Abstract
One of the major factors associated with global change is the ever-increasing concentration of atmospheric CO2. Although the stimulating effects of elevated CO2 (eCO2) on plant growth and primary productivity have been established, its impacts on the diversity and function of soil microbial communities are poorly understood. In this study, phylogenetic microarrays (PhyloChip) were used to comprehensively survey the richness, composition and structure of soil microbial communities in a grassland experiment subjected to two CO2 conditions (ambient, 368 p.p.m., versus elevated, 560 p.p.m.) for 10 years. The richness based on the detected number of operational taxonomic units (OTUs) significantly decreased under eCO2. PhyloChip detected 2269 OTUs derived from 45 phyla (including two from Archaea), 55 classes, 99 orders, 164 families and 190 subfamilies. Also, the signal intensity of five phyla (Crenarchaeota, Chloroflexi, OP10, OP9/JS1, Verrucomicrobia) significantly decreased at eCO2, and such significant effects of eCO2 on microbial composition were also observed at the class or lower taxonomic levels for most abundant phyla, such as Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes and Acidobacteria, suggesting a shift in microbial community composition at eCO2. Additionally, statistical analyses showed that the overall taxonomic structure of soil microbial communities was altered at eCO2. Mantel tests indicated that such changes in species richness, composition and structure of soil microbial communities were closely correlated with soil and plant properties. This study provides insights into our understanding of shifts in the richness, composition and structure of soil microbial communities under eCO2 and environmental factors shaping the microbial community structure.
Keywords: soil microbial community, PhyloChip, elevated CO2, microbial composition, microbial community structure
Introduction
The concentration of atmospheric CO2 has risen by approximately 36% since the mid-19th century, largely because of human activities, such as fossil fuel combustion and land use. With the current rate of increase of 1.9 p.p.m./year, it is projected to reach 700 p.p.m. by the end of this century, which may have major consequences on carbon cycling and the functioning of terrestrial ecosystems (IPCC, 2007). The stimulating effects of elevated CO2 (eCO2) on plant growth and primary productivity are well-established (Reich et al., 2001; Ainsworth and Long, 2005; Luo et al., 2006). For example, eCO2 has been found to increase plant growth (Curtis and Wang, 1998), enhance fine root production (Hungate et al., 1997) and augment soil carbon allocation (Zak et al., 1993; Hu et al., 2001). However, the influence of eCO2 on soil microbial communities remains poorly understood and controversial (Walther et al., 2002; Parmesan and Yohe, 2003; Heath et al., 2005; Carney et al., 2007; Drigo et al., 2007, 2009, 2010; Gruber and Galloway, 2008; Heimann and Reichstein, 2008; Lesaulnier et al., 2008; Austin et al., 2009; Ge et al., 2010; He et al., 2010b). Also, the plant growth stimulation observed under eCO2 may be transient (Drake et al., 1997; DeLucia et al., 1999) possibly because of the depletion of available nitrogen (N) (Luo et al., 2004; Reich et al., 2006). In addition, multiple global change factors, such as eCO2, elevated O3, warming and/or precipitation, may interact to alter soil microbial community diversity, composition, structure and function (Chung et al., 2006; Castro et al., 2010). Therefore, understanding the diversity, composition and structure of soil microbial communities is necessary for us to assess how eCO2 modifies ecosystem properties and functional processes.
Soil may be the most complex of all microbial communities with extremely high diversity. For example, 1 g of soil contains thousands to millions of different bacterial, archaeal and eukaryotic species (Torsvik et al., 2002; Gans et al., 2005) interwoven in extremely complex food webs. Furthermore, most (>99%) of those microbes are as-yet uncultured (Whitman et al., 1998). Thus, characterizing the phylogenetic diversity of soil microbial communities and their responses to global change (for example, eCO2) will make a significant contribution to understanding soil ecosystems.
Conventional molecular biology approaches have demonstrated that soil microbial diversity generally increased (Mitchell et al., 2003; Janus et al., 2005; Sonnemann and Wolters, 2005; Jossi et al., 2006; Lesaulnier et al., 2008), decreased (Horz et al., 2004) or remained unchanged (Barnard et al., 2004; Ebersberger et al., 2004; Loy et al., 2004; Chung et al., 2006; Gruter et al., 2006; Lipson et al., 2006; Drigo et al., 2007, 2009; Austin et al., 2009; Ge et al., 2010) in response to eCO2. The apparent discrepancy of microbial responses to eCO2 could be partially due to real differences among various ecosystems, but could also be due to differences in the methodologies used, such as terminal restriction-fragment length polymorphism, denaturing gradient gel electrophoresis, 16S rRNA-based sequencing, enzyme activities and phospholipid fatty acids. For example, it is possible that some methods may not be sensitive enough to resolve the differences caused by eCO2 at the community level.
Recently, 16S rRNA gene-based microarray technologies have been used to obtain more comprehensive information on microbial community diversity, composition, structure and dynamics. PhyloChip (G2) consists of 506 944 probe features, and of these features, 297 851 are oligonucleotide perfect match (PM) or mismatch match (MM) probes for 16S rRNA genes (Brodie et al., 2006, 2007). PhyloChip has been used to detect microorganisms in a variety of environments, such as contaminated sites (Brodie et al., 2006; Rastogi et al., 2010), air (Brodie et al., 2007), water (Hery et al., 2010), soil (Cruz-Martinez et al., 2009; DeAngelis et al., 2009; Yergeau et al., 2009; Teixeira et al., 2010), microbial fuel cell (Wrighton et al., 2008) and Huanglongbing pathogen-infected citrus (Sagaram et al., 2009). In addition, several studies demonstrated that PhyloChip could detect many more bacterial taxa as compared with the 16S rRNA gene-based clone library approach (DeSantis et al., 2007; La Duc et al., 2009; Rastogi et al., 2010), suggesting that PhyloChip provides more comprehensive surveys of microbial diversity, composition and structure.
The objectives of this study were to: (i) survey the richness and composition of soil microbial communities; (ii) examine the effects of eCO2 on the richness, composition and structure of soil microbial communities and (iii) link soil geochemistry and plant properties with the microbial community composition and structure using PhyloChip (Brodie et al., 2006, 2007). For these purposes, this study was conducted in a constructed grassland ecosystem subjected to CO2 manipulation for 10 years by using the free-air CO2 enrichment (FACE) technology. The results showed that eCO2 significantly altered the richness, composition and structure of soil microbial communities, especially for particular microbial populations, at the operational taxonomic unit (OTU) level. Such microbial population changes were closely correlated with soil and plant properties.
Materials and methods
The following is a summary of the methods used in this study. More detailed information is provided in Supplementary Data-A.
Site and sampling
This study was conducted within the BioCON (Biodiversity, CO2 and Nitrogen) experiment site (http://www.biocon.umn.edu/) located at the Cedar Creek Ecosystem Science Reserve in Minnesota, USA. The main BioCON field experiment has a total of 296 plots with three treatments: CO2 (ambient (aCO2), 368 p.p.m. versus elevated, 560 p.p.m.), N (ambient versus 4 g N per m2 per year) and plant diversity (1, 4, 9 or 16 species) (Reich et al., 2001). In this study, soil samples from 24 plots (12 replicates from aCO2, 12 replicates from eCO2 and all with 16 species and ambient N supply) were collected in July 2007 when they had been exposed to aCO2 or eCO2 for 10 years, and each sample was composited from five soil cores at a depth of 0–15 cm for analysis of soil properties or DNA extraction. Additional information about the BioCON experimental site, and plant groups and species, is provided in Supplementary Data-A.
Plant and soil analyses
The aboveground and belowground biomass, plant C and N concentrations, soil pH, volumetric soil moisture, total soil C and N concentrations, and in situ net N mineralization and net nitrification were measured as described previously (Reich et al., 2001, 2006; He et al., 2010b).
DNA extraction, purification and quantitation
Soil DNA was extracted by freeze-grinding mechanical lysis as described previously (Zhou et al., 1996). DNA quality was assessed by the ratios of 260/280 and 260/230 nm and final DNA concentrations were quantified by the PicoGreen method (Ahn et al., 1996).
PhyloChip analysis
The second generation of the PhyloChip (PhyloTech, San Francisco, CA, USA), which has 8741 OTUs and 842 subfamilies with 297 851 probes, was used for this study (Brodie et al., 2006). PhyloChip analysis included three major steps: (i) Amplification of 24 soil genomic DNAs using universal 16S rRNA primers (27F/1492R for bacteria and 4Fa/1492R for archaea); (ii) 500 ng of bacterial and 30 ng of archaeal PCR products were hybridized to each PhyloChip (Brodie et al., 2006, 2007) and (iii) hybridization data were preprocessed prior to statistical analysis as detailed in Supplementary Data-A. For eight of 12 eCO2 samples with less than 30 ng of archaeal PCR products, 10 μL of concentrated archaeal amplicons were used. A mixture of amplicons at known concentrations was added to each sample prior to fragmentation, which allows for standardization/normalization of PhyloChip data. Data obtained from the CEL files (produced from GeneChip Microarray Analysis Suite, version 5.1) were scaled by setting the mixture of internal standards (spike mix) mean intensity to 2500 to compensate for slight differences in probe responses on different chips. OTU reports were generated as described in Supplementary Data-A. Because setting a positive fraction (pf) cut-off can vary the number of passing OTUs (and hence affect the reported number of OTUs for each sample), several pf cut-off values (0.86, 0.88, 0.90, 0.92, 0.94) were evaluated by using the PhyloChip data analysis pipeline PhyloTrac (http://www.phylotrac.org/Home.html) and statistical methods as described under Materials and methods and Supplementary Data-A. A pf cut-off of 0.9 was determined to be a reasonable choice and used to generate the final OTU report used in this study.
