INTRODUCTION
HIV viral load testing is routinely used to monitor response to antiretroviral therapy (ART).1 Yet, reverse transcriptase by polymerase chain reaction (RT-PCR), which measures viral RNA, requires sophisticated infrastructure, technical expertise, and rigorous quality control. With simpler technology and lower cost, the ultrasensitive p24 antigen (Up24) assay uses a standard ELISA (enzyme-linked immunosorbent assay) format to detect p24 antigen.2 Two modifications increase sensitivity: heat denaturation causing dissociation of antigen-antibody complexes and tyramide-based amplification. Schupbach et al. further enhanced sensitivity with an improved virus lysis buffer.3
Schupbach et al. have shown that Up24 is a reliable marker for HIV-1 subtype B infection, disease progression, survival in advanced disease,4 and disease progression in early-stage disease.5 Additional studies have demonstrated the utility of Up24 to monitor response to ART.6–9 Evaluations of the assay in populations with predominantly non-B subtypes, have predicted mortality in Zimbabwe (subtype C)10 and Uganda (subtypes A, D, A/D recombinant).11 Investigators have found the Up24 assay accurate in samples from South Africa (subtype C)12 and Malawi (subtype C),13 but inferior to HIV RNA in Cote d’Ivoire (CRF02_AG),14 Burkina Faso (CRF06_cpx, CRF02_AG, subtypes A and G),15 and Senegal (CRF02_AG and subtype A).16 Of primary concern is limited sensitivity at lower viral levels.
Applications of special interest for use of a p24 assay in resource-limited settings have included early diagnosis in paediatric populations and recognition of acute HIV infection. Investigators using acid rather than heat for immune complex dissociation found that p24 antigenemia was associated with poor survival in HIV-1 infected Ugandan children, yet sensitivity to detect infection especially in the first weeks of life was low.17 Heat dissociation increased sensitivity. Subsequent studies of infants in Tanzania18 and Democratic Republic of Congo,19 incorporated heat dissociation and showed sensitivity of 98.7% and 100%, respectively. Recently, Fiscus et al. demonstrated sensitivity of 88% for the Up24 assay to diagnose acute HIV-1 infection in Malawi.20
Due to the potential cost savings (Up24, US$19 versus RT-PCR, US$60)14 and ongoing improvements to the Up24 assay, investigators continue to examine its accuracy, especially in populations infected with non-B subtypes and during ART. Development of a new virus dissociation buffer by the Swiss National Center for Retroviruses (SNCR) has optimized virus disruption and improved the sensitivity of the assay.3 The new buffer improves detection of particle-associated p24 from 250,000 copies/ml to 50,000 copies/ml, and detects continued presence of p24 reactivity in patients with HIV-1 RNA < 50 copies/ml in subtype B virus. Because the assay measures viral particles rather than nucleic acid, and mechanisms of viral pathogenicity involve protein mediators, Up24 may even have an advantage over conventional methods for ART monitoring.
In this population of Ugandans infected with predominantly HIV-1 A, D, and A/D recombinant subtypes,21 we compared the Up24 assay to RT-PCR. This study reports the agreement of Up24 with RT-PCR prior to ART and during ART in patients in Kampala, Uganda.
METHODS
Study population
We recruited HIV-1 infected persons attending the Infectious Diseases Institute for HIV/AIDS treatment at Makerere University College of Health Sciences in Kampala, Uganda. Participants were older than 18 years with serologic confirmation of HIV-1 infection, and either initiating ART or on ART. Five hundred fifty consecutive, eligible patients attending the clinic who provided written, informed consent were enrolled into an observational cohort between May 2004 and February 2005 as described previously.22 All participants with available samples were chosen; the number of samples was limited due to shipping restrictions. Of the 550 enrolled, 331 participants contributed a total of 394 blood samples that were evaluated by RT-PCR and Up24. Two hundred thirteen samples were from ART-naïve participants prior to treatment. And, 181 samples were from ART-experienced participants collected during treatment.
