO activity (nmol CO2 fixed min−1 mg−1 protein) |
|---|---|---|---|---|
| BW-2 | Chemoautotrophic | S2O32− /HCO3− CO2 | O2 | 4.6±0.8 |
| Extract heated to 100 °C for 10 min | Chemoautotrophic | S2O32− /HCO3− CO2 | O2 | None detected |
| SS-5 | Chemoautotrophic | S2O32− /HCO3−·CO2 | O2 | 15.1±4.2 |
| Extract heated to 100 °C for 10 min | Chemoautotrophic | S2O32− /HCO3− CO2 | O2 | None detected |
| MV-1 | Chemoautotrophic | S2O32− /HCO3− CO2 | O2 | 3.3±1.4 |
| Extract heated to 100 °C for 10 min | Chemoautotrophic | S2O32− /HCO3− CO2 | O2 | None detected |
| Escherichia coli | Heterotrophic | Tryptone-yeast extract | O2 | None detected |
Abbreviation: RubisCO, ribulose-1,5-bisphosphate carboxylase/oxygenase.
Cell-free extract of Candidatus Magnetovibrio blakemorei strain MV-1 was used as a positive control (Bazylinski et al., 2004) and cell-free extract of Escherichia coli grown heterotrophically was used as a negative control. RubisCO activity was ribulose-1,5-bisphosphate (RUDP)-dependent and no activity was observed in the absence of RUDP. Values represent mean±s.d. of triplicate analyses.
Whole-cell PCR of the strains, using primers specific for the various forms of RubisCO genes, resulted in the amplification of partial sequences (812 bp) of putative form I RubisCO, ‘green-like' cbbL genes from both BW-2 and SS-5. When these cbbL sequences were used in a BLAST search using the Genbank database, the most similar sequences to those we obtained were from chemolithoautotrophic species of the Betaproteobacteria class including the Fe(II) oxidizer Sideroxydans lithotrophicus (77.6% identity with BW-2) and Cupriavidus metallidurans (86.4% identity with SS-5).
In addition, a partial sequence (797 bp) of a form II RubisCO cbbM gene was obtained from strain SS-5, but not from strain BW-2. The most similar sequence to this gene fragment was from the cbbM gene of Thiobacillus denitrificans (87.3% identity).
When resazurin was replaced with phenol red in thiosulfate-containing growth medium for both MTB strains, the medium turned from pink to yellow particularly around the band of cells, suggesting that the strains oxidize thiosulfate to sulfuric acid. In experiments where 2 m thiosulfate was supplied to the cells, the thiosulfate was virtually depleted at the band of cells (0.025±0.013 m and 0.011±0.005 m thiosulfate remaining after growth by BW-2 and SS-5, respectively). If cells completely oxidize thiosulfate to sulfate then the concentration of sulfate recovered should be about double, ∼4 m, of what was supplied. This is what was observed (4.23±0.03 m and 4.15±0.06 m at the bands of cells of BW-2 and SS-5, respectively).
Attempts to grow BW-2 heterotrophically on a variety of organic compounds were unsuccessful. Cells of strain SS-5 appeared to show low levels of growth on succinate.
Nitrogenase activity
When grown in semisolid O2-gradient medium lacking NH4+ and NO3−, cells of BW-2 exhibited nitrogenase activity by the reduction of C2H2 to C2H4 (Table 2). The rate was relatively low compared with cultures of Candidatus Magnetovibrio blakemorei strain MV-1 used as a positive control (Bazylinski and Williams, 2007). The reason for this low rate is unclear, but activity was completely abolished when the fixed nitrogen sources, NH4+ and NO3−, were present in the growth medium. The total cell protein of cultures grown with NH4+ were about three times higher than those grown with NO3− or without either compound, consistent with the strain's ability to use NH4+ as a nitrogen source (Table 2). The fact that NO3− inhibited nitrogenase activity suggests that it can also be utilized as a nitrogen source by this strain although it did not appear to stimulate growth (Table 2).
Table 2. Nitrogenase activity based on C2H2 reduction to C2H4 by the magnetotactic gammaproteobacterial strains BW-2 and SS-5 grown autotrophically in semisolid (O2)-gradient medium with thiosulfate as the electron donor.
| Organism | Fixed N source | Total cell protein per culture (μg) | Rate of C2H2 reductiona | Duration of assay (hour) |
|---|---|---|---|---|
| MV-1 | None | 85.5±8.0 | 2.0±0.7 | 5.5 |
| BW-2 | None | 21.2±1.2 | 0.7±0.1 | 17.5 |
| NH4Cl (4 mm) | 59.1±9.8 | None detected | 17.5 | |
| NaNO3 (4 mm) | 18.4±4.4 | None detected | 17.5 | |
| SS-5 | None | Not determined | None detected | 22 |
Abbreviations: C2H2, acetylene; C2H4, ethylene.
