Abstract
Investigation of bacteria involved in pathogenesis of necrotizing enterocolitis (NEC) is limited by infant fragility, analysis restricted to feces, use of culture-based methods and lack of clinically-relevant animal models. This study used a unique preterm piglet model to characterize spontaneous differences in microbiome composition of NEC-predisposed regions of gut. Preterm piglets (n = 23) were cesarean-delivered and nurtured for 30 h over which time 52% developed NEC. Bacterial DNA from ileal content, ileal mucosa and colonic mucosa were PCR amplified, subjected to terminal restriction fragment length polymorphism (TRFLP) analysis and targeted 16s rDNA qPCR. Preterm ileal mucosa was specifically bereft in diversity of bacteria compared to ileal content and colonic mucosa. Preterm ileum was restricted to representation by only Proteobacteria, Firmicutes, Cyanobacteria and Chloroflexi. In piglets with NEC, ileal mucosa was uniquely characterized by increases in number of Firmicutes and diversity of phyla to include Actinobacteria and uncultured bacteria. Five specific TRFLP profiles, corresponding in closest identity to Clostridium butyricum, C. neonatale, C. proteolyticum, Streptomyces spp. and Leptolyngbya spp., were significantly more prevalent or observed only among samples from piglets with NEC. Total numbers of Clostridium spp. and C. butyricum were significantly greater in samples of NEC ileal mucosa but not ileal content or colonic mucosa. These results provide strong support for ileal mucosa as a focus for investigation of specific dysbiosis associated with NEC and suggest a significant role for Clostridium spp., and members of the Actinobacteria and Cyanobacteria in the pathogenesis of NEC in preterm piglets.
Key words: microbiome, Clostridium spp., animal model, NEC, necrotizing, intestine, profile
Introduction
Necrotizing enterocolitis (NEC) is a tragic complication of prematurity wherein approximately 10%1,2 of very low birth weight infants (<1,500 g) develop inexorable clinical signs of abdominal distension, bloody stools and mucosal necrosis of the ileum or proximal colon that oftentimes culminates in perforation. An estimated 30% of infants with NEC die.2 Despite being first described over a century ago, mortality from NEC has not changed perceptibly since 1997,1 until recently when probiotics were recognized as significantly protective against NEC.3
Bacterial colonization of the premature gut is recognized as the firebrand of NEC. How these conditions interact to instigate necrosis of the intestine in these infants is unknown, but the role of bacteria appears to be essential. Progress in characterization of the bacteria involved and their role in disease pathogenesis has been severely limited by the fragility of these infants and lack of clinically-relevant animal models of NEC. Consequently, all bacteriological studies of NEC infants to date have relied on analysis of feces4–8 or nasoduodenal tube aspirates4 as a surrogate for the offending microbiota, despite compelling evidence of immense variation in bacterial community composition along and within the lumen of the GI tract.9,10 Accordingly, more insightful data is likely to be gained by characterizing bacteria within sections of bowel afflicted by NEC such as the ileum; a location difficult to evaluate in human infants.
Until recently, all knowledge of NEC-associated bacteria was based on classical culture of feces, blood or peritoneal fluid from these infants.5–8 A recurring theme among many of these studies is the frequent presence of Clostridium spp. on the one hand or Enterobacteriaceae on the other. Since recognition that roughly 80% of all microbial members cannot be cultivated outside the intestinal tract,10 only 4 studies5–8 have applied modern molecular techniques to identifying the bacterial community composition of feces from infants with NEC. While inarguably state-of-the-art, the collective results of these studies appear as dichotomous in their findings as those of prior fecal culture studies in providing no consensus of the bacterial community characteristic of NEC. These findings could be interpreted to mean that there is no unifying microbiota associated of NEC. However, studies on human infants with NEC continue to unavoidably suffer from the limitations of fecal analysis and remain confounded by variables with recognized impact on the intestinal bacterial community composition including differences in mode of delivery, gestational age, type of enteral nutrition, concurrent disease and treatment with antibiotics.11
Originally described by researchers in Denmark,12 the piglet is the only animal model to spontaneously develop full-scale clinical NEC that is contingent on the same biological conditions that predispose human infants including prematurity, fortuitous bacterial colonization and feeding of enteral formula.12,13 Using this unique model, the present study is the first to perform a molecular-based analysis of the surface mucosal and lumen bacterial communities directly associated with NEC lesions. Piglets were delivered preterm by cesarean section, fed formula by means of an orogastric tube and nurtured akin to human infants for 30 h over which time 52% of piglets developed clinical, gross pathological and histological disease consistent with NEC.
Our findings show that the preterm ileum is uniquely bereft in number and diversity of mucosa-associated bacteria and is a focus of dysbiosis associated with NEC. Specifically, we observed an increase in bacterial diversity of NEC ileal mucosa characterized by increases in the number of Firmicutes and presence of Actinobacteria and uncharacterized bacteria. Five specific TRFLP profiles corresponding in closest identity to Clostridium butyricum, Clostridium neonatale, Clostridium proteolyticum and members of the Actinobacteria and Cyanobacteria were significantly more prevalent or observed only among samples from piglets with NEC. We hypothesize that this bacterial consortium has unique metabolic characteristics that are held in common with other bacterial communities implicated in the pathogenesis of NEC and that the preterm piglet model is likely to represent a powerful tool with which to characterize the metagenomic and metabolomic signature of NEC ileum.
Results
Preterm piglets delivered by cesarean section and fed infant formula develop clinical, radiographic, gross pathologic and histopathologic signs of NEC.