Statistical analysis
Pre-processed PhyloChip data were further analyzed by different statistical methods: (i) Response ratio (Luo et al., 2006); (ii) detrended correspondence analysis of the microbial community structure; (iii) analysis of similarities (Clarke, 1993), non-parametric multivariate analysis of variance (ADONIS) (Anderson, 2001) and multi-response permutation procedure (Mielke and Berry, 2001; McCune and Grace, 2002) were used to analyze differences of microbial communities by using the Binomial index (Anderson and Millar, 2004); (iv) Mantel test and canonical correspondence analysis for linking the functional structure of microbial communities to plant or soil variables; and (v) partial Mantel test and partial canonical correspondence analysis for co-variation analysis of soil and plant variables (Zhou et al., 2008; He et al., 2010b).
Results
Effects of eCO2 on plant and soil properties
The plant productivity measured by biomass of aboveground, roots and fine roots was significantly (P<0.05) stimulated by eCO2 (Supplementary Table S1), which is consistent with previous studies in this site (Reich et al., 2001; Adair et al., 2009). Also, the whole-plot total N (g m−2) and legume biomass significantly (P<0.05) increased at eCO2, but the percentages of nitrogen (N) in the whole-plot plant biomass, aboveground biomass and belowground biomass significantly (P<0.05) decreased (Supplementary Table S1). In addition, the aboveground carbon/nitrogen (C/N) ratio significantly (P<0.05) increased (Supplementary Table S1), probably because of an increase in plant biomass and a decrease in the aboveground N concentration. Similarly, eCO2 significantly (P<0.05) increased soil pH and soil moisture (at depths of 0–17, 42–59 and 83–100 cm). However, no significant (P>0.05) changes in soil carbon, nitrogen, C/N ratio, or rates of ammonification, nitrification or net N mineralization, were observed (Supplementary Table S2). The significant differences in plant characteristics and soil properties suggest that the diversity, composition and structure of soil bacterial communities may be shifted in response to eCO2.
Richness of soil microbial communities in response to eCO2
The richness of soil microbial communities was examined by PhyloChip. A total of 2269 OTUs were detected at least in three samples, accounting for 26% OTUs on the PhyloChip. An average of 1916 OTUs were detected at aCO2, which was significantly (P=0.0281) higher than an average of 1864 OTUs detected at eCO2 (Table 1). All detected OTUs were taxonomically derived from two archaeal phyla and 43 bacterial phyla, 55 classes, 99 orders, 164 families and 190 subfamilies; most phylotypes were detected at both aCO2 and eCO2, with few detected only at aCO2 or eCO2 (Table 2). At the phylum level, among a total of 2269 OTUs detected, 1002 OTUs were derived from Proteobacteria, a phylum with the highest number of detectable OTUs, followed by Firmicutes with 384, Actinobacteria with 289, Bacteroidetes with 162 and Acidobacteria with 76 OTUs (Table 1). Also, based on the number of OTUs detected in each phylum, two phyla had significantly (P<0.05) lower numbers of OTUs detected at eCO2 than at aCO2, including Chloroflexi (P=0.003) and OP10 (P=0.007) (Table 1). The results indicate that the richness of soil microbial communities was decreased at eCO2.
Table 1. Numbers of OTUs detected by PhyloChip in major phyla under aCO2 and eCO2 conditions.
| Phylum | OTUs on PhyloChip |
No. of OTUs detected by PhyloChip |
|||
|---|---|---|---|---|---|
| Total (%) | aCO2 | eCO2 | P (t-test) | ||
| Crenarchaeota | 79 | 12 (15.1) | 12.00±0.00 | 12.00±0.00 | 1.000 |
| Euryarchaeota | 224 | 2 (0.9) | 1.00±0.00 | 1.00±0.00 | 1.000 |
| Acidobacteria | 98 | 76 (77.6) | 67.67±6.56 | 64.25±5.96 | 0.195 |
| Actinobacteria | 810 | 289 (35.6) | 233.17±20.32 | 225.92±17.24 | 0.356 |
| Bacteroidetes | 880 | 162 (18.4) | 123.42±14.39 | 119.67±20.00 | 0.603 |
| Chlorobi | 21 | 11 (52.4) | 9.42±1.78 | 8.83±1.59 | 0.406 |
| Chloroflexi | 117 | 44 (37.6) | 37.58±4.58 | 31.33±4.75 | 0.003 |
| Cyanobacteria | 202 | 51 (25.2) | 45.42±3.37 | 43.00±3.19 | 0.085 |
| Firmicutes | 2012 | 384 (19.1) | 312.25±30.54 | 300.75±25.39 | 0.327 |
| Gemmatimonadetes | 15 | 9 (60.0) | 8.67±0.49 | 8.58±0.51 | 0.689 |
| Natronoanaerobium | 7 | 5 (71.4) | 4.00±0.74 | 3.42±0.79 | 0.076 |
| Nitrospira | 29 | 8 (27.6) | 7.3±0.98 | 6.5±1.73 | 0.161 |
| OP10 | 12 | 7 (58.3) | 5.67±1.07 | 4.58±0.67 | 0.007 |
| OP9/JS1 | 12 | 5 (41.6) | 4.25±1.14 | 3.5±1.17 | 0.125 |
| Planctomycetes | 182 | 26 (14.3) | 20.00±4.09 | 17.25±2.22 | 0.053 |
| Proteobacteria | 3170 | 1002 (31.6) | 849.75±70.62 | 837.58±85.72 | 0.708 |
| Spirochaetes | 150 | 36 (24.0) | 33.75±2.93 | 32.67±2.99 | 0.380 |
| Synergistes | 19 | 5 (26.3) | 5.00±0.00 | 5.00±0.00 | 1.000 |
| TM7 | 45 | 9 (20.0) | 8.83±0.58 | 8.25±1.29 | 0.166 |
| Verrucomicrobia | 78 | 36 (46.1) | 28.33±3.92 | 25.75±3.05 | 0.085 |
| Others (<5 OTUs) | 250 | 53 (21.2) | 46.35±9.37 | 37.67±6.35 | 0.173 |
| Unclassified | 329 | 37 (11.2) | 32.92±1.83 | 31.25±3.14 | 0.126 |
| Total | 8741 | 2269 (26.0) | 1916.6±52.03 | 1864.1±57.25 | 0.028 |
Abbreviations: aCO2, ambient CO2; eCO2, elevated CO2; OTU, operational taxonomic unit.
Boldface indicates significantly changed phylotypes or all detected OTUs.
Table 2. Phylotypes detected by PhyloChip at different taxonomic levels.
| Domain | Phylum | Class | Order | Family | Subfamily | |
|---|---|---|---|---|---|---|
| Total no. detected phylotypes | 2 | 45 | 55 | 99 | 164 | 190 |
| Shared at aCO2 and eCO2 | 2 | 44 | 52 | 97 | 163 | 188 |
| Only detected at aCO2 | 0 | 1 | 2 | 1 | 1 | 2 |
| Only detected at eCO2 | 0 | 0 | 1 | 1 | 1 | 0 |
Overall taxonomic composition and structure of soil microbial communities in response to eCO2
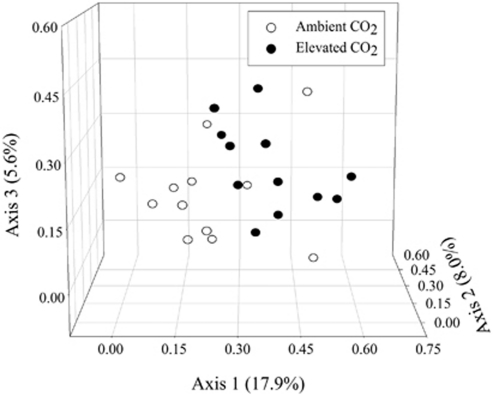
To examine if eCO2 affects the taxonomic composition and structure of soil microbial communities, detrended correspondence analysis was performed for PhyloChip signal intensity data. Overall, the majority of the samples from eCO2 and aCO2 were distributed in different parts of the data space, although there was some overlap. Eight of 12 aCO2 samples were separated well from the eCO2 samples, but four other aCO2 samples seemed to be clustered closer to the eCO2 than the aCO2 samples (Figure 1). Based on the Binomial index (Anderson and Millar, 2004), three non-parametric, multivariate statistical tests, analysis of similarities, ADONIS and multi-response permutation procedure, showed significant (P=0.007, 0.046 and 0.018, respectively) differences between microbial communities at aCO2 and eCO2. The results indicated that the overall taxonomic composition and structure of soil microbial communities was altered at eCO2.