Cultures of Candidatus Magnetovibrio blakemorei strain MV-1 were used as a positive control (Bazylinski and Williams, 2007). Experiments were performed in triplicate for each growth condition, and results are reported as the mean±s.d.
nmol C2H4 produced min−1 mg−1 cell protein. Rates were determined from linear regression calculations. For Candidatus Magnetovibrio blakemorei strain MV-1, r⩾0.9826 (n=5); for strain BW-2, r⩾0.9830 (n=5).
Cells of SS-5 did not exhibit nitrogenase activity as evidenced by the production of C2H4 from C2H2 in the standard medium with cysteine as the reducing agent although cells grew in this medium. When cysteine was replaced by dithiothreitol (1.3 m) as the reducing agent, growth was observed only when a fixed nitrogen source (for example, NH4+) was provided, suggesting that cells of this strain utilize cysteine as a nitrogen source and cannot fix N2.
Motility and magnetotaxis
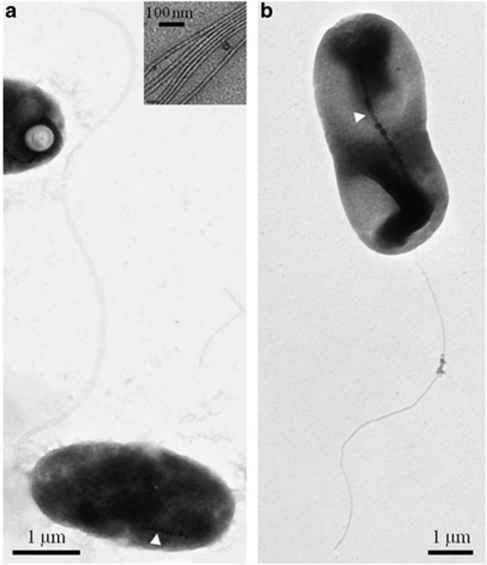
Cells of both strains are motile and magnetotactic (Supplementary Videos S1 and S2); BW-2 possesses a polar unsheathed bundle of seven flagella, whereas SS-5 possesses one unsheathed polar flagellum (Figure 2). The average swimming speed for cells of strain BW-2 in fresh cultures was 92±22 μm s−1 (n=56), whereas strain SS-5 had an average swimming speed of 49±8 μm s−1 (n=58).
Figure 2.
TEM images of negatively stained (1% uranyl acetate) cells of the magnetotactic Gammaproteobacteria. (a) TEM image of cells of strain BW-2 showing chain of magnetite magnetosomes (at white arrow) and a polar bundle of flagella. Inset shows that the flagellar bundles consist of seven individual flagellum. (b) TEM of cells of strain SS-5 showing chain of magnetosomes (at white arrow) and a single polar flagellum.
Magnetosomes of strains BW-2 and SS-5
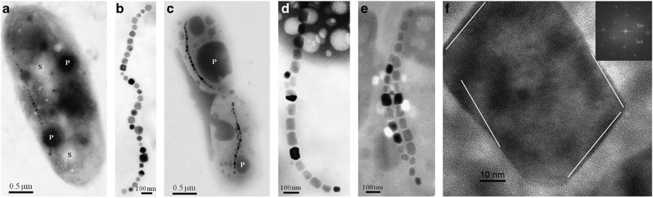
Cells of both strains BW-2 and SS-5 biomineralize a chain of octahedral crystals of magnetite in their magnetosomes (Figures 3b and d) although they differ in that those of SS-5 crystals are elongated (Figure 3d) and that some cells of strain SS-5 synthesize two parallel chains (Figure 3e). Cells grown in semisolid O2-gradient medium with thiosulfate as the electron donor contained 30±9 (n=46) and 20±7 (n=45) magnetosomes per cell for strains BW-2 and SS-5, respectively. Magnetite crystals of BW-2 averaged 67±16 × 63±15 nm in size with a shape factor of 0.94±0.04 (n=189). For SS-5, the average length of the crystals was 86±27 nm and the width 63±19 nm with a shape factor of 0.74±0.07 (n=171). Crystals of both strains fit in the theoretical single-magnetic-domain size range (Butler and Banerjee, 1975) as for mature magnetosome magnetite crystals from MTB (Bazylinski and Frankel, 2004) (Supplementary Figure 1).