A total of 29 piglets were delivered by cesarean section from 2 sows at 106 d of gestation. Six piglets failed to thrive and died or were euthanized within 4 h of birth. Among the 23 surviving piglets, 12 (52%) developed NEC based on clinical (n = 11), radiographic (n = 8), gross pathologic (n = 11) and histopathologic (n = 12) criteria an average of 24.2 ± 1.2 h after initiation of enteral feeding (range, 16–30 h) (Fig. 1). Only one piglet developing NEC survived to the designated 30 h endpoint of the study. All other piglets developing NEC required humane euthanasia (n = 8) or died unexpectedly (n = 3) due to acute onset of severe abdominal distension and cardiopulmonary failure. Eleven piglets were determined to not have NEC based on the presence of none (n = 8) or only 1 diagnostic criteria (n = 3) and all of these piglets survived to the endpoint of the study. Aside from changes associated with the per-acute onset of NEC, we observed no predictive differences in sex, body weight, rectal temperature, pulse or respiratory rate, pre-feeding gastric residual volume, mentation or fecal consistency between piglets that did or did not develop NEC.
Figure 1.
Criteria for diagnosis of NEC in preterm, cesarean-derived and formula-fed piglets. Criteria for NEC included severe abdominal distension, gaseous distention of the intestines (A; closed arrow = gas in portal vein; open arrow = orogastric feeding tube), hemorrhagic discoloration and sloughing or ulceration of the small or large bowel and atrophy, necrosis and sloughing of epithelium (B).
NEC is associated with small intestinal villous atrophy and sloughing of the intestinal epithelium.
As a means to quantify the magnitude of intestinal mucosal injury observed in piglets diagnosed with NEC, light microscopic measures of mucosal atrophy and denudation were performed on histological specimens of distal ileal and proximal colonic mucosa. Specimens from piglets with NEC were characterized by significant atrophy of ileal villi and sloughing of intestinal epithelium from both the ileal and colonic mucosa (Fig. S1).
NEC lesions are not characterized by significant differences in the gram stain identity, location along the crypt-villus axis or linear density of ileal mucosa-associated bacteria.
To determine if ileal lesions from piglets with NEC differed significantly in the Gram stain identity, villus versus crypt location or linear density of mucosa-associated bacteria compared to piglets without NEC, two-dimensional counts of bacteria were performed on Gram stained histological specimens from piglets with and without NEC. No significant or insightful differences were observed between specimens from piglets with or without NEC (Table S1). In both groups, the villous epithelium was characterized by significantly greater numbers and/or linear density of Gram negative compared to Gram positive bacteria. In NEC, Gram positive and negative bacteria tended to associate in greater density with the denuded versus epithelialized surface of the villi, however this did not reach statistical significance.
The intestinal tract of preterm piglets is initially colonized by a low diversity of mucosa-associated and lumen-dwelling bacteria.
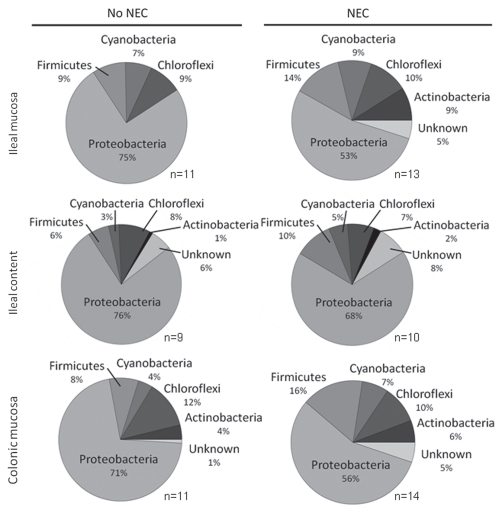
As a means to further characterize differences in the bacterial community composition between piglets with and without NEC, TRFLP analysis was performed using DNA extracted from mucosal scrapings of the terminal ileum, proximal colon and distal small intestinal content from each piglet. From total DNA, a 1.5 kb fragment of the bacterial 16S ribosomal RNA gene was amplified and digested using 3 restriction enzymes. The resulting fragment profiles were tentatively assigned to specific phylogenetic bacterial groups. Fragments were not produced using samples of ileal and colonic mucosa from piglets (n = 6) that failed to thrive and died within 4 h of birth, suggesting a lack of significant bacterial colonization of the intestine within the first few hours of birth (data not shown). For piglets that did not develop NEC, a total of 71 unique TRFLP profiles were identified. In general, a significantly greater number of profiles were observed in the ileal content and colonic mucosa compared to the ileal mucosa of preterm piglets (Table 1). The TRFLP profiles were identified as representative of 4 core bacterial phyla including the Proteobacteria, Firmicutes, Cyanobacteria and Chloroflexi. Compared to ileal mucosa, ileal lumen content and colonic mucosa also contained profiles tentatively assigned to Actinobacteria as well as profiles predictive of uncultured bacteria of undetermined phylotypes (Fig. 2). A group of core TRFLP profiles were detected most frequently among samples from piglets independent of the presence or absence of NEC. Table 2 lists the closest bacterial phylotypes assigned to those profiles and their phylum.
Table 1.
Number and diversity of unique TRFLP patterns identified in ileal mucosa, ileal lumen content and colonic mucosa of piglets that did or did not develop NEC
| Location | No NEC | NEC | ||||||||
| No. samples | Unique TRFLP patterns | Diversity† index | No. samples | Unique TRFLP patterns | Diversity† index | |||||
| Mean ± SD | Range | Total | Mean ± SD | Mean ± SD | Range | Total | Mean ± SD | |||
| Ileal mucosa | 11 | 5.1 ± 3.2 | 2–12 | 15 | 1.5 ± 0.6 | 13 | 9.5 ± 4.4** | 0–14 | 38 | 2.2 ± 0.5* |
| Ileal content | 9 | 11.8 ± 5.8** | 6–23 | 51 | 2.4 ± 0.4 | 10 | 9.9 ± 6.5 | 1–20 | 42 | 2.0 ± 0.9 |
| Colonic mucosa | 11 | 10.0 ± 6.3* | 0–21 | 39 | 2.2 ± 0.6 | 14 | 11.6 ± 4.0 | 4–17 | 40 | 2.4 ± 0.4 |
*p < 0.05, **p < 0.01 versus same parameter for no NEC. †Shannon-Wiener diversity index was calculated as follows: H' = − ∑(pi)(log2 pi), here p is the number of unique phyla relative to the sum of all unique phyla in the sample.