Laboratory Testing
We compared HIV viral load by RT-PCR (Amplicor HIV-1 Monitor 1.5 assay, Roche Molecular Systems, Inc., Branchburg, NJ) versus the HIV-1 p24 Core Profile ELISA boosted by the ELAST ELISA Amplification System (PerkinElmer Life Sciences, Boston, MA). Each assay was performed according to manufacturer’s instructions. The standard Roche Amplicor Monitor HIV-1 RNA assay was used on viral RNA extracted from peripheral blood samples as described elsewhere.23 RT-PCR assays were performed by the Makerere University-Johns Hopkins University CoreLab, which is College of American Pathologists (CAP) certified. Values for RT-PCR ranged from 2.6 log10 copies/ml (399 copies/ml) to 6.0 log10 copies/ml (1,000,000 copies/ml).
For the ultrasensitive p24 antigen (Up24) assay, a plasma sample of 50 µl of plasma was incubated with 25 µl of a mixture containing 30 mM of Tris/HCl (pH 7.2), 450 mM of NaCl, 1.5% Triton X-100, 1.5% deoxycholic acid (sodium salt), 0.3% sodium dodecylsulfate, and 10 mM of EDTA at room temperature before boiling for 5 min on a dry heat block preheated to 100°C.24 After cooling, duplicate 250-µl aliquots were tested with the HIV-1 Core Profile ELISA in combination with the ELAST ELISA Amplification System as described previously.25
For each sample, the averaged result of end point reading was recorded as picograms per millimeter if the concentration was above the diagnostic cutoff corresponding to the mean plus 3 standard deviations of 6 different HIV-negative control plasma samples run in every experiment. An additional negative control was run in all tests. HIV p24 concentrations were reported as femtograms of HIV-1 p24/ml of plasma. As has been reported previously, limit of detection is 1.9 log10 fg/ml.26 Values for Up24 ranged from 1.9 log10 fg/ml to 6.0 log10 fg/ml. The p24 assays were performed at the Johns Hopkins Medical Institutions.
Statistical Analysis
Statistical analyses were performed using SAS software (version 9.1.3, SAS Institute, Cary, NC). Values were log10 transformed. We compared p24 antigen level with RNA viral load by linear regression including slope and intercept, and Pearson correlation coefficient. We calculated sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) with 95% confidence intervals using viral RNA by RT-PCR as the gold standard.
Ethical Approval
This study protocol was approved by the Johns Hopkins Medicine Institutional Review Board, the Makerere University Faculty of Medicine Research and Ethics Committee, and the Uganda National Council of Science and Technology.
RESULTS
Patients and Baseline Characteristics
Two hundred thirteen ART-naïve participants each contributed one sample prior to ART. Of the 118 participants who were ART-experienced, 55 contributed one sample and 63 contributed two samples for a total of 181 samples collected on ART. Median viral load for the entire group of 394 samples was 4.9 log10 copies/ml (IQR, 2.6–5.5); 114 samples (29%) were undetectable by RT-PCR (≤2.6 log10 copies/ml). Median Up24 was 3.1 log10 fg/ml (IQR, 1.9–3.9) and 163 samples (41%) were undetectable (≤1.9 log10 fg/ml). For the 213 samples collected prior to ART, median viral load was 5.4 log10 copies/ml (IQR, 5.0–5.7) and median Up24 was 3.7 log10 fg/ml (IQR, 1.9–4.3). For the 181 samples collected during ART, both median viral load and median Up24 were undetectable.