Figure 3.
TEM images of inclusions within cells of strains BW-2 and SS-5. (a) TEM image of a whole unstained cell of strain BW-2 showing numerous sulfur- (S) and phosphorus-rich (P) inclusions as determined by EDX (data not shown) and a chain of magnetite magnetosomes. (b) High-magnification TEM image of a chain of magnetosomes from a cell of strain BW-2. The magnetosome crystals were identified as magnetite (data not shown) with octahedral crystal habits. (c) TEM image of a whole unstained cell of strain SS-5 showing phosphorus-rich (P) inclusions as determined by EDX (data not shown) and a chain of magnetite magnetosomes. (d, e) High-magnification TEM image of a chain and double chain, respectively, of magnetite magnetosomes from a cell of strain SS-5. (f) High-resolution TEM image of a magnetosome crystal observed in strain SS-5 identified as magnetite on the basis of its lattice spacings, with the electron beam parallel to a (110)-type crystallographic direction (consistent with the Fourier transform of the image in the upper right inset, indexed according to the magnetite structure). The white lines are parallel to (111)-type, octahedral faces, indicating that the crystal is a slightly distorted octahedron, elongated parallel to (111).
Phylogeny of the strains BW-2 and SS-5
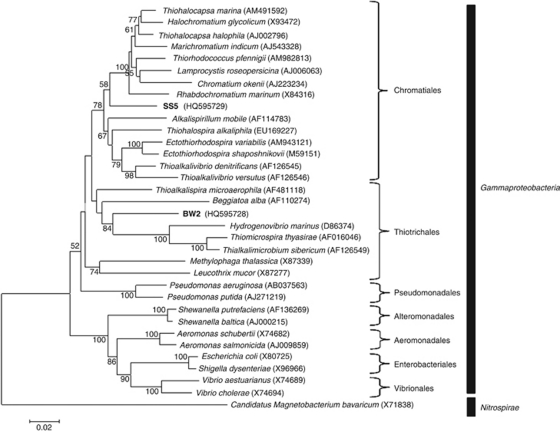
16S rRNA gene sequences of strains BW-2 and SS-5 clearly indicate that they phylogenetically belong to the Gammaproteobacteria class (Figure 4). A phylogenetic tree based on the 16S rRNA gene sequences of the strains shows that SS-5 lies within the order Chromatiales while BW-2 belongs to the order Thiotricales (Figure 4).
Figure 4.
Phylogenetic tree, based on 16S rRNA gene sequences, showing the positions of strains BW-2 and SS-5 in the Gammaproteobacteria class. Bootstrap values (higher than 50) at nodes are percentages of 1000 replicates. The 16S rRNA gene sequence of the magnetotactic bacterium Candidatus Magnetobacterium bavaricum, a member of the Nitrospirae phylum, was used to root the tree. GenBank accession numbers are given in parentheses. Bar represents 2% sequence divergence.
The highest 16S rRNA gene sequence identities to those of BW-2 and SS-5 were from sequences obtained directly from the environment belonging to as yet uncultured bacteria. For strain SS-5, the highest identity sequence (93.4%) was retrieved from a filamentous ectosymbiont on the tail of the marine oligochaete worm Tubificoides benedii collected from sulfidic mudflats at the Wadden Sea (Ruehland and Dubilier, 2010). For BW-2, the highest identity sequence (91.7%) was retrieved from a soil sample that was heavily contaminated with oil collected from the Jidong Oilfield near Bohai Bay, China (Liu et al., 2009). The three organisms in culture with the highest 16SrRNA gene sequence identities to SS-5 belong to the Chromatiales order and include Thiohalocapsa marina (93.0%), Thiohalocapsa halophila (92.5%) and Thiorhodococcus pfennigii (92.0%). The three organisms in culture with the highest 16S rRNA sequence identities to BW-2 are Thiohalospira alkaliphila (90.2% Chromatiales), Thioalkalispira microaerophila (89.9% Thiotrichales) and Thioalkalivibrio versutus (89.9% Chromatiales).