Figure 1.
Detrended correspondence analysis of PhyloChip data for both aCO2 and eCO2 samples. Only OTUs (a total of 2269) detected in three or more samples out of 12 at aCO2 or eCO2 were analyzed.
Relationships between microbial communities and soil and plant properties
To link the taxonomic structure of microbial communities with soil and plant properties, Mantel tests and canonical correspondence analysis were performed. By using the BioENV procedure (Clarke and Ainsworth, 1993), five plant variables, including total root biomass (TRB), species count from % cover (SCFPC), aboveground percentage carbon (APC), fine roots at a depth of 0–20 cm (FR0–20) and total biomass (TB), were selected from 24 plant parameters (Supplementary Table S1). Similarly, five soil variables, including soil %N at a depth of 10–20 cm (N10–20), soil C/N ratio at a depth of 10–20 cm (SCN10–20), soil pH, nitrification rate (mg kg−1 day−1) and net N mineralization rate (mg kg−1 day−1) were selected from 20 soil parameters (Supplementary Table S2).
Based on the above selected sets of plant and soil variables, partial Mantel tests were initially performed to correlate the microbial community measured by the signal intensity of all detected 2269 OTUs with those environmental factors, and such an analysis showed that the microbial community on the whole was not correlated significantly with the soil variables (P=0.091) or plant variables (P=0.227) (Table 3). Then, we examined the correlations of the plant or soil properties with specific microbial populations at different taxonomic levels (phylum, class, order, family and subfamily). At the phylum level, 14 phylotypes significantly (P<0.05) correlated with the soil or/and plant properties. For example, there was a significant (P=0.011) correlation between Crenarchaeota and the selected plant variables, and a significant (P=0.037) correlation between Proteobacteria and the selected soil variables (Supplementary Table S3). At the class level, 16 classes were significantly (P<0.05) correlated with the soil or/and plant characteristics. For example, γ-Proteobacteria and OP10 were significantly (P=0.002 and 0.015, respectively) correlated with the selected soil variables, whereas Cyanobacteria and Spirochaetes were significantly (P=0.027 and 0.045, respectively) correlated with the selected plant variables (Table 3). Also, there were significant (P=0.012 and 0.009) correlations between the microbial community and the selected plant properties for both archaeal classes, C1 and Thermoprotei, respectively (Table 3). Similarly, 48 families were detected to be correlated with the plant or soil properties (Supplementary Table S4). For example, the signal intensities of Anaplasmataceae from α-Proteobacteria and Spirochaetaceae from Spirochaetes had significant (P=0.019 and 0.046, respectively) correlations with the selected plant variables, and those of Enterobacteriaceae and Vibrionaceae from γ-Proteobacteria had significant (P=0.001 and 0.004, respectively) correlations with the selected soil variables, whereas Erysipelotrichaceae from Mollicutes was significantly correlated with both soil (P=0.038) and plant (P=0.025) properties (Supplementary Table S4). In addition, five unclassified classes and 29 unclassified families were significantly (P<0. 05) correlated with the selected soil or plant variables, respectively, suggesting that soil and plant factors may also largely shape taxonomically uncharacterized microorganisms (Table 3 and Supplementary Table S4).
Table 3. Relationships of microbial phylotypes (OTUs) detected at the class level by PhyloChip to soil and plant variables revealed by partial Mantel test.
|
In association with: Controlling |
Soila |
Plantb |
||||
|---|---|---|---|---|---|---|
|
Plantb |
Soila |
|||||
| Phylum | Class | OTU no. | rM | P | rM | P |
| All detected | 2269 | 0.166 | 0.091 | 0.082 | 0.227 | |
| Acidobacteria | Acidobacteria-4 | 10 | 0.075 | 0.272 | 0.283 | 0.046 |
| Bacteroidetes | KSA1 | 1 | 0.083 | 0.262 | 0.230 | 0.071 |
| Unclassified | 8 | 0.039 | 0.306 | 0.144 | 0.053 | |
| Caldithrix | Unclassified | 2 | 0.122 | 0.187 | 0.271 | 0.040 |
| Chlamydiae | Chlamydiae | 2 | 0.069 | 0.058 | −0.003 | 0.478 |
| Chlorobi | Chlorobia | 11 | 0.143 | 0.088 | 0.096 | 0.195 |
| Chloroflexi | Chloroflexi-3 | 2 | 0.168 | 0.106 | 0.192 | 0.069 |
| Chloroflexi-4 | 2 | 0.138 | 0.127 | 0.213 | 0.063 | |
| Dehalococcoidetes | 7 | 0.062 | 0.289 | 0.243 | 0.065 | |
| Unclassified | 6 | −0.022 | 0.495 | 0.309 | 0.059 | |
| Coprothermobacteria | Unclassified | 1 | 0.112 | 0.196 | 0.194 | 0.07 |
| Crenarchaeota | C1 | 12 | 0.077 | 0.258 | 0.455 | 0.012 |
| Thermoprotei | 2 | 0.085 | 0.261 | 0.455 | 0.009 | |
| Cyanobacteria | Cyanobacteria | 48 | 0.147 | 0.145 | 0.315 | 0.027 |
| Deferribacteres | Deferribacer | 1 | 0.053 | 0.098 | −0.057 | 0.919 |
| Dictyoglomi | Dictyoglomi | 1 | 0.158 | 0.174 | 0.423 | 0.029 |
| DSS1 | Unclassified | 1 | 0.174 | 0.095 | 0.213 | 0.067 |
| Firmicutes | Catabacter | 7 | 0.131 | 0.162 | 0.249 | 0.060 |
| Symbiobacteria | 2 | 0.154 | 0.144 | 0.354 | 0.023 | |
| Unclassified | 17 | 0.124 | 0.220 | 0.245 | 0.063 | |
| Lentisphaerae | Unclassified | 3 | 0.178 | 0.084 | 0.212 | 0.065 |
| Marinegroup-A | mgA-1 | 2 | 0.299 | 0.054 | 0.555 | 0.014 |
| OD1 | OP11-5 | 1 | 0.172 | 0.038 | 0.111 | 0.152 |
| OP10 | Unclassified | 4 | 0.316 | 0.015 | 0.049 | 0.298 |
| OP3 | Unclassified | 3 | 0.132 | 0.076 | 0.040 | 0.282 |
| OP8 | Unclassified | 1 | −0.177 | 0.920 | 0.329 | 0.039 |
| OP9JS1 | OP9 | 2 | 0.121 | 0.138 | 0.195 | 0.064 |
| Proteobacteria | Gammaproteobacteria | 330 | 0.326 | 0.002 | 0.031 | 0.357 |
| Unclassified | 9 | 0.027 | 0.392 | 0.181 | 0.060 | |
| Spirochaetes | Spirochaetes | 36 | 0.092 | 0.212 | 0.257 | 0.045 |
| SR1 | Unclassified | 1 | 0.072 | 0.265 | 0.452 | 0.020 |
| Synergistes | Unclassified | 5 | 0.085 | 0.218 | 0.260 | 0.030 |
| Thermodesulfobacteria | Thermodesulfobacteria | 1 | 0.049 | 0.349 | 0.249 | 0.056 |
| Thermotogae | Thermotogae | 1 | 0.182 | 0.078 | 0.103 | 0.184 |
| TM6 | Unclassified | 1 | 0.090 | 0.264 | 0.416 | 0.035 |
| TM7 | Unclassified | 4 | 0.161 | 0.101 | 0.227 | 0.068 |
| Unclassified | Unclassified | 37 | 0.160 | 0.150 | 0.218 | 0.068 |
| WS3 | Unclassified | 2 | 0.055 | 0.303 | 0.166 | 0.089 |
Abbreviation: OTU, operational taxonomic unit.
Selected soil variables include soil %N at a depth of 10–20 cm (N10–20), soil %C and N at a depth of 10–20 cm (SCN10–20), soil pH, nitrification (mg kg−1 day−1) and net N mineralization (mg kg−1 day−1).
Selected plant variables include total root biomass (TRB), species count from % cover (SCFPC), aboveground percentage carbon (APC), fine roots at a depth of 0–20 cm (FR0–20) and total biomass (TB).
Only classes with P-values <0.10 to soil or plant variables have been listed.
Boldface indicates significantly changed phylotypes or all detected OTUs.