Ultrasensitive p24 antigen versus RT-PCR
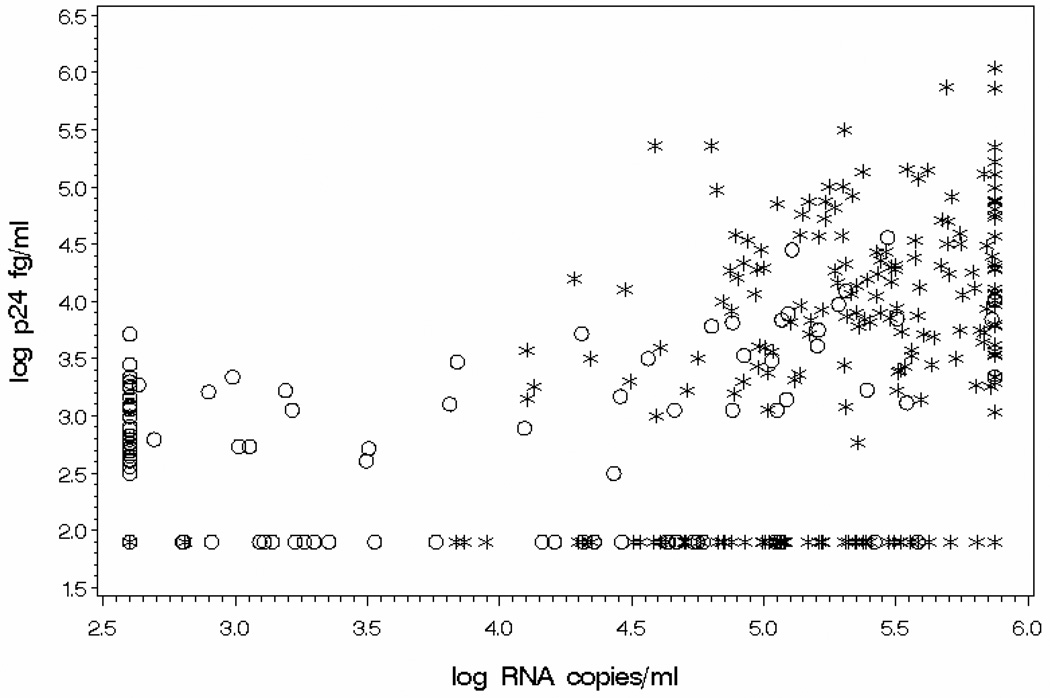
Figure 1 illustrates the scatterplot of log10 Up24 antigen versus log10 RT-PCR for HIV-1 viral load. Regression analysis demonstrated a slope of 0.50 and intercept of 0.87. Pearson correlation coefficient was 0.58 (p < 0.0001) and r2 was 0.33. Linear regression analysis was done for two subgroups of samples, those collected from participants naive to ART and those collected from participants on ART. For the 213 samples collected from ART-naïve participants, slope was 0.80, intercept was −0.70, the correlation coefficient was 0.39, and r2 was 0.15. For the 181 samples taken while participants were on ART, slope was 0.33, intercept was 1.39, the correlation coefficient was 0.47, and r2 was 0.22.
Figure 1.
Correlation analysis of ultrasensitive p24 antigen assay (log10 fg/ml) and reverse transcriptase by polymerase chain reaction RT-PCR (log10 copies/ml) for HIV-1 viral load. N=394. Up24 antigen = 0.87 + 0.50 * RNA; r2 = 0.33; Pearson correlation coefficient = 0.58 (P-value < 0.0001).
“*” represents samples collected from participant naïve to ART, N=213.
“○” represents samples collected from participant on ART, N=181.
Up24 values ranged from 1.9 log10 fg/ml to 6.0 log10 fg/ml. RT-PCR values ranged from 2.6 log10 copies/ml to 6.0 log10 copies/ml.
Sensitivity of the Up24 assay to detect viral load ≥ 400 copies/ml was 69% (Table 1) and specificity was 67%. Positive predictive value (PPV) of Up24 for viral load ≥ 400 copies/ml was 84% and negative predictive value (NPV) was 47%. In those 213 participants tested prior to ART, the sensitivity of Up24 was 73%, while both specificity and PPV were 100%. In those 181 participants tested while receiving ART, sensitivity and specificity were 58% and 66%, respectively.
Table 1.
Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of ultrasensitive heat-denatured p24 antigen assay (Up24) results for viral load by reverse transcriptase-polymerase chain reaction (RT-PCR) in samples collected from participants prior to antiretroviral therapy (ART) and during ART (CI, confidence interval).
| Sensitivity n/N* (%, 95% CI) |
Specificity n/N^ (%, 95% CI) |
PPV n/N¶ (%, 95% CI) |
NPV n/N§ (%, 95% CI) |
|
|---|---|---|---|---|
| Not on ART (N=213) |
153/211 73 (66–79) |
2/2 100 |
153/153 100 |
2/60 3 (0–8) |
| On ART (N=181) |
40/69 58 (46–70) |
74/112 66 (57–75) |
40/78 51 (40–62) |
74/103 72 (63–81) |
| All participants (N=394) |
193/280 69 (64–74) |
76/114 67 (62–72) |
193/231 84 (80–88) |
76/163 47 (42–52) |
Number of samples positive by RT-PCR (>2.6 log10 copies/ml) and Up24 (>1.9 log10 fg/ml)/total positive by RT-PCR (>2.6 log10 copies/ml).
Number of samples negative by RT-PCR (≤2.6 log10 copies/ml) and Up24 (≤1.9 log10 fg/ml)/total negative by RT-PCR (≤2.6 log10 copies/ml).
Number of samples positive by RT-PCR (>2.6 log10 copies/ml) and Up24 (>1.9 log10 fg/ml)/total positive by Up24 (>1.9 log10 fg/ml).
Number of samples negative by RT-PCR (≤2.6 log10 copies/ml) and Up24 (≤1.9 log10 fg/ml)/total negative by Up24 (≤1.9 log10 fg/ml).
The percentage of samples found to be detectable by Up24 varied according to HIV viral load by RT-PCR (p<0.0001) (Table 2). Sensitivity of Up24 was 90%, 80%, 68%, 62%, and 45% to detect viral load of >500,000; 250,000–500,000; 100,000–250,000; 50,000–100,000; and 400–50,000 copies/ml, respectively. Of 114 samples deemed undetectable by RT-PCR (<400 copies/ml), the Up24 assay detected 33% (95% CI, 25–42%) and the Up24 median value was undetectable at 1.9 log10 fg/ml (IQR, 1.9–2.7). Of these 114 samples, 112 were contributed by participants on ART. For samples deemed undetectable by Up24 (N=163), the RT-PCR median value was 3.1 log10 copies/ml (IQR, 2.6–4.9).
Table 2.
HIV-1 viral load as measured by ultrasensitive heat-denatured HIV-1 p24 antigen assay (Up24) (log10 fg/ml) compared to reverse transcriptase-polymerase chain reaction analysis (RT-PCR) (copies/ml).
| HIV-1 viral load by RT- PCR (copies/ml) |
< 400 | 400– 50,000 |
50,000– 100,000 |
100,000– 250,000 |
250,000– 500,000 |
>500,000 |
| Number of samples tested | 114 | 62 | 37 | 72 | 50 | 59 |
| Number detected by Up24 | 38 | 28 | 23 | 49 | 40 | 53 |
| Median Up24 (log10 fg/ml) (Interquartile range) |
1.9 (1.9–2.7) |
1.9 (1.9–3.2) |
3.4 (1.9–4.2) |
3.6 (1.9–4.2) |
3.9 (3.2–4.4) |
4.1 (3.5–4.7) |
| Sensitivity of Up24* (95% CI) |
33% (25–42%) |
45% (33–58%) |
62% (47–78%) |
68% (57–79%) |
80% (69–91%) |
90% (82–98%) |
Number of samples detected by Up24/number of samples tested.
Χ2, test for trend, p-value < 0.0001.
DISCUSSION
Specificity of Up24 in those with undetectable viral load was 67% and remained unchanged (66%) in the subset of those participants on ART. Therefore, we found that the Up24 assay misclassified 38 of 112 (34%) on ART as falsely positive according to RT-PCR results. This disagreement with the RT-PCR results may limit the clinical utility of the Up24 to determine whether a patient receiving ART is fully suppressed.