Discussion
Before this work, all magnetite-producing MTB have been found to be phylogenetically associated with the Alpha- and Deltaproteobacteria classes of the Proteobacteria phylum and the Nitrospirae phylum (Amann et al., 2007). Here we report the isolation of two novel magnetite-producing MTB that are unequivocally phylogenetically affiliated with the Gammaproteobacteria. Strain SS-5 belongs to the order Chromatiales, the bacterium with the highest 16S rRNA gene sequence identity in culture being T. marina (93.0%). Although the 16S rRNA gene sequences with the highest identity to that of strain BW-2 belong to species of the Chromatiales (highest sequence identity T. alkaliphila (90.2%)), strain BW-2 appears to phylogenetically lie well within the order Thiotrichales. Based on the divergence of the 16 rRNA genes of BW-2 and SS-5, these new MTB unequivocally represent new genera in the Gammaproteobacteria class that have a 16S rRNA gene sequence identity between them of 87.9%.
The discovery of magnetotactic Gammaproteobacteria is significant for a number of reasons. The mam, mms, and mtx genes are known to encode for a number of magnetosome membrane proteins and other proteins that appear to be involved in mineralizing magnetite magnetosomes and constructing the magnetosome chain (Schüler, 2008). These genes are organized mainly as clusters within a potentially mobile magnetosome gene island (MAI) in many MTB including those of the Alpha- and Deltaproteobacteria (Jogler et al., 2009) and the Nitrospirae (Jogler et al., 2011). For example, in Magnetospirillum gryphiswaldense, the magnetosome genes fall within a 35-kb stretch of DNA that is flanked by a larger 130-kb segment containing mobile elements and tRNA genes (Ullrich et al., 2005), characteristics typical of a gene island (Reiter and Palm, 1990). Other MTB lack some of these latter features (for example, Candidatus Magnetococcus marinus strain MC-1; Schübbe et al. (2009)). Some spontaneous non-magnetotactic mutants of M. gryphiswaldense that do not biomineralize magnetite have been shown to have the MAI deleted (Schübbe et al., 2003). These findings together with the observed distribution of magnetite-driven magnetotaxis within the Alpha- and Deltaproteobacteria classes and the Nitrospirae strongly suggests that individual MTB in these groups acquired the genes for magnetite magnetosome biomineralization through horizontal gene transfer of the MAI (Jogler et al., 2009, 2011). If mam, mms and mtx genes are present in these new and other magnetotactic Gammaproteobacteria, they might have similarly acquired magnetite magnetosomes genes. However, degenerate primers designed for nine mam genes including mamA, mamB, mamJ, mamK, mamM, mamO, mamP, mamQ and mamT, from conserved sequences of known Magnetospirillum species (Alphaproteobacteria) were ineffective at amplifying mam gene sequences from either strain BW-2 or SS-5, although the primers were effective in amplifying mam gene sequences from newly isolated Magnetospirillum strains (data not shown). This is most likely due to the fact that the sequence of these genes were not conserved enough at the nucleotide level to amplify the target sequences although it does not exclude the possibility that these genes are completely absent from the genomes of the new gammaproteobacterial MTB. We believe the latter scenario is unlikely based on the fact that the primers were also ineffective with non-Magnetospirillum alphaproteobacterial MTB such as Candidatus Magnetovibrio blakemorei strain MV-1 and others.
Interestingly, the magnetite crystals found in the gammaproteobacterial strains described here have octahedral crystal habits, one strain producing elongated octahedral habits. Crystals with similar habits have previously been found only in the Alphaproteobacteria (Jogler and Schüler, 2009) but not in MTB of the Deltaproteobacteria and the Nitrospirae that solely produce bullet-shaped crystals (Amann et al., 2007; Lefèvre et al., 2010). These latter groups appear to possess less mam genes than MTB of the Alphaproteobacteria (Abreu et al., 2011; Jogler et al., 2011) that synthesize cuboctahedral and elongated prismatic crystals of magnetite and possess a greater diversity of mam genes (Abreu et al., 2011). Thus, based on the presence of these same magnetite crystal morphologies in the new magnetotactic Gammaproteobacteria, we predict that a similar diversity of mam genes to those found in the magnetotactic Alphaproteobacteria is also present in the MTB of this group.