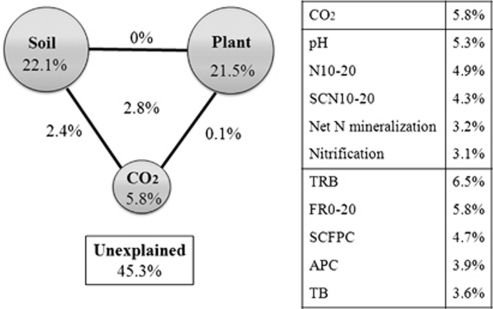
Variation partition analysis (Ramette and Tiedje, 2007) was then used to assess the contribution of CO2, soil and plant properties to the taxonomic structure of microbial communities with the same selected variables (Figure 2). When the plant and soil variables were held constant, there was a significant (P=0.037) correlation between community structure and CO2; when plant variables and CO2 were held constant, there was a significant (P=0.048) correlation between community structure and soil variables; and when soil variables and CO2 were held constant, the plant variables did not show a significant (P=0.082) correlation with microbial community. The single variable CO2 was able to independently explain 5.8% of the variation observed, which was the second largest contributor based on all 11 individual variables. Five soil variables could independently explain 22.1% of the variation, and five plant variables could explain 21.5% of the variation (Figure 2). Also, the interactions between CO2 and soil variables, CO2 and plant variables, and soil and plant variables, and among CO2, soil and plant variables, were 0.1%, 2.4%, 0.0% and 2.8%, respectively (Figure 2). In addition, 45.3% of the variation remained unexplained (Figure 2). The above statistical analyses suggest that CO2 has a direct effect on the microbial community, and that both soil and plant properties are almost equally important for shaping microbial communities through indirect CO2 effects in this grassland ecosystem.
Figure 2.
Variation partition analysis of the effects of CO2, soil and plant variables on the phylogenetic structure of soil microbial communities. The BioENV procedure was used to identify common sets of soil and plant variables important to the microbial community. The same sets of soil or plant variables were used for variation partition analysis and partial Mantel tests (Table 3; Supplementary Table S3 and Supplementary Table S4). The concentrations of CO2 are 368 p.p.m. for ambient and 560 p.p.m. for elevated environments; soil variables included soil %N at a depth of 10–20 cm (N10–20), soil C and N ratio at a depth of 10–20 cm (SCN10–20), soil pH, nitrification rate (mg kg−1 day−1) and net N mineralization rate (mg kg−1 day−1); plant variables included total root biomass (TRB), species count from % cover (SCFPC), aboveground percentage carbon (APC), fine roots at a depth of 0–20 cm (FR0–20) and total biomass (TB).
Significantly changed and unique OTUs
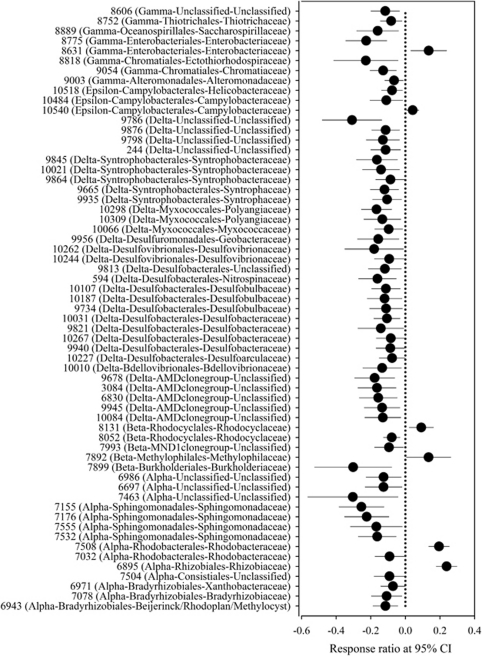
To examine effects of eCO2 on microbial community composition, both significantly changed and unique OTUs were identified. Among 2269 OTUs detected, 2075 were shared by aCO2 and eCO2 samples, and 194 unique OTUs were only detected at aCO2 (123) or eCO2 (71), respectively. For those shared OTUs, a response ratio was calculated for each OTU based on its signal intensity. A total of 194 OTUs were significantly (P<0.05) decreased and only 13 were significantly (P<0.05) increased at eCO2 (Table 4). Most phyla, including relatively abundant ones (for example, Crenarchaeota, Acidobacteria, Chloroflexi, Planctomycetes, Verrucomicrobia), did not have OTUs detected with increased signal intensities at eCO2, and only a few phyla (for example, Actinobacteria, Bacteroidetes, Firmicutes, Proteobacteria) had OTUs with both increased and decreased signal intensities at eCO2 (Table 4), which is consistent with the general trend that the richness decreased at eCO2. The signal intensities of 6 and 56 OTUs were significantly (P<0.05) increased and decreased, respectively, in Proteobacteria. Specifically, two OTUs were increased and 12 decreased in α-Proteobacteria, 2 and 2 in β-Proteobacteria, 1 and 2 in ɛ-Proteobacteria, and 1 and 7 in γ-Proteobacteria, respectively, but all 31 OTUs derived from δ-Proteobacteria were decreased at eCO2 (Figure 3). Similarly, 29 and 2 (OTU3497 and OTU3254) OTUs were significantly (P<0.05) decreased and increased, respectively, in Firmicutes, which were mostly derived from two classes, Clostridia and Bacilli (Supplementary Figure S1). Among 194 unique OTUs, 123 and 71 were from aCO2 and eCO2, respectively, and those OTUs were largely derived from the most abundant phyla, such as Proteobacteria, Firmicutes and Actinobacteria (Table 4 and Supplementary Table S5). The analysis of significantly changed and unique OTUs further confirms that the phylogenetic composition of soil microbial communities changed in response to eCO2.
Table 4. Numbers of shared OTUs detected at both aCO2 and eCO2 based on the hybridization signal intensity and unique OTUs detected only at aCO2 or eCO2.
| Phylum |
Shared OTUs |
Unique OTUs |
||
|---|---|---|---|---|
| Decrese–eCO2 | Increase–eCO2 | aCO2 | eCO2 | |
| Crenarchaeota | 12 | 0 | 0 | 0 |
| Euryarchaeota | 1 | 1 | 1 | 1 |
| Acidobacteria | 15 | 0 | 3 | 2 |
| Actinobacteria | 10 | 1 | 26 | 17 |
| Bacteroidetes | 4 | 2 | 8 | 9 |
| Chlorobi | 1 | 0 | 0 | 0 |
| Chloroflexi | 11 | 0 | 5 | 0 |
| Cyanobacteria | 4 | 1 | 4 | 0 |
| Firmicutes | 29 | 2 | 25 | 11 |
| Gemmatimonadetes | 2 | 0 | 0 | 0 |
| Natronoanaerobium | 0 | 0 | 1 | 0 |
| Nitrospira | 4 | 0 | 0 | 0 |
| OP10 | 2 | 0 | 2 | 0 |
| OP9/JS1 | 3 | 0 | 0 | 0 |
| Planctomycetes | 9 | 0 | 6 | 1 |
| Proteobacteria | 55 | 6 | 33 | 29 |
| Spirochaetes | 2 | 0 | 0 | 0 |
| Synergistes | 1 | 0 | 0 | 0 |
| TM7 | 0 | 0 | 0 | 0 |
| Verrucomicrobia | 16 | 0 | 4 | 1 |
| Others (<5 OTUs) | 6 | 0 | 3 | 0 |
| Unclassified | 7 | 0 | 2 | 0 |
| Total | 194 | 13 | 123 | 71 |
Abbreviations: aCO2, ambient CO2; eCO2, elevated CO2; OTU, operational taxonomic unit.
Increase–CO2 or Decrease–eCO2 indicates the signal intensity of an OTU was significantly higher or lower at eCO2, respectively.
Figure 3.
Significantly changed OTUs in the phylum of Proteobacteria at eCO2 by using the response ratio method (Luo et al., 2006) at 95% confidence interval.