The specificity of 67% may be due to an uncovering of p24 antigen made possible by the virus disruption buffer developed by the Swiss National Center for Retroviruses (SNCR) and may represent “aggregated viral protein originating from damaged or destroyed particles.”3 Using the SNCR buffer, Schupbach et al. observed persistent p24 reactivity in 34% of patients with undetectable RNA on ART.9 This may also represent viral replication in reservoirs other than plasma. Newer technologies, such as ultrasensitive nanoparticle technology, may elucidate mechanisms that explain this persistent p24 reactivity.27,28
The sensitivity of the Up24 to detect viral load deemed detectable by RT-PCR is greater in those not receiving ART compared to those receiving ART. This is due to the greater prevalence of higher viral loads in those untreated. The Up24 assay detected 90% of viral loads > 500,000 copies/ml, yet the sensitivity of Up24 decreased to 45% at viral loads ranging from 400 to 50,000 copies/ml. This occurred despite the use of the SNCR buffer, which has been shown to improve antigen-antibody dissociation and increase sensitivity.3,20,29 In this population of 331 individuals with predominantly A, D, and A/D recombinant subtypes,21 the Up24 assay generated 27% false negative results in those not on ART, which increased to 42% for those on ART with lower viral loads. Our findings are similar to the results of studies that were conducted in Cote d’Ivoire (HIV-1 CRF02_AG)14 and Burkina Faso (CRF06_cpx, CRF02_AG, subtypes A and G)15 which included patients on ART with lower viral load levels.
Because Up24 measures viral proteins and RT-PCR measures viral RNA, we do not anticipate perfect agreement. Our correlation coefficient was 0.55. Other groups have also reported correlation coefficients much less than one.4,5,30 This can be explained by the fact that the p24 antigen is ‘a structural component of the retroviral particle [that exists] as a soluble protein in plasma, as a protein bound in immune complexes, or as a component of a subviral particle.’31 Whereas, HIV-1 RNA is a particle-associated molecule.
Incomplete dissociation of antigen-antibody complexes is one putative mechanism explaining less than perfect sensitivity. Investigators have illustrated two scenarios. High levels of antibody concentrations may cause re-association after neutralization when antigens bind to capture antibody.31 Or, high-affinity antibodies that are resistant to heat dissociation may cause incomplete release of antigen.32
To address the question of whether Up24 reliably detected HIV-1 in non-B subtypes, Ribas et al. evaluated plasma samples containing isolates classified by gag/env heteroduplex mobility assay as group M subtypes: A, B, C, D, F, G, H; various recombinant strains; and one group O virus.26 The Up24 was positive in 110 of 120 (92%) samples with a mean of 3.59 log10 fg/ml. Three samples from one individual infected with subtype A were not detectable by Up24, but when retested with the INNOTEST HIV antigen mAb test (Innogenetics, Ghent, Belgium), the samples were positive. Similarly, Burgisser et al. noted underestimation of p24 antigenaemia in patients infected with subtypes A, C, and D both on and off ART.33 The predominance of A, D, and A/D recombinant subtypes in our study population may have contributed to the diminished sensitivity of Up24 at lower viral loads.
Prado et al. tested 51 patients during a trial of structured treatment interruptions in Barcelona, Spain using the commercial buffer and found reduced sensitivity as viral load declined (82%, RNA > 100,000 copies/ml; 39%, 10,000 < RNA< 100,000 copies/ml; 1%, 50 < RNA < 10,000 copies/ml).32 They suggested that incomplete dissociation due to high-affinity immune complexes contributed to the limited sensitivity of the Up24. And, as p24 antigen levels decreased with increasing erythrocyte concentrations, HIV-1 immune complexes may have bound to erythrocytes and reduced viral protein in plasma.
Many studies have demonstrated that the p24 antigen is a marker of HIV-1 infection that predicts disease progression and death. And, in HIV-1 subtype B, Up24 can be used to monitor response to ART. Furthermore, recent developments in nanoparticle technology indicate that the compromised specificity of the assay may be an artifact of the technical limitation of RT-PCR. Our results suggest that the diagnostic accuracy of Up24 antigen assay to monitor response to ART in predominantly A, D, and A/D recombinant subtypes is limited by its sensitivity specifically at lower viral loads. As well, there was some disagreement between the Up24 and RT-PCR for detection of negative samples, which may limit the use of the Up24 to detect viral suppression.