Figure 2.
Phylum-associated unique TRFLP profiles expressed as percent of total unique TRFLP profiles identified by disease condition (NEC or No NEC) and sampling site (ileal mucosa, ileal lumen content and colonic mucosa) of piglets. The analysis shows a very highly diverse population of Proteobacteria, represented by the number of unique patterns associated with this phylum, compared to the rest of detected phyla. n = number of piglets.
Table 2.
Prevalence of unique TRFLP patterns identified are core representatives of the microbiome of the ileal mucosa, ileal lumen content and colonic mucosa of piglets independent of the development of necrotizing enterocolitis (NEC)
| Phylum | Class | Closest relative | Percentage of samples in which each pattern was identified | |||||
| Ileal mucosa | Ileal content | Colonic mucosa | ||||||
| No NEC n = 11 |
NEC n = 13 |
No NEC n = 9 |
NEC n = 10 |
No NEC n = 11 |
NEC n = 14 | |||
| Chloroflexi | Anaerolineae/Chloroflexi | Uncultured/Chloroflexi | 45.5 | 69.2 | 66.7 | 60.0 | 63.6 | 71.4 |
| Cyanobacteria | Cyanobacteria | Nostoc | 36.4 | 46.2 | 22.2 | 40.0 | 18.2 | 21.4 |
| Firmicutes | Clostridia | Clostridium tyrobutyricum | 36.4 | 46.2 | 22.2 | 50.0 | 18.2 | 21.4 |
| Clostridium algidicarnis | 9.1 | 7.7 | 0.0 | 0.0 | 9.1 | 35.7 | ||
| Proteobacteria | Gammaproteobacteria | Alkanindiges | 72.7 | 76.9 | 88.9 | 60.0 | 72.7 | 71.4 |
| Uncultured/Alkanindiges/Acinetobacter | 0.0 | 0.0 | 11.1 | 0.0 | 36.4 | 21.4 | ||
| Uncultured/Microbulbife | 0.0 | 0.0 | 0.0 | 0.0 | 45.5 | 42.9 | ||
| Neptuniibacter | 18.2 | 46.2 | 11.1 | 20.0 | 27.3 | 57.1 | ||
| Uncultured/Photobacterium/Hyphomicrobium sp. | 27.3 | 53.8 | 11.1 | 20.0 | 27.3 | 64.3 | ||
| Klebsiella/Salmonella/Erwinia | 100.0 | 92.3 | 55.6 | 70.0 | 90.9 | 100.0 | ||
| Serratia | 0.0 | 0.0 | 22.2 | 0.0 | 36.4 | 28.6 | ||
| Uncultured/Methylococcaceae | 9.1 | 23.1 | 11.1 | 20.0 | 54.5 | 64.3 | ||
| Betaproteobacteria | Uncultured/Nitrosomonas/Nitrospira | 90.9 | 92.3 | 88.9 | 100 | 63.6 | 100 | |
| Uncultured/Gallionellaceae/Rhodocyclaceae | 9.1 | 15.4 | 11.1 | 0.0 | 54.5 | 21.4 | ||
n = number of samples. Patterns represented were identified in ≥ 3 piglets within a given location per disease condition.
NEC is associated with increased diversity of phylotypes that associate with the ileal mucosa.
In piglets that developed NEC, a total of 77 unique TRFLP profiles were identified. The main difference between piglets with NEC compared to those without NEC was a significant increase in diversity of the ileal mucosa microbiota (Table 1). Compared to piglets without NEC, the ileal mucosa of piglets with NEC showed profiles tentatively assigned to Actinobacteria and unknown uncultured bacteria. As such, the ileum of piglets with NEC more closely resembled, in proportions of represented phyla, those characteristic of ileal lumen content and colonic mucosa (Fig. 2). In addition to the appearance of patterns associated to Actinobacteria and unknown phyla, the ileal mucosa of piglets with NEC had a significant increase in the number of patterns belonging to the Firmicutes. No significant differences in diversity of the ileal lumen content or colonic mucosa microbiota were observed between piglets that developed NEC compared to those that did not (Table 1).
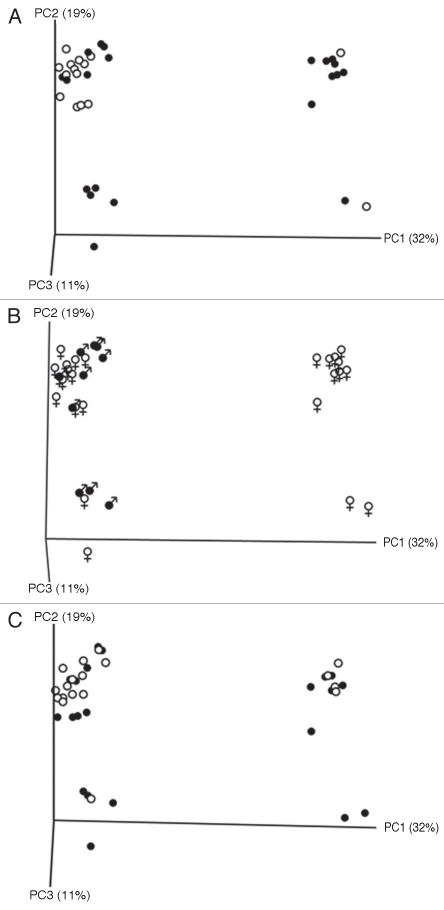
Principal Coordinate Analysis of TRFLP data, illustrated that the first three principal coordinate components accounted for 62% of the variation between all samples. Identification of individual samples on the basis of NEC diagnosis and sex of the piglets, but not sample site, revealed close associations with variation observed within the intestinal microbiome (Fig. 3).
Figure 3.