Significantly changed microbial populations at eCO2
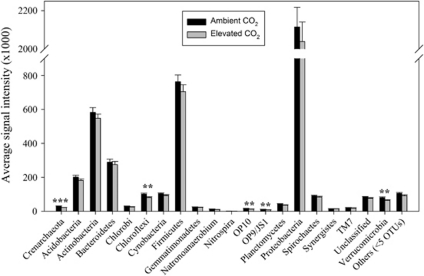
To understand what specific microbial populations may be affected by eCO2, we mapped OTUs detected to microbial populations at the phylum or lower levels, and significantly changed populations were identified by response ratio based on PhyloChip hybridization signal intensity. At the phylum level, five phyla, including one archaeal phylum (Crenarchaeota) and four bacterial phyla (Chloroflexi, OP10, OP9/JS1, Verrucomicrobia), showed significantly (P<0.05) decreased signal intensities, but most abundant phyla (for example, Proteobacteria, Firmicutes, Actinobacteria, Bacteroidetes, Acidobacteria) remained unchanged at eCO2 (Figure 4). A further examination of those significantly changed phyla showed that those changes occurred in some specific microbial groups at the class or lower levels. In the phylum of Chloroflexi, a significant decrease of signal intensities at eCO2 was observed in three orders (Chloroflexi-1a, 1b, 1f) of the class Anaerolineae, and the class of Dehalococcoidetes, whereas the signal intensities of other classes (for example, Chloroflexi-3, Chloroflexi-4, Thermomicrobia) did not change significantly (Supplementary Figure S2A). In the phylum of Verrucomicrobia, all significant changes appeared to occur in the order of Verrucomicrobiales from the class of Verrucomicrobiae, in which three families (Verrucomicrobiaceae, Verrucomicrobia subdivision-3, Verrucomicrobia subdivision-7) and an unclassified phylotype had significantly (P<0.01) decreased signal intensities, although the other two families (Verrucomicrobia subdivision-5 and Xiphinematobacteraceae) detected did not show significant changes in signal intensity at eCO2 (Supplementary Figure S2B). Crenarchaeota is an archaeal phylum showing significantly (P<0.01) decreased signal intensities at eCO2, and such decreases were observed in three orders (C1a, C1b, Cenarchaeales) from two classes (C1 and Thermoprotei) (Supplementary Figure S3A). In addition, significant decreases of signal intensity were seen in two less characterized phyla (OP10 and OP9/JS1), with one from an unclassified class in OP10 (Supplementary Figure S3B) and the other from an unclassified order of JS1class in OP9/JS1 (Supplementary Figure S3C).
Figure 4.
Average PhyloChip hybridization signal intensities for aCO2 and eCO2 samples at the phylum level. Significance was tested by response ratios (Luo et al., 2006). ***P<0.01; **P<0.05.
Although significant changes were not observed at eCO2 for the most abundant phyla at the phylum level based on summed intensities, such significances were detected at the class or lower taxonomic levels for some phyla. In the phylum Proteobacteria, the signal intensity of the AMD clone order of δ-Proteobacteria was significantly (P<0.05) decreased although no significant changes were detected at the class level (Supplementary Figure S4). In the phylum Firmicutes, the signal intensities of the family Syntrophomonadaceae in the order of Clostridiales and an unclassified order in the class of Clostridia significantly (P<0.05) decreased at eCO2, as did an unclassified phylotype (P<0.01), although no significant changes were observed in other phytolypes (for example, Bacilli, Mollicutes) (Supplementary Figure S5). Also, three groups of Actinobacteria showed significantly decreased signal intensities at eCO2, which included the order Acidimicrobiaceae and an unclassified phylotype in the class Acidimicrobiales; the order Bifidobacteriaceae in the class Bifidobacteriales and an unclassified phylotype at the phylum level, and interestingly, no significant changes in signal intensity were observed in the largest class Actinomycetales (Supplementary Figure S6). In addition, no significant changes were observed in the phylum of Bacteroidetes (Supplementary Figure S7) or Acidobacteria even at the class, order or family level (Supplementary Figure S8). Those results indicate that eCO2 significantly affected some specific microbial populations at different taxonomic levels, such as phylum, class, order and family, and those phylotypes generally appeared to have decreased signal intensities at eCO2.
Discussion
The long-term sustainability of ecosystem productivity requires detailed knowledge of its biodiversity coupled to profound understanding of its functioning. To better understand the implications of eCO2 on microbial communities, we used PhyloChip to comprehensively survey the richness, composition and structure of soil microbial communities in the BioCON grassland. Our results showed that eCO2 significantly altered the microbial community diversity, composition and structure, especially for particular microbial populations at the OTU level. Such microbial population changes were significantly correlated with soil and plant properties.
This study provides a comprehensive survey of the microbial richness and composition of grassland soil microbial communities. Previous studies with 16S rRNA-based analyses using clone libraries (Janssen, 2006; Lesaulnier et al., 2008), microarrays (for example, PhyloChip) (Cruz-Martinez et al., 2009; DeAngelis et al., 2009; Yergeau et al., 2009), pyrosequencing (Roesch et al., 2007; Fulthorpe et al., 2008; Campbell et al., 2010; Eilers et al., 2010; Uroz et al., 2010) and other approaches (Drigo et al., 2007, 2008, 2009, 2010; Feng et al., 2009) show that soil microbial communities are highly diverse and complex. In this study, 2269 OTUs affiliated 45 phyla, 55 classes, 99 orders, 164 families and 190 subfamilies were detected. Proteobacteria was the most well-represented phylum, with α-, β-, δ-, γ- and ɛ-Proteobacteria, as well as unclassified classes, detected. This group of bacteria has considerable morphological, physiological and metabolic diversity, which are of great importance to global carbon, nitrogen and sulfur cycling (Kersters et al., 2006). Firmicutes were detected as the second most prevalent phylum in terms of the number of OTUs, and Bacilli, Clostridia and Mollicutes were found to be major classes in this grassland ecosystem. Some previous cloning analyses may underestimate this group as cells or spores are known to be difficult to lyse during DNA extraction (Janssen, 2006). The members of the Actinobacteria phylum are a group of Gram-positive bacteria that have an important role in organic matter turnover and carbon cycling, such as decomposition of cellulose and chitin, and Actinomycetales and Acidimicrobiales were found to be major classes in the BioCON site. A previous study showed that long-term organic and inorganic amendments significantly altered the Actinobacterial community structure but not its diversity (Piao et al., 2008). Bacteroidetes are the fourth most prevalent group of bacteria detected in this study, with three major classes (Bacteroidetes, Flavobacteria, Sphingobacteria). Acidobacteria are among the most dominant phyla in soil-borne microbial communities, and generally are classified into eight classes (Handelsman, 2004). The diversity of Acidobacteria in soil was recently examined using different approaches, and a higher proportion of Acidobacteria was observed in bulk soil than in rhizosphere soil (Kielak et al., 2008). Therefore, this study provides a comprehensive survey of the richness and composition of soil microbial communities at this grassland ecosystem.
Elevated atmospheric CO2 may affect soil microbial communities in both direct and indirect ways. In a previous study, soil CO2 flux increased 0.57 mmol m−2 s−1 or 16% on average at eCO2 conditions as compared with aCO2 conditions in the BioCON site (Craine et al., 2001). Also, a recent study in the same site showed that the abundance of key genes involved in microbial C and N fixation, and labile C degradation, was significantly increased at eCO2 (He et al., 2010b). Those results suggest that eCO2 may directly impact soil microbial community structure and function. However, as CO2 concentrations in the pore space of soil generally are between 2000 and 38 000 p.p.m., much higher than those in the atmosphere even under aCO2 condition, the direct effects of eCO2 on soil microbial communities may be negligible compared with potential indirect effects, such as increased plant carbon inputs to soil and changes in soil properties (Drigo et al., 2008). The data presented here reflect this idea: CO2 alone explained 5.8% of the total variation of microbial community structure, compared with soil variables at 22.1% and plant variables at 21.5%. Also, eCO2 significantly increased plant productivity, whole-plot total N, soil pH and soil moisture, and decreased whole-plot plant N, aboveground and belowground N concentrations. The results suggest eCO2 may directly and indirectly affect soil microbial communities, and the indirect effects appear to make more of a contribution to shaping the soil microbial communities.
Effects of eCO2 on plant and soil properties are expected to modify taxonomic microbial community composition and structure, and regulate ecosystem functioning. First, it is indicated by differential responses of soil microbial populations to eCO2. Increases in soil carbon, coupled to an increase in cellulolytic and chitinolytic activities, were noted to alter the availability of soil substrates for microbial metabolism (Larson et al., 2002; Phillips et al., 2002). Previous studies showed increases in the abundance of Actinobacteria and Bacteroidetes at eCO2 (Sait et al., 2006; Lesaulnier et al., 2008). However, we did not see significant changes in the total signal intensities for Actinobacteria, Bacteroidetes or other most abundant phyla at the phylum level in response to eCO2. In α-Proteobacteria, a significant increase in the abundance of OTUs related to Rhodobium and a significant decrease in the abundance of OTUs related to Bradyrhizobium were detected in the trembling aspen FACE study (Lesaulnier et al., 2008), and a recent study showed a stimulation of purple phototrophic α- and β-Proteobacteria in a flooded paddy soil by eCO2 (Feng et al., 2009), which are generally consistent with this study. Also, the preference of Acidobacteria in bulk soil has been suggested to be a result of the oligotrophic lifestyle for many members of this phylum (Fierer et al., 2007). A higher input of organic matter into soil at eCO2 may be favorable for carbon polymer-degrading or fast-growing microorganisms, which presumably outcompete Acidobacteria. If true, the signal intensity of Acidobacteria may remain unchanged or decrease at eCO2, which was observed in this study. In addition, a decrease in the abundance of Crenarchaea and Verrucomicrobia at eCO2 was observed previously (Lesaulnier et al., 2008), which is consistent with our observation in this study. Members of Verrucomicrobia are reported to be negatively impacted by soil moisture (Buckley and Schmidt, 2001), which has been shown to increase at eCO2 in the BioCON site (Reich et al., 2001; He et al., 2010b) and other sites (Zavaleta et al., 2003), which is largely due to reduced stomatal conductance of plants (Kandeler et al., 2008). Second, significant correlations are observed between environmental factors and microbial communities. A negative impact of increased concentrations of organic matter on the growth of Acidobacteria was observed previously (Stevenson et al., 2004), and consistently, Acidobacteria-4 populations were found to be correlated significantly with plant variables in this study. Also, more carbon input into soil may affect autotrophic populations, such as Chloroflexis and Cyanobacteria. Indeed, a decrease in the signal intensity of those phyla was observed in this study. pH has been considered an important factor affecting the diversity and structure of soil microbial communities (Fierer and Jackson, 2006), and our Mantel analysis showed a significant correlation between soil properties, including pH and γ-Proteobacteria or OP10. In this study, soil pH was ∼6.2 in aCO2 plots and it significantly increased to ∼6.5 in eCO2 plots, suggesting a possible shift of microbial community composition and structure. Therefore, our results indicate that both soil and plant properties, such as soil pH, moisture and plant biomass, significantly affect the microbial richness, composition and structure, which may determine or modify ecosystem functioning.