ACKNOWLEDGMENTS
Support for this project was provided by the Academic Alliance for AIDS Care and Prevention in Africa; the Infectious Diseases Institute; the Bill and Melinda Gates Foundation; the Laboratory of Immunoregulation, Division of Intramural Research, National Institute of Allergy and Infectious Diseases; and from the National Institutes of Health, K-23 AI060384 research training award. The authors wish to thank the study participants. We also extend our gratitude to the clinical study team at the Infectious Diseases Institute who provided care to the participants including Jane Namukasa, the nurse counselor, Pauline Nabunya, and Diana Kizza.
Financial Support: Support for this project was provided by the Academic Alliance for AIDS Care and Prevention in Africa; the Infectious Diseases Institute; the Bill and Melinda Gates Foundation; the Laboratory of Immunoregulation, Division of Intramural Research, National Institute of Allergy and Infectious Diseases; and from the National Institutes of Health, K-23 AI060384 research training award.
Footnotes
These data were presented at the 43rd Annual Meeting of the Infectious Diseases Society of America in San Francisco, CA, USA, October 6–9, 2005 (Abstract #880).
None of the authors report a potential conflict of interest.
References
- 1.Hammer SM, Eron JJ, Jr, Reiss P, et al. Antiretroviral treatment of adult HIV infection: 2008 recommendations of the International AIDS Society-USA panel. JAMA. 2008;300:555–570. doi: 10.1001/jama.300.5.555. [DOI] [PubMed] [Google Scholar]
- 2.Fiscus SA, Cheng B, Crowe SM, et al. HIV-1 viral load assays for resource-limited settings. PLoS Med. 2006;3:e417. doi: 10.1371/journal.pmed.0030417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schupbach J, Tomasik Z, Knuchel M, et al. Optimized virus disruption improves detection of HIV-1 p24 in particles and uncovers a p24 reactivity in patients with undetectable HIV-1 RNA under long-term HAART. J Med Virol. 2006;78:1003–1010. doi: 10.1002/jmv.20655. [DOI] [PubMed] [Google Scholar]
- 4.Ledergerber B, Flepp M, Boni J, et al. Human immunodeficiency virus type 1 p24 concentration measured by boosted ELISA of heat-denatured plasma correlates with decline in CD4 cells, progression to AIDS, and survival: comparison with viral RNA measurement. J Infect Dis. 2000;181:1280–1288. doi: 10.1086/315366. [DOI] [PubMed] [Google Scholar]
- 5.Sterling TR, Hoover DR, Astemborski J, Vlahov D, Bartlett JG, Schupbach J. Heat-denatured human immunodeficiency virus type 1 protein 24 antigen: prognostic value in adults with early-stage disease. J Infect Dis. 2002;186:1181–1185. doi: 10.1086/343807. [DOI] [PubMed] [Google Scholar]
- 6.Boni J, Opravil M, Tomasik Z, et al. Simple monitoring of antiretroviral therapy with a signal-amplification-boosted HIV-1 p24 antigen assay with heat-denatured plasma. AIDS. 1997;11:F47–F52. doi: 10.1097/00002030-199706000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Respess RA, Cachafeiro A, Withum D, et al. Evaluation of an ultrasensitive p24 antigen assay as a potential alternative to human immunodeficiency virus type 1 RNA viral load assay in resource-limited settings. J Clin Microbiol. 2005;43:506–508. doi: 10.1128/JCM.43.1.506-508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schupbach J, Boni J, Flepp M, Tomasik Z, Joller H, Opravil M. Antiretroviral treatment monitoring with an improved HIV-1 p24 antigen test: an inexpensive alternative to tests for viral RNA. J Med Virol. 2001;65:225–232. doi: 10.1002/jmv.2024. [DOI] [PubMed] [Google Scholar]