Principal Coordinate Analysis plots of unique TRFLP patterns as grouped by disease condition (A; 22 no NEC [white] and 27 NEC [black] samples), sex of piglet (B; 36 female [white] and 11 male [black] samples) and sample location (C; 24 ileum [white] and 25 colon [black] mucosa).
Five specific TRFLP profiles are significantly more prevalent in piglets with NEC.
Five unique profiles were identified as being significantly more prevalent or observed only among samples from piglets with NEC (Table 3). Three of the identified profiles were tentatively assigned to Clostridia and their closest relatives were C. butyricum, C. proteolyticum and an uncultured bacterium (SJTU F0168; GenBank EF398279), which was later identified as Clostridium neonatale (GenBank AF275949), a novel species of Clostridium first isolated from the blood of neonates during an intensive care unit outbreak of NEC.14 All three Clostridium spp. phylotypes were largely absent in piglets without NEC and significantly overrepresented among samples of colonic mucosa from piglets with NEC. The remaining two TRFLP profiles were assigned to the classes Actinobacteria and Cyanobacteria and were significantly more prevalent among samples of ileal and colonic mucosa from piglets with NEC (Table 3). Irrespective of sampling site, two or more of these five TRFLP profiles were identified in 11/12 (92%) piglets with NEC (avg. no. of profiles = 3.3**, range, 0–5) and only 2/11 (18%) piglets that did not have NEC (avg. no. of profiles = 0.73, range, 0–3; **p < 0.01).
Table 3.
Unique TRFLP patterns identified as significantly more prevalent or observed only among samples from piglets with NEC
| Phylum | Class | Closest relative | Percentage of samples in which each pattern was identified | |||||
| Ileal mucosa | Ileal content | Colonic mucosa | ||||||
| No NEC n = 11 |
NEC n = 13 |
No NEC n = 9 |
NEC n = 10 |
No NEC n = 11 |
NEC n = 14 | |||
| Actinobacteria | Actinobacteria | Streptomyces | 0.0 | 61.5** | 11.1 | 20.0 | 18.2 | 28.6 |
| Cyanobacteria | Cyanobacteria | Leptolyngbya | 0.0 | 38.50.07,* | 11.1 | 10.0 | 18.2 | 64.30.06,* |
| Firmicutes | Clostridia | Clostridium butyricum | 0.0 | 23.1 | 0.0 | 10.0 | 0.0 | 42.9* |
| Clostridium proteolyticum | 0.0 | 23.1 | 11.1 | 10.0 | 9.1 | 50.00.08,* | ||
| Clostridium neonatale Uncultured bacterium SJTU F 01 68 (GenBank EF398297) | 0.0 | 23.1 | 0.0 | 10.0 | 0.0 | 42.9* | ||
n = number of samples. *p < 0.05 (except where indicated); **p < 0.01.
NEC is associated with significant increases in ileal mucosa-associated total clostridium spp. including C. butyricum and C. proteolyticum.
In addition to identifying TRFLP profiles that were common or differed significantly in prevalence between samples from piglets with or without NEC, we selected specific phylotypes for quantitative analysis by qPCR (Fig. 4). There was no significant difference in total Eubacteria between piglets with and without NEC; however, as expected, significantly greater quantities of total Eubacteria were present in samples of colonic mucosa and ileal content compared to those obtained from ileal mucosa (Fig. 4A). Among the unique TRFLP profiles identified as more prevalent among piglets with NEC (Table 3), concurrent significant increases in bacterial numbers were observed for total Clostridium spp. and C. butyricum in the ileal mucosa of piglets with NEC (Fig. 4B). In addition, the average number of C. proteolyticum and C. neonatale were increased by one order of magnitude (from 2.3 × 101 to 3.8 × 102 and 3.5 × 103 to 5 × 104, respectively) in the ileal mucosa of piglets with NEC.
Figure 4.
Quantitative PCR for total Eubacterial 16s rDNA (A; *p < 0.05 versus ileal mucosa) and Clostridium genus and species-specific 16s rDNA (B; *p < 0.05 versus no NEC). Number of samples = ileal mucosa (no NEC, 11; NEC, 13), ileal content (no NEC, 9; NEC, 10) and colonic mucosa (no NEC, 11; NEC, 14). Data represent median, 10th, 25th, 75th and 90th percentiles.
Among the core group of TRFLP profiles whose prevalence appeared to be unassociated with NEC (Table 2), we observed a significant decrease in the number of bacteria represented within the groups Klebsiella/Salmonella/Erwinia and Photobacterium/Hypomicrobium as well as species of the family Methylococcaceae in the colonic mucosa of piglets with NEC.
Discussion
The gastrointestinal tract is sterile at birth and rapidly colonized by bacteria from the mother and environment. Relative to healthy full-term infants, the fecal flora of preterm infants is distinguished by a lower number and diversity of bacteria.5,7,8,15 Findings in the present study mirror these observations. The number of unique profiles identified by TRFLP in our preterm piglets ranged from 0 to 23. By sampling regions of the gut specifically predisposed to development of NEC, we have additionally demonstrated that preterm ileal mucosa is uniquely bereft in both number and diversity of bacteria compared to the ileal content and colonic mucosa.
There is little consensus on the identity of bacterial communities that pioneer the intestinal tract of preterm infants. Using similar approaches as reported here, feces from preterm infants were represented by Proteobacteria, Firmicutes, Fusobacteria and Bacteroidetes.8 The bacterial community within the preterm piglet gut was also limited and represented by Proteobacteria and Firmicutes but alternative representation by Chloroflexi and Cyanobacteria. Moreover, among NEC-predisposed regions of the gut, additional bacterial communities were observed for ileal content and colonic mucosa that were lacking in ileal mucosa. These additional bacterial communities were represented by Actinobacteria and unknown uncultured bacteria. Whether these phyla are unique to the piglet model or our study environment is unknown because comparable samples of NEC-predisposed regions of gut from preterm human infants have yet to be characterized. It is important to point out that TRFLP does not quantify the number of bacteria within each of the represented phyla. Rather, these data impart information on the diversity of unique patterns identified within each phyla. In addition, resolution of these patterns to species-level identity is less than can be achieved with approaches such as phylochip arrays or pyrosequencing and depends on the choice of primer and restriction enzyme combination as well as the influence of potential bias at the level of experimental design, sampling and storage protocols, DNA isolation and PCR amplification.16 PCR primers targeting amplification of the bacterial 16S rRNA gene have shown different efficiencies and potential mismatches with newly discovered strains or environmental 16S rRNA gene sequences.17 The universal primers selected for TRFLP in this study detected a wide range of bacterial phylotypes; however, qPCR with phylotype-specific primers confirmed the limitations of the selected universal primers, which do not anneal equally well to all microbial 16S rDNA hence failing to amplify all bacteria present in the sample.