The central hypothesis of this study was that, at eCO2, an increase in plant biomass (Reich et al., 2006) and soil carbon inputs (Adair et al., 2009), and associated microenvironmental changes (Reich, 2009; He et al., 2010b), would stimulate microbial, especially bacterial growth, which would lead to significant changes in the richness, composition, structure and function of soil microbial communities. Previous studies of effects of eCO2 on soil microbial communities showed variable responses. For example, a study conducted at a trembling aspen FACE experiment site in Wisconsin, USA, showed an increase in heterotrophic decomposers and a decrease in nitrate reducers of the domain bacteria and archaea, although the total bacterial abundance did not change (Lesaulnier et al., 2008). By contrast, no detectable effects on microbial community structure, microbial activity, potential soil N mineralization or nitrification rates were observed at a sweetgum FACE experiment in Tennessee, USA (Austin et al., 2009). For testing our core hypothesis, this study had several strengths: (i) It was conducted at a well-designed BioCON experimental site, with 12 replicates for each CO2 condition, so that the effects of eCO2 on soil microbial communities could be robustly examined; (ii) PhyloChip is considered a powerful tool for a comprehensive survey of microbial richness and composition (DeAngelis et al., 2009; Rastogi et al., 2010), which may overcome the limitations of cloning-based approaches and (iii) this study was conducted in a grassland ecosystem with defined plant species, which minimizes the effects of plant diversity and composition on soil microbial communities. Indeed, consistently with some previous studies in grasslands (Schortemeyer et al., 1996; Drissner et al., 2007), our results indicated that eCO2 had significant effects on the richness, composition and structure of soil microbial communities. First, such changes are reflected in a decrease in richness, which generally agrees with previous studies (Begon et al., 1996; Hughes et al., 2001). Similarly, a significant decrease in richness was observed in some phyla (for example, Chloroflexi, OP10). Second, more OTUs were found to have decreased signal intensities and fewer OTUs with increased signals at eCO2 among shared OTUs, and more unique OTUs were detected at aCO2 than eCO2. Third, based on PhyloChip signal intensities, although some specific microbial populations, especially from the most abundant phyla, remained unchanged at the phylum level, significant changes were apparent at the class or lower levels, suggesting the microbial composition was altered at eCO2 at finer taxonomic scales. Finally, the taxonomic structure is different between aCO2 and eCO2 samples as shown by statistical analyses (for example, detrended correspondence analysis, analysis of similarities, ADONIS, multi-response permutation procedure). Recently, a study using a comprehensive functional gene array, GeoChip 3.0 (He et al., 2010a), also demonstrated that the functional composition and structure of soil microbial communities were significantly altered at eCO2 (He et al., 2010b), which may be due to eCO2-induced shifts in microbial populations. The results suggest that the richness, composition and structure of soil microbial communities shift in response to eCO2.
PhyloChip has been considered a powerful tool to comprehensively and rapidly analyze microbial communities. Specifically, as such a microarray-based technology has a defined probe set and targets known populations, it minimizes or eliminates sampling artifacts, including under-sampling, unequal sampling and random sampling (Zhou et al., 2008), making it a preferable approach for community-scale comparison of microbial communities, as has been demonstrated in this study. Like other high-throughput technologies, however, PhyloChip has its limitations. For example, PhyloChip only detects known sequences already present in a database at the time of probe design, so the G2 PhyloChip used in this study may not fully cover the species richness of soil microbial communities, and a follow-up study using the G3 PhyloChip could prove beneficial. To discover unknown 16S rRNA genes, future investigations may use high-quality, full-length sequencing as a complementary approach to further understand the taxonomic and phylogenetic diversity, composition, structure and function of the soil microbial communities in this grassland ecosystem.
Acknowledgments
This work was supported by the United States Department of Agriculture (Project 2007-35319-18305) through the NSF-USDA Microbial Observatories Program; the National Science Foundation under DEB-0716587, DEB-0620652, DEB-0322057, DEB-0080382, DEB-0218039, DEB-0219104, DEB-0217631, DEB-0716587, LTREB DEB-0716587 and LTER DEB-9411972 projects; the DOE Program for Ecosystem Research; the Minnesota Environment and Natural Resources Trust Fund (DE-FG96ER2291) and the DOE under contract DE-AC02-05CH11231 through the University of California and Lawrence Berkeley National Laboratory; and by NIH Grant U01-HG004866.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Adair EC, Reich P, Hobbie S, Knops J. Interactive effects of time, CO2, N, and diversity on total belowground carbon allocation and ecosystem carbon storage in a grassland community. Ecosystems. 2009;12:1037–1052. [Google Scholar]
- Ahn S, Costa J, Emanuel J. PicoGreen quantitation of DNA: effective evaluation of samples pre- or post-PCR. Nucleic Acids Res. 1996;24:2623–2625. doi: 10.1093/nar/24.13.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsworth EA, Long SP. What have we learned from 15 years of free-air CO enrichment (FACE)? A meta-analytic review of the responses of photosynthesis, canopy properties and plant production to rising CO2. New Phytol. 2005;165:351–372. doi: 10.1111/j.1469-8137.2004.01224.x. [DOI] [PubMed] [Google Scholar]
- Anderson MJ. A new method for non-parametric multivariate analysis of variance. Aust Ecol. 2001;26:32–46. [Google Scholar]
- Anderson MJ, Millar RB. Spatial variation and effects of habitat on temperate reef fish assemblages in northeastern New Zealand. J Exp Marine Biol Ecol. 2004;305:191–221. [Google Scholar]
- Austin EE, Castro HF, Sides KE, Schadt CW, Classen AT. Assessment of 10 years of CO2 fumigation on soil microbial communities and function in a sweetgum plantation. Soil Biol Biochem. 2009;41:514–520. [Google Scholar]
- Barnard R, Barthes L, Le Roux X, Leadley PW. Dynamics of nitrifying activities, denitrifying activities and nitrogen in grassland mesocosms as altered by elevated CO2. New Phytol. 2004;162:365–376. [Google Scholar]
- Begon M, Harper JL, Townsend CR. Ecology: Individuals Populations and Communities. Blackwell Publishing Ltd: Cambridge, MA, USA; 1996. [Google Scholar]
- Brodie EL, DeSantis TZ, Joyner DC, Baek SM, Larsen JT, Andersen GL, et al. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl Environ Microbiol. 2006;72:6288–6298. doi: 10.1128/AEM.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodie EL, DeSantis TZ, Parker JP, Zubietta IX, Piceno YM, Andersen GL. Urban aerosols harbor diverse and dynamic bacterial populations. Proc Natl Acad Sci USA. 2007;104:299–304. doi: 10.1073/pnas.0608255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley DH, Schmidt TM. Environmental factors influencing the distribution of rRNA from Verrucomicrobia in soil. FEMS Microbiol Ecol. 2001;35:105–112. doi: 10.1111/j.1574-6941.2001.tb00793.x. [DOI] [PubMed] [Google Scholar]
- Campbell BJ, Polson SW, Hanson TE, Mack MC, Schuur EAG. The effect of nutrient deposition on bacterial communities in Arctic tundra soil. Environ Microbiol. 2010;12:1842–1854. doi: 10.1111/j.1462-2920.2010.02189.x. [DOI] [PubMed] [Google Scholar]
- Carney MC, Hungate BA, Drake BG, Megonigal JP. Altered soil microbial community at elevated CO2 leads to loss of soil carbon. Proc Natl Acad Sci USA. 2007;104:4990–4995. doi: 10.1073/pnas.0610045104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro HF, Classen AT, Austin EE, Norby RJ, Schadt CW. Soil microbial community responses to multiple experimental climate change drivers. Appl Environ Microbiol. 2010;76:999–1007. doi: 10.1128/AEM.02874-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Zak D, Lilleskov E. Fungal community composition and metabolism under elevated CO2 and O3. Oecologia. 2006;147:143–154. doi: 10.1007/s00442-005-0249-3. [DOI] [PubMed] [Google Scholar]