- 9.Schupbach J, Gunthard H, Joos B, et al. HIV-1 p24 may persist during long-term highly active antiretroviral therapy, increases little during short treatment breaks, and its rebound after treatment stop correlates with CD4(+) T cell loss. J Acquir Immune Defic Syndr. 2005;40:250–256. doi: 10.1097/01.qai.0000181281.75670.56. [DOI] [PubMed] [Google Scholar]
- 10.Erikstrup C, Kallestrup P, Zinyama-Gutsire RB, et al. p24 as a predictor of mortality in a cohort of HIV-1-infected adults in rural Africa. J Acquir Immune Defic Syndr. 2008;48:345–349. doi: 10.1097/QAI.0b013e31817dc3d1. [DOI] [PubMed] [Google Scholar]
- 11.Spacek LA, Henson C, Gray RH, Wawer MJ, Serwadda D, Sewankambo N, Schupbach J, Laeyendecker O, Quinn TC. Evaluation of heat-denatured HIV-1 p24 antigen as an alternative, inexpensive marker for HIV-1 RNA viral load in non-B HIV-1 subtypes in sub-Saharan Africa. 10th Conference on Retroviruses and Opportunistic Infections. 2003 [Google Scholar]
- 12.Stevens G, Rekhviashvili N, Scott LE, Gonin R, Stevens W. Evaluation of two commercially available, inexpensive alternative assays used for assessing viral load in a cohort of human immunodeficiency virus type 1 subtype C-infected patients from South Africa. J Clin Microbiol. 2005;43:857–861. doi: 10.1128/JCM.43.2.857-861.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pascual A, Cachafeiro A, Funk ML, Fiscus SA. Comparison of an assay using signal amplification of the heat-dissociated p24 antigen with the Roche Monitor human immunodeficiency virus RNA assay. J Clin Microbiol. 2002;40:2472–2475. doi: 10.1128/JCM.40.7.2472-2475.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonard D, Rouet F, Toni TA, et al. Field evaluation of an improved assay using a heat-dissociated p24 antigen for adults mainly infected with HIV-1 CRF02_AG strains in Cote d'Ivoire, West Africa. J Acquir Immune Defic Syndr. 2003;34:267–273. doi: 10.1097/00126334-200311010-00002. [DOI] [PubMed] [Google Scholar]
- 15.Lombart JP, Vray M, Kafando A, et al. Plasma virion reverse transcriptase activity and heat dissociation-boosted p24 assay for HIV load in Burkina Faso, West Africa. AIDS. 2005;19:1273–1277. doi: 10.1097/01.aids.0000180098.58017.48. [DOI] [PubMed] [Google Scholar]
- 16.Ondoa P, Dieye TN, Vereecken C, et al. Evaluation of HIV-1 p24 antigenemia and level of CD8+CD38+ T cells as surrogate markers of HIV-1 RNA viral load in HIV-1-infected patients in Dakar, Senegal. J Acquir Immune Defic Syndr. 2006;41:416–424. doi: 10.1097/01.qai.0000209901.12750.d0. [DOI] [PubMed] [Google Scholar]
- 17.Guay LA, Hom DL, Kabengera SR, et al. HIV-1 ICD p24 antigen detection in ugandan infants: use in early diagnosis of infection and as a marker of disease progression. J Med Virol. 2000;62:426–434. doi: 10.1002/1096-9071(200012)62:4<426::aid-jmv6>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 18.Lyamuya E, Bredberg-Raden U, Massawe A, et al. Performance of a modified HIV-1 p24 antigen assay for early diagnosis of HIV-1 infection in infants and prediction of mother-to-infant transmission of HIV-1 in Dar es Salaam, Tanzania. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;12:421–426. doi: 10.1097/00042560-199608010-00014. [DOI] [PubMed] [Google Scholar]
- 19.De Baets AJ, Edidi BS, Kasali MJ, et al. Pediatric human immunodeficiency virus screening in an African district hospital. Clin Diagn Lab Immunol. 2005;12:86–92. doi: 10.1128/CDLI.12.1.86-92.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiscus SA, Pilcher CD, Miller WC, et al. Rapid, real-time detection of acute HIV infection in patients in Africa. J Infect Dis. 2007;195:416–424. doi: 10.1086/510755. [DOI] [PubMed] [Google Scholar]