The proteobacterial classes are thought to have emerged in the following order during evolution (Klenk et al., 1999; Gupta, 2000; Kersters et al., 2006; Emerson et al., 2007): Epsilon- and Deltaproteobacteria then the Alpha-, Beta- and Gammaproteobacteria. Therefore, it is MTB of the latter diverging classes that biomineralize the apparently more morphologically consistent, possibly more controlled crystals of magnetite. Abreu et al. (2011) suggested that these morphologies are partially a result of the extra magnetosome genes present in these organisms that may have evolved through gene mutation and/or gene duplication, and then were distributed between species of these groups by horizontal gene transfer. If mam, mms, and mtx genes are present in these new and other magnetotactic Gammaproteobacteria, determination of the gene sequences and gene arrangement might provide additional evidence for horizontal gene transfer of the MAI and to determine whether there are significant genetic similarities between the Gammaproteobacteria and Alphaproteobacteria classes as far as magnetite biomineralization is concerned, accounting for the synthesis of similar magnetite crystal habits. In addition, it may also be helpful in determining the origin and evolution of the MAI.
Our pure-culture studies revealed physiological similarities to other cultured MTB of the Alphaproteobacteria (Bazylinski et al., 2004; Bazylinski and Williams, 2007; Geelhoed et al., 2010). Both strains grow microaerobically as bands of cells at the OAI in O2-gradient cultures and chemolithoautotrophically utilizing thiosulfate, oxidizing it stoichiometrically to sulfate, or sulfide as an electron donor, and the Calvin–Benson–Bassham cycle for autotrophy as evidenced by the presence of RubisCO activity of cell-free extracts and fragments of different forms of RubisCO genes. Of the two strains, only SS-5 displays any potential for heterotrophic growth as cells appeared to show meager growth on succinate in the absence of thiosulfate and sulfide. Thus, strain BW-2 may be an obligate chemolithoautotroph as are many other Gammaproteobacteria (for example, T. alkaliphila) (Sorokin et al., 2008). These physiological findings are consistent with the models of magnetotaxis presented by Frankel et al. (1997) and Spring and Bazylinski (2000) in which magnetotaxis functions to make chemotaxis more efficient in aiding chemolithoautotrophic MTB in locating an optimal position in chemical/redox gradients (OAI) where electron donors (for example, reduced sulfur compounds) and acceptors (O2) overlap or are close to overlapping and are thus relatively easily accessible to the bacteria.
Like all cultured MTB tested (Bazylinski and Williams, 2007), strain BW-2 appears to be capable of dinitrogen fixation based on cell's ability to reduce acetylene to ethylene under microaerobic conditions where a fixed nitrogen source was absent from the growth medium with the exception of cysteine, the reducing agent, and nitrilotriacetic acid was used as a chelating agent in the mineral solution. Cells of strain SS-5 do not appear to possess this ability as nitrogenase activity was lacking under the same culture conditions, as well as when dithiothreitol, which does not contain nitrogen, replaced cysteine as the reducing agent. Although more intensive physiological studies are required before the strains can be formally described and named, our results shows that, like MTB in other phylogenetic groups, these Gammaproteobacteria show potential for carbon, sulfur, iron and nitrogen (strain BW-2) cycling in highly saline, non-marine natural habitats.
The discovery of MTB in inland, saline, aquatic environments is also an important finding because saline lakes are widespread on Earth (Hann, 2006). As MTB have never been reported in these environments, these locales have apparently not been extensively searched for magnetofossils, specific types of magnetite crystals representing putative remains of MTB in terrestrial and extraterrestrial materials (Jimenez-Lopez et al., 2010). Thus, it would be interesting to determine whether magnetite crystals get incorporated and preserved in saline evaporites such as the halite abundant in Badwater Basin (Schubert et al., 2009) as they appear to be in sedimentary carbonates in marine environments (McNeill et al., 1988; Sakai and Jige, 2006).