Investigators have long sought to identify changes in the number, composition or identity of fecal bacteria that correspond with development of NEC. Despite the advent of modern molecular approaches, there is considerable discrepancy between studies in the reported differences in bacterial communities between feces from infants with and without NEC. Compared to bacteria from preterm infants without NEC, fecal bacteria from those with NEC have ranged from indistinguishable6 to dominated by a particular group such as the proteobacteria8 or clostridia,7 or represented by specific species of bacteria.5 This variation may be attributed in part to unavoidable influences of the environment, mode of delivery, gestational age, type of enteral nutrition, concurrent disease or treatment with antibiotics.11 Alternatively, there may be no universal signature of NEC at the level of the microbiome.
Under the controlled conditions of the present study, NEC in preterm piglets was associated significantly and specifically with a spontaneous change in the bacterial community composition of ileal mucosa. In particular, the ileal mucosa of piglets with NEC acquired an increase in the number and diversity of bacterial phylotypes. Our analysis demonstrated increases in the number of Firmicutes and acquisition of Actinobacteria and bacteria of unknown identity. As such, the bacterial community of NEC ileal mucosa adopted a composition more similar to the ileal content and colonic mucosa of normal preterm piglets. It is tempting to interpret these observations as indicative of bacterial overgrowth of the ileal mucosa in infants with NEC. However, NEC ileum was not associated with an increase in total eubacteria as quantified by either 16S rDNA qPCR or linear density of bacteria in contact with the mucosa. Accordingly, these results more aptly support a change in composition of the ileal mucosaassociated microbiota rather than an increase in the number of total bacteria. This interpretation is further supported by a strong relationship between differences in the microbiota composition and presence or absence of NEC as demonstrated by PCoA. The microbiota composition was also suggested by PCoA to be related to gender, however there were too few male piglets in the study to draw this conclusion.
We next examined the community of TRFLP profiles from each NEC-predisposed location for differences in specific bacterial identities between piglets with and without NEC. Five specific TRFLP profiles were significantly more prevalent or observed only among samples from piglets with NEC. These corresponded in their closest identity to Clostridium butyricum, Clostridium neonatale, Clostridium proteolyticum, Streptomyces spp., and Leptolyngbya spp.. Two or more of these five TRFLP profiles were identified in 11/12 piglets with NEC compared to only 2/11 piglets that did not have NEC. With the exception of Streptomyces spp., these bacteria corresponded in membership to phyla significantly increased in representation (Firmicutes) or newly acquired (Actinobacteria) by the ileal mucosa of piglets with NEC. In addition to their overrepresentation among samples from piglets with NEC, the total number of Clostridium spp. and Clostridium butyricum in particular were significantly and specifically greater in samples of NEC ileal mucosa but not ileal content or colonic mucosa as determined by means of qPCR.
It remains unclear how C. butyricum, C. neonatale, C. proteolyticum and members of Actinobacteria and Cyanobacteria contributed to NEC in the present study. It is additionally unclear if these bacteria are a cause versus consequence of NEC in these piglets. Neither the Actinobacteria or Cyanobacteria have previously been associated with NEC, however members of each phylum have interesting metabolic, cytotoxic and antimicrobial attributes. A controversial role for clostridia in pathogenesis of NEC dates back to the 1970's.7,14,18–22 Clostridia are reported to be associated with NEC more frequently than any other group of bacteria. Interpretation of these studies however has been mired by inconsistent use of anaerobic cultures, the frequent isolation of clostridia from infants without NEC, variation in the species of clostridia associated with NEC, and frequent reports of NEC in the absence of clostridia. The advent of culture-independent analyses of fecal bacteria from human infants with NEC5–8 has not resolved this controversy. In one report, NEC was exclusively associated with the presence of C. perfringens7 while in another very few matches to the entire genus of clostridia were detected out of more than 10,000 16S rDNA fragment sequences.5 While it is possible that clostridia were confined to NEC-predisposed regions of gut rather than feces of these infants, it is far more probable that other populations of bacteria are also capable of instigating NEC, particularly members of the Enterobacteriaceae.8 It is noteworthy that C. butyricum isolated from infants with NEC is alone sufficient to induce disease in gnotobiotic chicken23 and quail models24 of NEC. A growing list of attributes of C. butyricum are being associated with NEC including neuraminidase activity,24,25 lactose fermentation,14,24 production of butyric acid26 and release of soluble cytotoxic factors.21
The preterm piglet may be particularly useful model for studying the pathophysiology of Clostridium spp.-associated NEC. The presence of clostridia is frequently associated with acute outbreaks and more lethal courses of NEC in human infants.14,18,19,21,27 This was also true of the present study in which all but one piglet with NEC had acute clinical disease and died or had to be euthanized prior to the 30 h endpoint of the study. TRFLP patterns from small intestinal mucosa consistent with Clostridium spp. were also exclusively demonstrated in preterm piglets with NEC in prior studies,12,13 however resolution of specific species was not possible due to inclusion of only one restriction enzyme.28
In light of the controlled conditions, comparability of the model system to human disease, and novel focus on spontaneous changes in the precocious microbiome of NEC-predisposed regions of gut, results of the present study provide strong support for the ileal mucosa as a focus for investigation of specific dysbioses associated with NEC. Further, our results support a significant role for colonization by Clostridium spp., and members of the Actinobacteria and Cyanobacteria in the pathogenesis of NEC in preterm piglets. Whether these bacteria represent primary pathogenic culprits or secondary consequences of NEC remains unclear. We hypothesize that this bacterial consortium has unique metabolic characteristics that are held in common with other bacterial communities implicated in the pathogenesis of NEC. The preterm piglet is likely to represent a powerful model system in which to characterize the metagenomic and metabolomic signature of NEC ileum.
