Abstract
The recent finding that the formation of staphylococcal enterotoxins in food is very different from that in cultures of pure Staphylococcus aureus sheds new light on, and brings into question, traditional microbial risk assessment methods based on planktonic liquid cultures. In fact, most bacteria in food appear to be associated with surfaces or tissues in various ways, and interaction with other bacteria through molecular signaling is prevalent. Nowadays it is well established that there are significant differences in the behavior of bacteria in the planktonic state and immobilized bacteria found in multicellular communities. Thus, in order to improve the production of high-quality, microbiologically safe food for human consumption, in situ data on enterotoxin formation in food environments are required to complement existing knowledge on the growth and survivability of S. aureus. This review focuses on enterotoxigenic S. aureus and describes recent findings related to enterotoxin formation in food environments, and ways in which risk assessment can take into account virulence behavior. An improved understanding of how environmental factors affect the expression of enterotoxins in foods will enable us to formulate new strategies for improved food safety.
Keywords: Staphylococcus aureus, staphylococcal enterotoxins, staphylococcal food poisoning, foodborne illness, virulence regulation, risk assessment
Introduction
The battle against bacterial foodborne diseases is facing new challenges due to rapidly changing patterns of human consumption, the globalization of the food market and climate change. Today, consumers want more natural food products that are less processed, without preservatives, with low salt, sugar or fat contents, but with an extended shelf-life and high quality.1 The demand for convenient, ready-to-eat food has also increased, and the food industry has developed new food processing techniques such as semi-prepared, minimally processed, chilled food in response to these demands.2,3 Convenience food offers a suitable growth environment for toxin-producing bacteria such as Staphylococcus aureus, which is able to grow and express virulence in a wide variety of foods such as milk products, mixed foods, meat and meat products, egg and egg products, cakes and ice cream.4 The European Food Safety Authority (EFSA) reported in 2009 that cheese followed by mixed or buffet meals were the two main food vehicles in verified outbreaks of food poisoning caused by staphylococcal toxins.5 As in all industries, there is a desire to minimize production costs, leading to the search for low-cost raw materials globally, resulting in ingredients from many countries being combined into one dish. Consequently, food is prepared, produced and stored differently, and the behavior of foodborne pathogens under these different conditions is not yet fully understood, potentially increasing the risk of foodborne illness.
To improve the production of microbiologically safe food for the consumer, data related to the physiology of foodborne pathogens in authentic food situations is required to complement existing knowledge on the growth and survival of planktonic bacteria in liquid cultures. In fact, recently reported data have shown that there are significant differences in the behavior of bacteria in the planktonic state and in actual food matrices.6–8 Knowledge about the effects of critical food-related factors on microbial responses such as virulence gene expression, lag phase duration, growth rate and extracellular virulence formation, will not only aid in the prevention of foodborne diseases, but also enable the advancement of quantitative microbial risk assessment (QMRA). A risk assessment consists of four steps: (1) hazard identification, (2) hazard characterization, (3) exposure assessment and (4) risk characterization. Steps 1 to 3 are combined to assess the health risk in relation to the specific risk questions addressed. The dose-response relationship is crucial in the hazard characterization step, while the ability to estimate the likelihood and amount of the hazardous material ingested is important in exposure assessment. The latter usually involves the use and development of predictive microbiology models for quantification of growth, inactivation and toxin production. The application of risk analysis frameworks and preventive approaches (e.g., hazard analysis and critical control points, HACCP) and the responsibility of food producers, are key principles in the new paradigm to address foodborne illnesses.9 A scientific assessment of the risk to health is the basis for the management of foodborne hazards in terms of the measures chosen to control risk to an appropriate level of protection (ALOP). The ALOP must be translated into a metric, a safety level, useful for setting limits that producers can relate to.10 Proposed metrics include Food Safety Objectives (FSO), Performance Objectives (PO), Performance Criteria (PC) and Microbial Criteria (MC).11 Regardless of the metric chosen, the key in this process is the development of a risk assessment procedure.12
This review focuses on S. aureus and describes recent findings related to enterotoxin expression, formation and regulation in food environments, and ways in which risk assessment can be improved by in situ virulence data. In general, the enterotoxin(s) are formed during S. aureus multiplication in food, but new findings show that bacterial growth and enterotoxin production may be decoupled in food products. Different metabolic regulatory systems involved in enterotoxin expression will be discussed, as well as recent risk assessment approaches.