- Clarke KR. Nonparametric multivariate analyses of changes in community structure. Aust J Ecol. 1993;18:117–143. [Google Scholar]
- Clarke KR, Ainsworth M. A method of linking multivariate community structure to environmental variables. Mar Ecol Prog Ser. 1993;92:205–219. [Google Scholar]
- Craine JM, Wedin DA, Reich PB. Grassland species effects on soil CO2 flux track the effects of elevated CO2 and nitrogen. New Phytol. 2001;150:425–434. [Google Scholar]
- Cruz-Martinez K, Suttle KB, Brodie EL, Power ME, Andersen GL, Banfield JF. Despite strong seasonal responses, soil microbial consortia are more resilient to long-term changes in rainfall than overlying grassland. ISME J. 2009;3:738–744. doi: 10.1038/ismej.2009.16. [DOI] [PubMed] [Google Scholar]
- Curtis PS, Wang X. A meta-analysis of elevated CO2 effects on woody plant mass, form, and physiology. Oecologia. 1998;113:299–313. doi: 10.1007/s004420050381. [DOI] [PubMed] [Google Scholar]
- DeAngelis KM, Brodie EL, DeSantis TZ, Andersen GL, Lindow SE, Firestone MK. Selective progressive response of soil microbial community to wild oat roots. ISME J. 2009;3:168–178. doi: 10.1038/ismej.2008.103. [DOI] [PubMed] [Google Scholar]
- DeLucia EH, Hamilton JG, Naidu SL, Thomas RB, Andrews JA, Finzi A, et al. Net primary production of a forest ecosystem with experimental CO2 enrichment. Science. 1999;284:1177–1179. doi: 10.1126/science.284.5417.1177. [DOI] [PubMed] [Google Scholar]
- DeSantis T, Brodie E, Moberg J, Zubieta I, Piceno Y, Andersen G. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microb Ecol. 2007;53:371–383. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- Drake BG, Gonzàlez-Meler MA, Long SP. MORE EFFICIENT PLANTS: a consequence of rising atmospheric CO2. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:609–639. doi: 10.1146/annurev.arplant.48.1.609. [DOI] [PubMed] [Google Scholar]
- Drigo B, Kowalchuk G, van Veen J. Climate change goes underground: effects of elevated atmospheric CO2 on microbial community structure and activities in the rhizosphere. Biol Fertil Soils. 2008;44:667–679. [Google Scholar]
- Drigo B, Kowalchuk GA, Yergeau E, Bezemer TM, Boschker HTS, Van Veen JA. Impact of elevated carbon dioxide on the rhizosphere communities of Carex arenaria and Festuca rubra. Glob Change Biol. 2007;13:2396–2410. [Google Scholar]
- Drigo B, Pijl AS, Duyts H, Kielak AM, Gamper HA, Houtekamer MJ, et al. Shifting carbon flow from roots into associated microbial communities in response to elevated atmospheric CO2. Proc Natl Acad Sci USA. 2010;107:10938–10947. doi: 10.1073/pnas.0912421107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drigo B, van Veen JA, Kowalchuk GA. Specific rhizosphere bacterial and fungal groups respond differently to elevated atmospheric CO2. ISME J. 2009;3:1204–1217. doi: 10.1038/ismej.2009.65. [DOI] [PubMed] [Google Scholar]
- Drissner D, Blum H, Tscherko D, Kandeler E. Nine years of enriched CO2 changes the function and structural diversity of soil microorganisms in a grassland. Eur J Soil Sci. 2007;58:260–269. [Google Scholar]
- Ebersberger D, Werrnbter N, Niklaus PA, Kandeler E. Effects of long term CO2 enrichment on microbial community structure in calcareous grassland. Plant Soil. 2004;264:313–323. [Google Scholar]
- Eilers KG, Lauber CL, Knight R, Fierer N. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol Biochem. 2010;42:896–903. [Google Scholar]
- Feng Y, Lin X, Wang Y, Zhang J, Mao T, Yin R, et al. Free-air CO2 enrichment (FACE) enhances the biodiversity of purple phototrophic bacteria in flooded paddy soil. Plant Soil. 2009;324:317–328. [Google Scholar]
- Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology. 2007;88:1354–1364. doi: 10.1890/05-1839. [DOI] [PubMed] [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulthorpe RR, Roesch LFW, Riva A, Triplett EW. Distantly sampled soils carry few species in common. ISME J. 2008;2:901–910. doi: 10.1038/ismej.2008.55. [DOI] [PubMed] [Google Scholar]
- Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- Ge Y, Chen C, Xu Z, Oren R, He J-Z. The spatial factor, rather than elevated CO2, controls the soil bacterial community in a temperate forest ecosystem. Appl Environ Microbiol. 2010;76:7429–7436. doi: 10.1128/AEM.00831-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber N, Galloway JN. An Earth-system perspective of the global nitrogen cycle. Nature. 2008;451:293–296. doi: 10.1038/nature06592. [DOI] [PubMed] [Google Scholar]
- Gruter D, Schmid B, Brandl H. Influence of plant diversity and elevated atmospheric carbon dioxide levels on belowground bacterial diversity. BMC Microbiol. 2006;6:68. doi: 10.1186/1471-2180-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman J. Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev. 2004;68:669–685. doi: 10.1128/MMBR.68.4.669-685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Deng Y, Van Nostrand JD, Tu Q, Xu M, Hemme CL, et al. GeoChip 3.0 as a high-throughput tool for analyzing microbial community composition, structure and functional activity. ISME J. 2010a;4:1167–1179. doi: 10.1038/ismej.2010.46. [DOI] [PubMed] [Google Scholar]
- He Z, Xu M, Deng Y, Kang S, Kellogg L, Wu L, et al. Metagenomic analysis reveals a marked divergence in the structure of belowground microbial communities at elevated CO2. Ecol Lett. 2010b;13:564–575. doi: 10.1111/j.1461-0248.2010.01453.x. [DOI] [PubMed] [Google Scholar]
- Heath J, Ayres E, Possell M, Bardgett RD, Black HIJ, Grant H, et al. Rising atmospheric CO2 reduces sequestration of root-derived soil carbon. Science. 2005;309:1711–1713. doi: 10.1126/science.1110700. [DOI] [PubMed] [Google Scholar]
- Heimann M, Reichstein M. Terrestrial ecosystem carbon dynamics and climate feedbacks. Nature. 2008;451:289–292. doi: 10.1038/nature06591. [DOI] [PubMed] [Google Scholar]
- Hery M, Sanguin H, Perez Fabiel S, Lefebvre X, Vogel TM, Paul E, et al. Monitoring of bacterial communities during low temperature thermal treatment of activated sludge combining DNA phylochip and respirometry techniques. Water Res. 2010;44:6133–6143. doi: 10.1016/j.watres.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Horz H-P, Barbrook A, Field CB, Bohannan BJM. Ammonia-oxidizing bacteria respond to multifactorial global change. Proc Natl Acad Sci USA. 2004;101:15136–15141. doi: 10.1073/pnas.0406616101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Chapin FS, Firestone MK, Field CB, Chiariello NR. Nitrogen limitation of microbial decomposition in a grassland under elevated CO2. Nature. 2001;409:188–191. doi: 10.1038/35051576. [DOI] [PubMed] [Google Scholar]
- Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BJM. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl Environ Microbiol. 2001;67:4399–4406. doi: 10.1128/AEM.67.10.4399-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hungate BA, Holland EA, Jackson RB, Chapin FS, Mooney HA, Field CB. The fate of carbon in grasslands under carbon dioxide enrichment. Nature. 1997;388:576–579. [Google Scholar]
- IPCC 2007Global Climate Projections Cambridge University Press: Cambridge, United Kingdom; p806 [Google Scholar]
- Janssen PH. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl Environ Microbiol. 2006;72:1719–1728. doi: 10.1128/AEM.72.3.1719-1728.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janus LR, Angeloni NL, Mccormack J, Rier ST, Tuchman NC, Kelly JJ. Elevated atmospheric CO2 alters soil microbial communities associated with trembling aspen (Populus tremuloides) roots. Microb Ecol. 2005;50:102–109. doi: 10.1007/s00248-004-0120-9. [DOI] [PubMed] [Google Scholar]
- Jossi M, Fromin N, Tarnawski S, Kohler F, Gillet F, Aragno M, et al. How elevated pCO2 modifies total and metabolically active bacterial communities in the rhizosphere of two perennial grasses grown under field conditions. FEMS Microbiol Ecol. 2006;55:339–350. doi: 10.1111/j.1574-6941.2005.00040.x. [DOI] [PubMed] [Google Scholar]