Materials and Methods
Piglet model of NEC.
Piglets were delivered by cesarean section from two sows at 106 d of gestation (term = 115 d). At this time, gastrointestinal maturation of the piglet is comparable to a pre-term human newborn. Sows were sedated (medetomidine, 20–50 µg/kg; ketamine, 5–20 µg/kg intramuscular) and anesthesia was induced with propofol (2–6 mg/kg intravenous) and maintained with sevofluorane. Piglets were delivered by flank laparotomy.
Piglets were stimulated by rubbing and provided 100% oxygen by face mask if needed until consistent respirations were observed. An orogastric feeding tube was placed percutaneously to exit through the cheek and each piglet was fed 10 ml/kg of warmed formula (Table S2) through the tube every 3 h. Piglets were identified by numbered collars and group-housed on heated blankets in a temperature and light-controlled and humidified laboratory animal facility. Piglets were monitored continuously during which time the rectal temperature, pulse and respiration rate, fecal consistency, mentation, gastric residual volume and severity of abdominal distension were recorded at each feeding. A 30 h study duration was chosen based on a prior report that 57% of pre-term piglets will develop NEC within 20–30 h of initiating oral feeding.12,29 Piglets developing clinical signs of NEC prior to and those remaining alive at 30 h were sedated with ketamine (15 mg IM) and then euthanized by intracardiac injection of pentabarbitone (65 mg/kg). All studies were approved by the Institutional Animal Care and Use Committee.
Diagnosis of NEC.
Piglets were diagnosed with NEC based on histopathology findings of severe villous atrophy, epithelial sloughing, and/or necrosis of ileal and colonic mucosa and the presence of 2 out of 3 supportive findings including pre-mortem clinical signs, radiographic findings and gross pathologic appearance consistent with NEC. In all cases, pre-mortem clinical signs of NEC were identified as acute and severe abdominal distension that was associated with rapid death or prompted humane euthanasia. A lateral and ventrodorsal abdominal radiograph of each piglet was taken after euthanasia and examined in blinded fashion by an ACVR certified veterinary radiologist for signs consistent with NEC based on the presence, amount and distribution of gas.30 Gross pathological evidence of NEC included gaseous distension of the small and large bowel, hemorrhagic discoloration of intestines and sloughing or ulceration of the intestinal mucosa.
Histomorphometry.
Immediately after euthanasia the intestines were removed and opened lengthwise. Full thickness sections of distal ileum and proximal colonic mucosa were gently rinsed in porcine Ringer's solution to remove macroscopic debris and fixed in 4% neutral buffered formalin. Strips of the mucosa were embedded in paraffin, sectioned at a thickness of 5 µm and stained with hematoxylin and eosin for light microscopy. For each piglet, 3–5 well-oriented villus-crypt units were selected for measurement using an ocular micrometer. Measurements included villus height, crypt depth, total linear surface of villus and linear surface of denuded villus, colonic mucosal thickness from the crypt base to surface and total length of epithelialized and denuded crypt plus surface epithelium. All measurements were performed by an investigator blinded to each sample identity.
Enumeration of mucosa-associated gram positive and gram negative bacteria.
The number of Gram positive and Gram negative bacteria in direct or contiguous contact with the epithelium were counted by an investigator blinded to each sample identity. Serial sections of ileal mucosa were stained with Hucker's crystal violet and Lugol's iodine, decolorized in acetone and counterstained with safranin. Digital photomicrographs of 3–5 well oriented villus-crypt units were captured at 40x magnification. Results were analyzed with respect to absolute numbers of bacteria, number of bacteria per epithelialized versus denuded basement membrane and number of bacteria per linear length of villus and crypt.
Sample collection for TRFLP analysis.
Following removal of the gastrointestinal tract, fluid residing within the lumen of the distal ileum of each piglet was collected into 15 ml conical vials. Full thickness sections of the distal ileum and proximal colon were gently rinsed in porcine Ringer's solution to remove macroscopic debris and then the mucosa was scraped from the seromuscular layers over ice using a glass microscope slide. Ileal fluid content and mucosal scrapings were stored at −80°C prior to DNA extraction.
DNA isolation.
Isolation of total DNA was performed using a commercial kit and Qiagen BioRobot Universal. Approximately 100 mg of sample was resuspended in 100 µl of 1x PBS with 8 mg lysozyme and 200 µl ATL buffer and incubated at 37°C for 30 minutes. Subsequently, samples were incubated 56°C overnight with 20 µl of proteinase K and then centrifuged at 800x g for 5 minutes. The supernatant was transferred to a Qiagen S-block and sonicated at 13 kHz in a VWR B2500A for 30 minutes at 65°C. Following sonication, the S-block was transferred to the bed of a Qiagen BioRobot Universal and DNA isolation was carried out using a customized isolation protocol.
16S rRNA gene PCR.
PCR was performed using Qiagen HotStar Hi-Fidelity PCR reagents and 100 ng genomic DNA per reaction. Amplification of the 16S rRNA gene was performed using the 16S universal primers 8F-Hex (5′-AGA GTT TGA TC(A/C) TGG CTC AG-3′),29 and 1492R (5′-GGT TAC CTT GTT ACG ACT T-3′),14 each at 100 µM. The thermal profile for amplification was initial denaturation at 95°C for 5 minutes, followed by 25 cycles of denaturation at 95°C for 1 minute, annealing at 50°C for 1 minute and extension at 72°C for 2 minutes, followed by a final 10 minute extension at 74°C. Each DNA sample was amplified in quadruplicate.