The Organism
S. aureus is a Gram-positive coccus occurring singly or in irregular clusters. The bacteria produce a carotenoid pigment resulting in golden-colored colonies, giving rise to the species epithet aureus (meaning golden). They are nonmotile and non-sporing chemoorganotrophics with both respiratory and fermentative metabolism.13,14 S. aureus is found in the nostrils and on the skin of warm-blooded animals, and the primary source of food contamination is the hands of food handlers.15,16 The organism can also be endemic in the processing environment.17 S. aureus has the ability to grow, and produce staphylococcal enterotoxins (SE), the causative agent of staphylococcal food poisoning (SFP), over an extensive range of temperature, pH, sodium chloride concentration and water activity (Table 1).4 The robustness of the organism permits its growth in many types of food, producing enterotoxins subsequently causing food poisoning. The bacteria can be killed through heat treatment of the food, but the enterotoxins are very heat resistant. Thus, although the bacteria are eliminated, the toxins will remain and can cause SFP.15
Table 1.
Factors affecting Staphylococcus aureus growth and enterotoxin formation4
| Factor | Optimal growth | Growth limits | Optimal SE production | SE production limit | Enterotoxin (s) reported affected | Notes to effect(s) on enterotoxin production | Examples of analysis of the specific factor in food products | References |
| Temperature | 35–41°C | 6–48°C | 34–40°C | 10–46°C | SEA, SEB, SEC, SED | Temperature seems to affect enterotoxin synthesis more that growth. | Milk Ham Egg products |
44, 141–144, 169 |
| pH | 6–7 | 4–10 | 7–8 | 5–9.6 | SEA, SEB, SEC, SED, SEE | Higher tolerance under aerobic compared with anaerobic growth conditions. Lactic acid particularly inhibits toxin formation. agr dependent regulation (SEC). |
Ham Sausage |
7, 75, 143–147, 169 |
| aw | 0.99 | 0.83 ≥ 0.99 | 0.99 | 0.86 ≥ 0.99 | SEA, SEB, SEC, SEH | SEB and SEC may be more sensitive than SEA and SEH. SEH enterotoxin production at aw: 0.97 > 1 > 0.95. |
Cured beef slurry Cured pork slurry Bacon Scrimp slurry Sausage |
85, 90, 96, 142, 147–152 |
| NaCl | 0% | 0–20% | 0% | <12% | SEA, SEB, SEC | Raises temperature limit for SEA production. Low osmolality increases enterotoxin production. SEB production seems more strongly inhibited than growth. |
Ham Sausage |
85, 88, 96, 144, 153–156, 169 |
| Oxygen | Aerobic | Anaerobic-aerobic | Aerobic | Anaerobic-aerobic | SEA, SEB, SEC, SEH | Increases yield of SEB up to 10-fold. 10% dissolved oxygen is optimal for SEB production. |
Ham Prawn Sausage |
98, 113, 157, 158, 159, 160, 161 |
| Redox potential (Eh) | >+200 mV | ≥200 to > +200 mv | >+200 mV | ≥100 to > +200 mv | - | - | - | - |
| Lactococcus lactis | - | - | - | - | sec, sel (sek, seg, seh) | Strongly reduces transcription of sec and sel and sightly sek, seg, seh | Cheese | 6, 107, 162 |
| sea | May favor the maintenance of sea in stationary phase. |
Staphylococcal food poisoning and enterotoxins.