Acknowledgments
This work was supported by US National Science Foundation (NSF) Grant EAR-0920718. We thank the staff at the National Park Service of Death Valley National Park. Samples were collected under Sampling Permit number DEVA-2010-SCI-0038 issued by the United States Department of the Interior National Park Service. NV was the recipient of an award from the NSF Research Experience for Undergraduates (REU) program A Broad View of Environmental Microbiology at the University of Nevada at Las Vegas (Award NSF-0649267).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abreu F, Cantão M, Nicolás M, Barcellos F, Morillo V, Paula de Almeida L, et al. 2011Common ancestry of iron oxide- and sulfide-based biomineralisation ISME Je-pub ahead of print 21 April 2011; doi: 10.1038/ismej.2011.35 [DOI] [PMC free article] [PubMed]
- Abreu F, Martins JL, Silveira TS, Keim CN, Lins de Barros HGP, Filho FJG, et al. ‘Candidatus Magnetoglobus multicellularis', a multicellular, magnetotactic prokaryote from a hypersaline environment. Int J Syst Evol Microbiol. 2007;57:1318–1322. doi: 10.1099/ijs.0.64857-0. [DOI] [PubMed] [Google Scholar]
- Amann R, Peplies J, Schüler D. Magnetoreception and Magnetosomes in Bacteria. Springer: Berlin, Germany; 2007. Diversity and taxonomy of magnetotactic bacteria; pp. 25–36. [Google Scholar]
- Bazylinski DA, Dean AJ, Schüler D, Phillips EJ, Lovley DR. N2-dependent growth and nitrogenase activity in the metal-metabolizing bacteria, Geobacter and Magnetospirillum species. Environ Microbiol. 2000;2:266–273. doi: 10.1046/j.1462-2920.2000.00096.x. [DOI] [PubMed] [Google Scholar]
- Bazylinski DA, Dean AJ, Williams TJ, Kimble-Long L, Middleton SL, Dubbels BL. Chemolithoautotrophy in the marine, magnetotactic bacterial strains MV-1 and MV-2. Arch Microbiol. 2004;182:373–387. doi: 10.1007/s00203-004-0716-y. [DOI] [PubMed] [Google Scholar]
- Bazylinski DA, Frankel RB. Magnetosome formation in prokaryotes. Nat Rev Microbiol. 2004;2:217–230. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- Bazylinski DA, Williams TJ.2007Ecophysiology of magnetotactic bacteriaIn: Schüler D (ed)Magnetoreception and Magnetosomes in Bacteria Springer: Berlin, Germany; 37–75. [Google Scholar]
- Beudeker RF, Cannon GC, Kuenen JG, Shively JM. Relations between D-ribulose-1,5-bisphosphate carboxylase, carboxysomes and CO2 fixing capacity in the obligate chemolithotroph Thiobacillus neapolitanus grown under different limitations in the chemostat. Arch Microbiol. 1980;124:185–189. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Butler RF, Banerjee SK. Theoretical single-domain grain size range in magnetite and titanomagnetite. J Geophys Res. 1975;80:4049–4058. [Google Scholar]
- Catling DC. A chemical model for evaporites on early Mars: Possible sedimentary tracers of the early climate and implications for exploration. J Geophys Res. 1999;104:16453–16469. [Google Scholar]
- Dundas I. Was the environment for primordial life hypersaline. Extremophiles. 1998;2:375–377. doi: 10.1007/s007920050081. [DOI] [PubMed] [Google Scholar]
- Emerson D, Rentz JA, Lilburn TG, Davis RE, Aldrich H, Chan C, et al. A novel lineage of proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PloS One. 2007;8:e667. doi: 10.1371/journal.pone.0000667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel RB, Bazylinski DA, Johnson MS, Taylor BL. Magneto-aerotaxis in marine coccoid bacteria. Biophys J. 1997;73:994–1000. doi: 10.1016/S0006-3495(97)78132-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity GM, Bell JA, Lilburn TG.2005Class III. Gammaproteobacteria class. NovIn: Brenner DJ, Krieg NR, Staley JT, Garrity GM (eds)Bergey's Manual of Systematic Bacteriology2nd edn, vol. 2Springer: New York; p1 [Google Scholar]
- Geelhoed JS, Kleerebezem R, Sorokin DY, Stams AJM, van Loosdrecht MCM. Reduced inorganic sulfur oxidation supports autotrophic and mixotrophic growth of Magnetospirillum strain J10 and Magnetospirillum gryphiswaldense. Environ Microbiol. 2010;12:1031–1040. doi: 10.1111/j.1462-2920.2009.02148.x. [DOI] [PubMed] [Google Scholar]