TRFLP analysis.
PCR reactions were purified using a QIAquick PCR Purification kit, eluted in 100 µl of buffer and stored at −20°C. Each sample was digested with restriction enzymes RsaI, HhaI and MspI (New England Biolabs) according to manufacturer's instructions. The digests were cleaned using a Qiagen Nucleotide Removal kit. Samples (5 µl) were combined with a master mix containing 4.5 µl of Hi-Di Formamide and 0.5 µl of Map Marker size standard MM-1000 ROX and incubated at 94°C for 3 minutes for denaturing prior to fragment detection using an ABI 3130xl capillary sequencer. Peaks were identified using GeneMapper 4.0 default analysis method detection parameters with a minimum peak height of 50 RFUs. Following peak detection, peaks falling outside of the size standard (50–1,000 bp) were removed and fragments with a relative peak area ratio (Pi) of ≥1% were considered for further analysis. The cleaned fragment files were uploaded to the Phylogenetic Assignment Tool (PAT) website (https://secure.limnology.wisc.edu/trflp/) and profile identification was carried out using a pattern database created from an in silico digest of the RDP database (http://rdp.cme.msu.edu/). The output files were compressed using a custom Microsoft Access Database macro. Phylogenetic assignment of the compressed pattern set was carried out using the RDP classifier website with default matching parameters. In all samples a number of profiles could not be assigned to specific phylogenetic groups and were considered as “unknown”. These were detected by T-RFLP analysis, identified by the PAT, but without phylogenetic information assigned based on the sequence. Diversity indices Shannon (H), Simpson (D) and Evenness (E) values were calculated as follows; (H = -∑(pi)(log2 pi), D = ∑(p/R)2) and E = H′/H′ max where p is the number of unique OTUs assigned to an identified class and R is the richness of OTUs detected in the sample. Statistical analyses were carried out using transformed Evenness values to establish significance. Non-discriminate analysis of samples was carried out using a QIIME module specifically developed for TRFLP analysis.
Quantitative (q)PCR.
All qPCR reactions were carried out in triplicate in an ABI 7500 Fast thermocycler and analyzed using the ABI 7500 Software version 2.0.1. Amplicon numbers were calculated during the annealing step. Standard curves for each primer set (Table S3) were generated from known concentrations of PCR products. Following a melting curve to determine sample quality, amplicon numbers were calculated by comparison of CT values between the standards and each sample.
Statistical analyses.
Numerical data were analyzed for normality (Kolmogorov-Smirnov) and variance (Levene median) and tested for significance using parametric or non-parametric tests as appropriate (SigmaStat; Jandel Scientific, San Jose CA). For all analyses, p ≤ 0.05 was considered significant.
Acknowledgements
This work was supported in part by grants from the North Carolina Translational and Clinical Sciences Institute (R40903) and the Center for Comparative Medicine and Translational Research, North Carolina State University. The authors thank Jack Odle, Korinn Saker, Donna Hardin, Anthony Blikslager, Vanessa Cook, Elizabeth Lennon, Kellet Babb, Connie Engle, Chris Moses, Lin Xi, Kathryn Price, Tina Herfel, Christopher Lassiter and Megan Cruze for outstanding technical and resource support. The Microbiome Core Facility is supported by grant number 5-P30-DK034987-26 from the National Institutes of Health.
Abbreviations
- NEC
necrotizing enterocolitis
- qPCR
quantitative polymerase chain reaction
- TRFLP
terminal restriction fragment length polymorphism
- PCoA
principal coordinate analysis
- 16S rDNA
16S ribosomal RNA gene
Disclosure of Potential Conflict of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
References
- 1.Fanaroff AA, Stoll BJ, Wright LL, Carlo WA, Ehrenkranz RA, Stark AR, et al. Trends in neonatal morbidity and mortality for very low birthweight infants. Am J Obstet Gynecol. 2007;196:1471–1478. doi: 10.1016/j.ajog.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgibbons SC, Ching Y, Yu D, Carpenter J, Kenny M, Weldon C, et al. Mortality of necrotizing enterocolitis expressed by birth weight categories. J Pediatr Surg. 2009;44:1072–1075. doi: 10.1016/j.jpedsurg.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Alfaleh K, Anabrees J, Bassler D. Probiotics reduce the risk of necrotizing enterocolitis in preterm infants: a meta-analysis. Neonatology. 2010;97:93–99. doi: 10.1159/000235684. [DOI] [PubMed] [Google Scholar]
- 4.Hoy CM, Wood CM, Hawkey PM, Puntis JW. Duodenal microflora in very-low-birth-weight neonates and relation to necrotizing enterocolitis. J Clin Microbiol. 2000;38:4539–4547. doi: 10.1128/jcm.38.12.4539-4547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mshvildadze M, Neu J, Shuster J, Theriaque D, Li N, Mai V. Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J Pediatr. 156:20–25. doi: 10.1016/j.jpeds.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Millar MR, Linton CJ, Cade A, Glancy D, Hall M, Jalal H. Application of 16S rRNA gene PCR to study bowel flora of preterm infants with and without necrotizing enterocolitis. J Clin Microbiol. 1996;34:2506–2510. doi: 10.1128/jcm.34.10.2506-2510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de la Cochetiere MF, Piloquet H, des Robert C, Darmaun D, Galmiche JP, Roze JC. Early intestinal bacterial colonization and necrotizing enterocolitis in premature infants: the putative role of Clostridium. Pediatr Res. 2004;56:366–370. doi: 10.1203/01.PDR.0000134251.45878.D5. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Hoenig JD, Malin KJ, Qamar S, Petrof EO, Sun J, et al. 16S rRNA gene-based analysis of fecal microbiota from preterm infants with and without necrotizing enterocolitis. Isme J. 2009;3:944–954. doi: 10.1038/ismej.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swidsinski A, Loening-Baucke V, Verstraelen H, Osowska S, Doerffel Y. Biostructure of fecal microbiota in healthy subjects and patients with chronic idiopathic diarrhea. Gastroenterology. 2008;135:568–579. doi: 10.1053/j.gastro.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 10.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 12.Sangild PT, Siggers RH, Schmidt M, Elnif J, Bjornvad CR, Thymann T, et al. Diet- and colonization-dependent intestinal dysfunction predisposes to necrotizing enterocolitis in preterm pigs. Gastroenterology. 2006;130:1776–1792. doi: 10.1053/j.gastro.2006.02.026. [DOI] [PubMed] [Google Scholar]
- 13.Bjornvad CR, Thymann T, Deutz NE, Burrin DG, Jensen SK, Jensen BB, et al. Enteral feeding induces diet-dependent mucosal dysfunction, bacterial proliferation and necrotizing enterocolitis in preterm pigs on parenteral nutrition. Am J Physiol Gastrointest Liver Physiol. 2008;295:1092–1103. doi: 10.1152/ajpgi.00414.2007. [DOI] [PubMed] [Google Scholar]
- 14.Alfa MJ, Robson D, Davi M, Bernard K, Van Caeseele P, Harding GK. An outbreak of necrotizing enterocolitis associated with a novel clostridium species in a neonatal intensive care unit. Clin Infect Dis. 2002;35:101–105. doi: 10.1086/341929. [DOI] [PubMed] [Google Scholar]
- 15.Magne F, Abely M, Boyer F, Morville P, Pochart P, Suau A. Low species diversity and high interindividual variability in faeces of preterm infants as revealed by sequences of 16S rRNA genes and PCR-temporal temperature gradient gel electrophoresis profiles. FEMS Microbiol Ecol. 2006;57:128–138. doi: 10.1111/j.1574-6941.2006.00097.x. [DOI] [PubMed] [Google Scholar]
- 16.von Wintzingerode F, Gobel UB, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 17.Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, et al. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–799. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Howard FM, Flynn DM, Bradley JM, Noone P, Szawatkowski M. Outbreak of necrotising enterocolitis caused by Clostridium butyricum. Lancet. 1977;2:1099–1102. doi: 10.1016/s0140-6736(77)90546-3. [DOI] [PubMed] [Google Scholar]
- 19.Kosloske AM, Ulrich JA, Hoffman H. Fulminant necrotising enterocolitis associated with Clostridia. Lancet. 1978;2:1014–1016. doi: 10.1016/s0140-6736(78)92337-1. [DOI] [PubMed] [Google Scholar]
- 20.Blakey JL, Lubitz L, Campbell NT, Gillam GL, Bishop RF, Barnes GL. Enteric colonization in sporadic neonatal necrotizing enterocolitis. J Pediatr Gastroenterol Nutr. 1985;4:591–595. doi: 10.1097/00005176-198508000-00017. [DOI] [PubMed] [Google Scholar]
- 21.Sturm R, Staneck JL, Stauffer LR, Neblett WW., 3rd Neonatal necrotizing enterocolitis associated with penicillin-resistant, toxigenic Clostridium butyricum. Pediatrics. 1980;66:928–931. [PubMed] [Google Scholar]
- 22.Stark PL, Lee A. Clostridia isolated from the feces of infants during the first year of life. J Pediatr. 1982;100:362–365. doi: 10.1016/s0022-3476(82)80430-7. [DOI] [PubMed] [Google Scholar]
- 23.Popoff MR, Szylit O, Ravisse P, Dabard J, Ohayon H. Experimental cecitis in gnotoxenic chickens monoassociated with Clostridium butyricum strains isolated from patients with neonatal necrotizing enterocolitis. Infect Immun. 1985;47:697–703. doi: 10.1128/iai.47.3.697-703.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bousseboua H, Le Coz Y, Dabard J, Szylit O, Raibaud P, Popoff MR, Ravisse P. Experimental cecitis in gnotobiotic quails monoassociated with Clostridium butyricum strains isolated from patients with neonatal necrotizing enterocolitis and from healthy newborns. Infect Immun. 1989;57:932–936. doi: 10.1128/iai.57.3.932-936.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seger R, Joller P, Bird GW, Wingham J, Wuest J, Kenny A, et al. Necrotising enterocolitis and neuraminidase-producing bacteria. Helv Paediatr Acta. 1980;35:121–128. [PubMed] [Google Scholar]
- 26.Waligora-Dupriet AJ, Dugay A, Auzeil N, Nicolis I, Rabot S, Huerre MR, Butel MJ. Short-chain fatty acids and polyamines in the pathogenesis of necrotizing enterocolitis: Kinetics aspects in gnotobiotic quails. Anaerobe. 2009;15:138–144. doi: 10.1016/j.anaerobe.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Dittmar E, Beyer P, Fischer D, Schafer V, Schoepe H, Bauer K, Schlosser R. Necrotizing enterocolitis of the neonate with Clostridium perfringens: diagnosis, clinical course and role of alpha toxin. Eur J Pediatr. 2008;167:891–895. doi: 10.1007/s00431-007-0614-9. [DOI] [PubMed] [Google Scholar]
- 28.Moyer CL, Tiedje JM, Dobbs FC, Karl DM. A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature. Appl Environ Microbiol. 1996;62:2501–2507. doi: 10.1128/aem.62.7.2501-2507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent AD, Smith DJ, Benson BJ, Triplett EW. Web-based phylogenetic assignment tool for analysis of terminal restriction fragment length polymorphism profiles of microbial communities. Appl Environ Microbiol. 2003;69:6768–6776. doi: 10.1128/AEM.69.11.6768-6776.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Epelman M, Daneman A, Navarro OM, Morag I, Moore AM, Kim JH, et al. Necrotizing enterocolitis: review of state-of-the-art imaging findings with pathologic correlation. Radiographics. 2007;27:285–305. doi: 10.1148/rg.272055098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.