Food safety is an important issue throughout the world, and is one of the WHO's 13 strategic objectives for 2008–2013. A study on the impact of food-related illness has recently been published in the USA and, due to a number of serious incidents in recent years, the US Food and Drug Administration (FDA) has stepped up efforts to improve the traceability of contaminated products.18 In Europe, the EFSA reported a total of 5,550 outbreaks of foodborne illness in 2009, affecting almost 49,000 people and causing 46 deaths. Among these, 293 outbreaks were caused by Staphylococcus spp and bacterial toxins (produced by Bacillus, Clostridium and Staphylococcus) were the fourth most common causative agent in foodborne outbreaks.5
SFP is a foodborne intoxication that develops in people who ingest food that has been improperly prepared or stored. The severity of the illness depends on the amount of food ingested, the amount of toxin in the ingested food and the general health of the victim.16 SFP can be caused by as little as 20–100 ng of enterotoxin.19 After ingestion, symptoms appear rapidly and abruptly, consistent with diseases caused by preformed toxins. The symptoms include copious vomiting, diarrhea, abdominal pain or nausea.13,14 Ingested bacteria do not produce toxin, and the symptoms therefore normally wear off within 24 h.
To date, 21 SEs or enterotoxin-like proteins (SEls) have been identified and designated SEA to SElV (Table 2).20,21 While SEs are the toxins that induce emesis, the related SEls either lack emetic activity or have not yet been tested for this.22 The genes encoding the different enterotoxins are carried and disseminated by different mobile genetic elements, i.e., prophages, plasmids, pathogenicity islands (SaPIs), enterotoxin gene cluster (egc) and the staphylococcal cassette chromosome (SCC).23–40 Enterotoxins are short, extracellular proteins that are water-soluble. They are most commonly described as very stable, and are resistant to heat as well as degrading enzymes.15,41,42 However, some cases have been reported where the toxins disappeared. Recently, SEA and SED were found to decrease in boiled ham after a period of accumulation,7,8 and a number of earlier studies have reported the disappearance of SEA in broth, minced food and raw and pasteurized milk.43,44 The apparent decrease in enterotoxin levels could simply be an analytical artifact, such as loss of serological recognition using immuno-based methods such as ELISA, which is a technique commonly used to detect enterotoxins. However, it has also been proposed that proteases produced by lactic acid bacteria (LAB) cause the decrease in SEA levels, or that SEA becomes cell-associated and is, therefore, not detected.45,46 Furthermore, it has been reported that the expression of genes encoding potential proteases were increased upon acid shock of S. aureus.47 These findings suggest that S. aureus, or other organisms present in the surrounding environment, e.g., LAB, could cause the decrease in enterotoxin level observed under certain conditions, possibly through extracellular protease activities. The enterotoxins, which are classified as superantigens, display the common characteristics of this group, i.e., pyrogenicity, immune suppression and a mitogenic effect on T cells.15 Superantigens can also cause toxic shock syndrome, a serious condition characterized by rashes, hypovolemic shock and respiratory distress syndrome.48 The majority of reported SFP outbreaks are associated with the classical enterotoxins, SEA-SEE; staphylococcal enterotoxin A (SEA) being considered the most common cause of SFP.49–51
Table 2.
The staphylococcal enterotoxins
| Enterotoxin | Variant | ORF length (bp) | Mature length (aa) | Molecular weight (Da) | Genetic backbone | References |
| SEA | 774 | 233 | 27,100 | Prophage | 15, 25, 52, 163 | |
| SEA1 | 774 | 233 | 27,100 | Prophage | ||
| SEA2 | 774 | 233 | 27,100 | Prophage | ||
| SEB | 801 | 239 | 28,336 | SaPI | 15, 59 | |
| SEC | SaPI | 15, 28, 31, 38, 60, 164 | ||||