- 21.Herbeck JT, Lyagoba F, Moore SW, et al. Prevalence and genetic diversity of HIV type 1 subtypes A and D in women attending antenatal clinics in Uganda. AIDS Res Hum Retroviruses. 2007;23:755–760. doi: 10.1089/aid.2006.0237.A. [DOI] [PubMed] [Google Scholar]
- 22.Kamya MR, Mayanja-Kizza H, Kambugu A, et al. Predictors of long-term viral failure among ugandan children and adults treated with antiretroviral therapy. J Acquir Immune Defic Syndr. 2007;46:187–193. doi: 10.1097/QAI.0b013e31814278c0. [DOI] [PubMed] [Google Scholar]
- 23.Schockmel GA, Yerly S, Perrin L. Detection of low HIV-1 RNA levels in plasma. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;14:179–183. doi: 10.1097/00042560-199702010-00013. [DOI] [PubMed] [Google Scholar]
- 24.Schupbach J, Boni J, Bisset LR, et al. HIV-1 p24 antigen is a significant inverse correlate of CD4 T-cell change in patients with suppressed viremia under long-term antiretroviral therapy. J Acquir Immune Defic Syndr. 2003;33:292–299. doi: 10.1097/00126334-200307010-00002. [DOI] [PubMed] [Google Scholar]
- 25.Schupbach J, Flepp M, Pontelli D, Tomasik Z, Luthy R, Boni J. Heat-mediated immune complex dissociation and enzyme-linked immunosorbent assay signal amplification render p24 antigen detection in plasma as sensitive as HIV-1 RNA detection by polymerase chain reaction. AIDS. 1996;10:1085–1090. [PubMed] [Google Scholar]
- 26.Ribas SG, Ondoa P, Schupbach J, van der GG, Fransen K. Performance of a quantitative human immunodeficiency virus type 1 p24 antigen assay on various HIV-1 subtypes for the follow-up of human immunodeficiency type 1 seropositive individuals. J Virol Methods. 2003;113:29–34. doi: 10.1016/s0166-0934(03)00219-2. [DOI] [PubMed] [Google Scholar]
- 27.Kim EY, Stanton J, Korber BT, et al. Detection of HIV-1 p24 Gag in plasma by a nanoparticle-based bio-barcode-amplification method. Nanomed. 2008;3:293–303. doi: 10.2217/17435889.3.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tang S, Zhao J, Storhoff JJ, et al. Nanoparticle-Based biobarcode amplification assay (BCA) for sensitive and early detection of human immunodeficiency type 1 capsid (p24) antigen. J Acquir Immune Defic Syndr. 2007;46:231–237. doi: 10.1097/QAI.0b013e31814a554b. [DOI] [PubMed] [Google Scholar]
- 29.Jennings C, Fiscus SA, Crowe SM, et al. Comparison of two human immunodeficiency virus (HIV) RNA surrogate assays to the standard HIV RNA assay. J Clin Microbiol. 2005;43:5950–5956. doi: 10.1128/JCM.43.12.5950-5956.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nadal D, Boni J, Kind C, et al. Prospective evaluation of amplification-boosted ELISA for heat-denatured p24 antigen for diagnosis and monitoring of pediatric human immunodeficiency virus type 1 infection. J Infect Dis. 1999;180:1089–1095. doi: 10.1086/315012. [DOI] [PubMed] [Google Scholar]
- 31.Schupbach J. Measurement of HIV-1 p24 antigen by signal-amplification-boosted ELISA of heat-denatured plasma is a simple and inexpensive alternative to tests for viral RNA. AIDS Rev. 2002;4:83–92. [PubMed] [Google Scholar]
- 32.Prado JG, Shintani A, Bofill M, Clotet B, Ruiz L, Martinez-Picado J. Lack of longitudinal intrapatient correlation between p24 antigenemia and levels of human immunodeficiency virus (HIV) type 1 RNA in patients with chronic hiv infection during structured treatment interruptions. J Clin Microbiol. 2004;42:1620–1625. doi: 10.1128/JCM.42.4.1620-1625.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgisser P, Vernazza P, Flepp M, et al. Performance of five different assays for the quantification of viral load in persons infected with various subtypes of HIV-1. Swiss HIV Cohort Study. J Acquir Immune Defic Syndr. 2000;23:138–144. doi: 10.1097/00126334-200002010-00005. [DOI] [PubMed] [Google Scholar]