- Gupta RS. The phylogeny of proteobacteria: relationships to other eubacterial phyla and eukaryotes. FEMS Microbiol Rev. 2000;24:367–402. doi: 10.1111/j.1574-6976.2000.tb00547.x. [DOI] [PubMed] [Google Scholar]
- Hahn MW. The microbial diversity of inland waters. Curr Opin Biotechnol. 2006;17:256–261. doi: 10.1016/j.copbio.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NY. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hunt CB. Death Valley: Geology, Ecology, Archaeology. University of California Press: Berkeley; 1975. [Google Scholar]
- Jimenez-Lopez C, Romanek CS, Bazylinski DA. Magnetite as a prokaryotic biomarker: a review. J Geophys Res-Biogeo. 2010;115:G00G03. [Google Scholar]
- Jogler C, Kube M, Schübbe S, Ullrich S, Teeling H, Bazylinski DA, et al. Comparative analysis of magnetosome gene clusters in magnetotactic bacteria provides further evidence for horizontal gene transfer. Environ Microbiol. 2009;15:1267–1277. doi: 10.1111/j.1462-2920.2009.01854.x. [DOI] [PubMed] [Google Scholar]
- Jogler C, Schüler D. Genomics, genetics, and cell biology of magnetosome formation. Ann Rev Microbiol. 2009;63:501–521. doi: 10.1146/annurev.micro.62.081307.162908. [DOI] [PubMed] [Google Scholar]
- Jogler C, Wanner G, Kolinko S, Niebler M, Amann R, Petersen N, et al. 2011Conservation of proteobacterial magnetosome genes and structures in an uncultivated member of the deep-branching Nitrospira phylum Proc Natl Acad Sci USA 1081134–1139.doi: 10.1073/pnas.1012694108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersters K, De Vos P, Gillis M, Swings J, Vandamme P, Stackebrandt E. Introduction to the proteobacteria. Prokaryotes. 2006;5:3–37. [Google Scholar]
- Klenk H-P, Meier TD, Durovic P, Schwass V, Lottspeich F, Dennis PP, et al. RNA polymerase of Aquifex pyrophilus: implications for the evolution of the bacterial rpoBC operon and extremely thermophilic bacteria. J Molec Evol. 1999;48:528–541. doi: 10.1007/pl00006496. [DOI] [PubMed] [Google Scholar]
- Knauth LP. Salinity history of the Earth′s early ocean. Nature. 1998;395:554–555. doi: 10.1038/26879. [DOI] [PubMed] [Google Scholar]
- Lane DJ.199116S/23S rRNA sequencingIn: Stackebrandt E, Goodfellow M (eds)Nucleic Acid Techniques in Bacterial Systematics John Wiley & Sons: New York; 115–175. [Google Scholar]
- Langevin Y, Poulet F, Bibring JP, Gondet B. Sulfates in the north polar region of Mars detected by OMEGA/Mars Express. Science. 2005;307:1584–1586. doi: 10.1126/science.1109091. [DOI] [PubMed] [Google Scholar]
- Lefèvre CT, Abreu F, Schmidt ML, Lins U, Frankel RB, Hedlund BP, et al. Moderately thermophilic magnetotactic bacteria from hot springs in Nevada. Appl Environ Microbiol. 2010;76:3740–3743. doi: 10.1128/AEM.03018-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Zhang Y, Ding R, Li D, Gao Y, Yang M. Comparison of archaeal and bacterial community structures in heavily oil-contaminated and pristine soils. J Biosci Bioeng. 2009;108:400–407. doi: 10.1016/j.jbiosc.2009.05.010. [DOI] [PubMed] [Google Scholar]
- Mancinelli RL, Fahlen TF, Landheim R, Klovstad MR. Brines and evaporites: analogs for Martian life. Adv Space Res. 2004;33:1244–1246. [Google Scholar]
- Manz W, Amann R, Ludwig W, Wagner M, Schleifer KH. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. System Appl Microbiol. 1992;15:593–600. [Google Scholar]
- McNeill DF, Ginsburg RN, Chang S-BR, Kirschvink JL. Magnetostratigraphic dating of shallow-water carbonates from San Salvador, the Bahamas. Geology. 1988;16:8–12. [Google Scholar]
- Pósfai M, Buseck PR, Bazylinski DA, Frankel RB. Iron sulfides from magnetotactic bacteria: structure, composition, and phase transitions. Am Mineral. 1998;83:1469–1481. [Google Scholar]
- Reese BK, Anderson MA, Amrhein C. Hydrogen sulfide production and volatilization in a polymictic eutrophic saline lake, Salton Sea, California. Sci Total Environ. 2008;406:205–218. doi: 10.1016/j.scitotenv.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Reiter WD, Palm P. Identification and characterization of a defective SSV1 genome integrated into a tRNA gene in the archaebacterium Sulfolobus sp. B12. Mol Gen Genet. 1990;221:65–71. doi: 10.1007/BF00280369. [DOI] [PubMed] [Google Scholar]