- Kandeler E, Mosier AR, Morgan JA, Milchunas DG, King JY, Rudolph S, et al. Transient elevation of carbon dioxide modifies the microbial community composition in a semi-arid grassland. Soil Biol Biochem. 2008;40:162–171. [Google Scholar]
- Kersters K, De Vos P, Gillis M, Swings J, Vandamme P, Stackebrandt E.2006Introduction to the ProteobacteriaIn: Dwarkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds)The Prokaryotes3rd ednSpringer: New York; 53–37. [Google Scholar]
- Kielak A, Pijl AS, van Veen JA, Kowalchuk GA. Phylogenetic diversity of Acidobacteria in a former agricultural soil. ISME J. 2008;3:378–382. doi: 10.1038/ismej.2008.113. [DOI] [PubMed] [Google Scholar]
- La Duc MT, Osman S, Vaishampayan P, Piceno Y, Andersen G, Spry JA, et al. Comprehensive census of bacteria in clean rooms by using DNA microarray and cloning methods. Appl Environ Microbiol. 2009;75:6559–6567. doi: 10.1128/AEM.01073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson JL, Zak DR, Sinsabaugh RL. Extracellular enzyme activity beneath temperate trees growing under elevated carbon dioxide and ozone. Soil Sci Soc Am J. 2002;66:1848–1856. [Google Scholar]
- Lesaulnier C, Papamichail D, McCorkle S, Ollivier B, Skiena S, Taghavi S, et al. Elevated atmospheric CO2 affects soil microbial diversity associated with trembling aspen. Environ Microbiol. 2008;10:926–941. doi: 10.1111/j.1462-2920.2007.01512.x. [DOI] [PubMed] [Google Scholar]
- Lipson DA, Blair M, Barron-Gafford G, Grieve K, Murthy R. Relationships between microbial community structure and soil processes under elevated atmospheric carbon dioxide. Microb Ecol. 2006;51:302–314. doi: 10.1007/s00248-006-9032-1. [DOI] [PubMed] [Google Scholar]
- Loy A, Kusel K, Lehner A, Drake HL, Wagner M. Microarray and functional gene analyses of sulfate-reducing prokaryotes in low-sulfate, acidic fens reveal cooccurrence of recognized genera and novel lineages. Appl Environ Microbiol. 2004;70:6998–7009. doi: 10.1128/AEM.70.12.6998-7009.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Hui D, Zhang D. Elevated CO2 stimulates net accumulations of carbon and nitrogen in land ecosystems: a meta-analysis. Ecology. 2006;87:53–63. doi: 10.1890/04-1724. [DOI] [PubMed] [Google Scholar]
- Luo Y, Su B, Currie WS, Dukes JS, Finzi A, Hartwig U, et al. Progressive nitrogen limitation of ecosystem responses to rising atmospheric carbon dioxide. BioScience. 2004;54:731–739. [Google Scholar]
- McCune B, Grace JB. Analysis of Ecological Communities. MJM Software Design: Gleneden Beach, OR; 2002. [Google Scholar]
- Mielke PW, Berry KJ.2001Permutation Methods: A Distance Function Approach2nd edn.Springer-Verlag: New York, NY [Google Scholar]
- Mitchell E, Gilbert D, Buttler A, Amblard C, Grosvernier P, Gobat J.2003Structure of Microbial Communities in Sphagnum Peatlands and Effect of Atmospheric Carbon dioxide Enrichmentvol. 46. Springer-Verlag: New York, LLC; 187–199. [DOI] [PubMed] [Google Scholar]
- Parmesan C, Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421:37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- Phillips RL, Zak DR, Holmes WE, White DC. Microbial community composition and function beneath temperate trees exposed to elevated atmospheric carbon dioxide and ozone. Oecologia. 2002;131:236–244. doi: 10.1007/s00442-002-0868-x. [DOI] [PubMed] [Google Scholar]
- Piao Z, Yang L, Zhao L, Yin S. Actinobacterial community structure in soils receiving long-term organic and inorganic amendments. Appl Environ Microbiol. 2008;74:526–530. doi: 10.1128/AEM.00843-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette A, Tiedje JM. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc Natl Acad Sci USA. 2007;104:2761–2766. doi: 10.1073/pnas.0610671104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi G, Osman S, Vaishampayan P, Andersen G, Stetler L, Sani R. Microbial diversity in uranium mining-impacted soils as revealed by high-density 16S microarray and clone library. Microb Ecol. 2010;59:94–108. doi: 10.1007/s00248-009-9598-5. [DOI] [PubMed] [Google Scholar]
- Reich PB. Elevated CO2 reduces losses of plant diversity caused by nitrogen deposition. Science. 2009;326:1399–1402. doi: 10.1126/science.1178820. [DOI] [PubMed] [Google Scholar]
- Reich PB, Hobbie SE, Lee T, Ellsworth DS, West JB, Tilman D, et al. Nitrogen limitation constrains sustainability of ecosystem response to CO2. Nature. 2006;440:922–925. doi: 10.1038/nature04486. [DOI] [PubMed] [Google Scholar]
- Reich PB, Knops J, Tilman D, Craine J, Ellsworth D, Tjoelker M, et al. Plant diversity enhances ecosystem responses to elevated CO2 and nitrogen deposition. Nature. 2001;410:809–812. doi: 10.1038/35071062. [DOI] [PubMed] [Google Scholar]
- Roesch LF, Fulthorpe RR, Riva A, Casella G, Hadwin AK, Kent AD, et al. Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J. 2007;1:283–290. doi: 10.1038/ismej.2007.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagaram US, DeAngelis KM, Trivedi P, Andersen GL, Lu S-E, Wang N. Bacterial diversity analysis of Huanglongbing pathogen-infected citrus using PhyloChip arrays and 16S rRNA gene clone library sequencing. Appl Environ Microbiol. 2009;75:1566–1574. doi: 10.1128/AEM.02404-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sait M, Davis KER, Janssen PH. Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl Environ Microbiol. 2006;72:1852–1857. doi: 10.1128/AEM.72.3.1852-1857.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schortemeyer M, Hartwig UA, Hendrey GR, Sadowsky MJ. Microbial community changes in the rhizospheres of white clover and perennial ryegrass exposed to Free Air Carbon dioxide Enrichment (FACE) Soil Biol Biochem. 1996;28:1717–1724. [Google Scholar]
- Sonnemann I, Wolters V. The microfood web of grassland soils responds to a moderate increase in atmospheric CO2. Glob Change Biol. 2005;11:1148–1155. [Google Scholar]
- Stevenson BS, Eichorst SA, Wertz JT, Schmidt TM, Breznak JA. New strategies for cultivation and detection of previously uncultured microbes. Appl Environ Microbiol. 2004;70:4748–4755. doi: 10.1128/AEM.70.8.4748-4755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira LCRS, Peixoto RS, Cury JC, Sul WJ, Pellizari VH, Tiedje J, et al. Bacterial diversity in rhizosphere soil from Antarctic vascular plants of Admiralty Bay, maritime Antarctica. ISME J. 2010;4:989–1001. doi: 10.1038/ismej.2010.35. [DOI] [PubMed] [Google Scholar]
- Torsvik V, Ovreas L, Thingstad TF. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science. 2002;296:1064–1066. doi: 10.1126/science.1071698. [DOI] [PubMed] [Google Scholar]
- Uroz S, Buée M, Murat C, Frey-Klett P, Martin F. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ Microbiol Rep. 2010;2:281–288. doi: 10.1111/j.1758-2229.2009.00117.x. [DOI] [PubMed] [Google Scholar]
- Walther G-R, Post E, Convey P, Menzel A, Parmesan C, Beebee TJC, et al. Ecological responses to recent climate change. Nature. 2002;416:389–395. doi: 10.1038/416389a. [DOI] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KC, Agbo P, Warnecke F, Weber KA, Brodie EL, DeSantis TZ, et al. A novel ecological role of the Firmicutes identified in thermophilic microbial fuel cells. ISME J. 2008;2:1146–1156. doi: 10.1038/ismej.2008.48. [DOI] [PubMed] [Google Scholar]
- Yergeau E, Schoondermark-Stolk SA, Brodie EL, Dejean S, DeSantis TZ, Goncalves O, et al. Environmental microarray analyses of Antarctic soil microbial communities. ISME J. 2009;3:340–351. doi: 10.1038/ismej.2008.111. [DOI] [PubMed] [Google Scholar]
- Zak D, Pregitzer K, Curtis P, Teeri J, Fogel R, Randlett D. Elevated atmospheric CO2 and feedback between carbon and nitrogen cycles. Plant Soil. 1993;151:105–117. [Google Scholar]
- Zavaleta ES, Thomas BD, Chiariello NR, Asner GP, Shaw MR, Field CB. Plants reverse warming effect on ecosystem water balance. Proc Natl Acad Sci USA. 2003;100:9892–9893. doi: 10.1073/pnas.1732012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Kang S, Schadt CW, Garten CT, Jr Spatial scaling of functional gene diversity across various microbial taxa. Proc Natl Acad Sci USA. 2008;105:7768–7773. doi: 10.1073/pnas.0709016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.