- Ruehland C, Dubilier N. Gamma- and epsilonproteobacterial ectosymbionts of a shallow-water marine worm are related to deep-sea hydrothermal vent ectosymbionts. Environ Microbiol. 2010;12:2312–2326. doi: 10.1111/j.1462-2920.2010.02256.x. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sakai S, Jige M. Characterization of magnetic particles and magnetostratigraphic dating of shallow-water carbonates in the Ryukyu Islands, northwestern Pacific. Island Arc. 2006;15:468–475. [Google Scholar]
- Schübbe S, Kube M, Scheffel A, Wawer C, Heyen U, Meyerdierks A, et al. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J Bacteriol. 2003;185:5779–5790. doi: 10.1128/JB.185.19.5779-5790.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübbe S, Williams TJ, Xie G, Kiss HE, Brettin TS, Martinez D, et al. Complete genome sequence of the chemolithoautotrophic marine magnetotactic coccus strain MC-1. Appl Environ Microbiol. 2009;75:4835–4852. doi: 10.1128/AEM.02874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert BA, Lowenstein TK, Timofeeff MN. Microscopic identification of prokaryotes in modern and ancient halite, Saline Valley and Death Valley, California. Astrobiology. 2009;9:467–482. doi: 10.1089/ast.2008.0282. [DOI] [PubMed] [Google Scholar]
- Schüler D. The biomineralization of magnetosomes in Magnetospirillum gryphiswaldense. Int Microbiol. 2002;5:209–214. doi: 10.1007/s10123-002-0086-8. [DOI] [PubMed] [Google Scholar]
- Schüler D. Genetics and cell biology of magnetosome formation in magnetotactic bacteria. FEMS Microbiol Rev. 2008;32:654–672. doi: 10.1111/j.1574-6976.2008.00116.x. [DOI] [PubMed] [Google Scholar]
- Simmons SL, Edwards KJ. Unexpected diversity in populations of the many-celled magnetotactic prokaryote. Environ Microbiol. 2007;9:206–215. doi: 10.1111/j.1462-2920.2006.01129.x. [DOI] [PubMed] [Google Scholar]
- Simmons SL, Sievert SM, Frankel RB, Bazylinski DA, Edwards KJ. Spatiotemporal distribution of marine magnetotactic bacteria in a seasonally stratified coastal salt pond. Appl Environ Microbiol. 2004;70:6230–6239. doi: 10.1128/AEM.70.10.6230-6239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin DY, Tourova TP, Muyzer G, Kuenen GJ. Thiohalospira halophila gen. nov., sp. nov. and Thiohalospira alkaliphila sp. nov., novel obligately chemolithoautotrophic, halophilic, sulfur-oxidizing gammaproteobacteria from hypersaline habitats. Int J Syst Evol Microbiol. 2008;58:1685–1692. doi: 10.1099/ijs.0.65654-0. [DOI] [PubMed] [Google Scholar]
- Spiridonova EM, Berg IA, Kolganova TV, Ivanovski RN, Kuznetsov BB, Turova TP. An oligonucleotide primer system for amplification of the ribulose-1,5-bisphosphate carboxylase/oxygenase genes of bacteria of various taxonomic groups. Microbiology (Mikrobiologiya) 2004;73:316–325. [PubMed] [Google Scholar]
- Spring S, Bazylinski DA. Magnetotactic bacteria. Prokaryotes. 2000;2:842–862. [Google Scholar]
- Squyres SW, Grotzinger JP, Arvidson RE, Bell JF, 3rd, Calvin W, Christensen PR, et al. In situ evidence for an ancient aqueous environment at Meridiani Planum, Mars. Science. 2004;306:1709–1714. doi: 10.1126/science.1104559. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tourova TP, Kovaleva OL, Sorokin DY, Muyzer G. Ribulose-1,5-bisphosphate carboxylase/oxygenase genes as a functional marker for chemolithoautotrophic halophilic sulfur-oxidizing bacteria in hypersaline habitats. Microbiology. 2010;156:2016–2025. doi: 10.1099/mic.0.034603-0. [DOI] [PubMed] [Google Scholar]
- Ullrich S, Kube M, Schübbe M, Reinhardt R, Schüler D. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J Bacteriol. 2005;187:7176–7184. doi: 10.1128/JB.187.21.7176-7184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KP, Gillespie JJ, Sobral BW, Nordberg EK, Snyder EE, Shallom JM, et al. Phylogeny of Gammaproteobacteria. J Bacteriol. 2010;192:2305–2314. doi: 10.1128/JB.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe RS, Thauer RK, Pfennig NA. A “capillary racetrack” method for isolation of magnetotactic bacteria. FEMS Microbiol Ecol. 1987;45:31–35. [Google Scholar]
- Wolin EA, Wolin MJ, Wolfe RS. Formation of methane by bacterial extracts. J Biol Chem. 1963;238:2882–2886. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.