| SEC1 | 801 | 239 | 27,531 | SaPI | ||
| SEC2 | 801 | 239 | 27,531 | |||
| SEC3 | 801 | 239 | 27,563 | SaPI | ||
| SECbov | 816a | 271b | 27,618 | SaPI | ||
| SECsheepc | 27,517 | |||||
| SED | 777 | 228 | 26,360 | Plasmid | 15, 24 | |
| SEE | 774 | 230 | 26,425 | Prophage | 15, 27 | |
| SElG | 777 | 233 | 27,043 | egc | 15, 29, 54, 57, 165 | |
| SElG2 | 729a | 242b | Prophage | |||
| SElGV | 777 | 233 | 26,985 | egc | ||
| SElH | 726 | 218 | 25,210 | sccd | 15 | |
| SEI | 729 | 218 | 24,298 | egc | 15, 29, 32 | |
| SEIv | 729a | 242b | egc | |||
| SElJ | 806 | 245 | 28,565 | Plasmid | 15, 34, 39 | |
| SElK | 729 | 219 | 25,539 | SaPI | 15, 38, 54, 57, 166 | |
| SElK2 | 729a | 242b | Prophage | |||
| SElL | 723 | 215 | 24,593 | SaPI | 15, 28, 38 | |
| SElM | 722 | 217 | 24,842 | egc | 15, 29 | |
| SElN | 720 | 227 | 26,067 | egc | 15, 29, 32 | |
| SElNvc | egc | |||||
| SElO | 783 | 232 | 26,777 | egc | 15, 29 | |
| SElP | 783a | 260e | 27,000 | Prophage | 31, 48, 167 | |
| SElQ | 729a | 242e | 25,207 | SaPI | 38, 166 | |
| SER | 600a | 259e | 27,049 | Plasmid | 34, 168 | |
| SES | 774a | 257e | 26,217 | Plasmid | 168 | |
| SET | 651a | 216 | 22,614 | Plasmid | 168 | |
| SElU | 786a | 261e | 27,100 | egc | 32, 48 | |
| SElUv | 771 | 256e | egc | |||
| SElV | 720 | 239b | egc | 21 |
ORF, open reading frame.
ORF obtained from the National Center for Biotechnology Information, NCBI, www.ncbi.nlm.nih.gov.ludwig.lub.lu.se/gene, March 25, 2010
precursor aa length from NCBI, www.ncbi.nlm.nih.gov.ludwig.lub.lu.se/sites/entrez?db=Protein&itool=toolbar, March 25, 2010
ORF length and aa sequence not found in NCBI.
R. Cao, unpublished data.
precursor aa length.
Regulation of Enterotoxin Formation
The classical enterotoxins (SEA-SEE).
Prophage-encoded enterotoxins (sea and see). The sea gene is carried by a polymorphic family of temperate bacteriophages.25 The bacteriophage is inserted into the bacterial chromosome as a prophage and behaves like part of the bacterial genome. However, under environmental stress conditions, such as mild food preservation conditions, the prophage can be induced to replicate the phage genome and release new bacteriophages.52 Today, at least six completely sequenced S. aureus strains containing different sea-carrying prophages, Φ252B, ΦMu3, ΦMu50A, ΦNM3, ΦSa3ms and ΦSa3mw, have been found, all of which frequently carry the genes for enterotoxin A, staphylokinase and the complement inhibitor.31,53–56 It was recently demonstrated that the transcription of sea is linked to some extent to the lifecycle of the SEA-encoding prophage,57 in contrast to many other non-phage encoded enterotoxin genes such as seb, sec and sed. The polymorphic nature of the prophages has been found to affect the amount of SEA produced by the bacterial strain carrying the prophage.26 Sequence analysis of the sea gene and its neighboring genomic regions have further indicated that SEA-producing strains can be grouped into two major groups, SEA1 and SEA2.52 The endogenous promoter region, P1, immediately upstream of sea, is found in both groups.26 In addition, there may also be a second phage-related latent promoter, P2, shown to express sea after prophage induction.57 We have observed that S. aureus strains producing high amounts of SEA belong to the SEA1 group, and that strains producing low amounts of SEA belong to the SEA2 group (unpublished data). Furthermore, a subgroup of the SEA1 strains was also found to be associated with a stress-induced boost in SEA production as the second phage-related promoter, P2, was activated. Enterotoxin E (SEE) is the toxin most similar to SEA, having 90% amino acid identity.48 The see gene is situated on a defective prophage, in contrast to the prophage encoding sea and see expression appears to be unaffected by bacterial growth.58
agr-regulated enterotoxins (seb, sec and sed). The seb gene is carried on the S. aureus pathogenicity island, SaPI3,59 while enterotoxin C (SEC) exists in multiple variants, C1, C3, Cbov, which are situated on SaPI4, SaPIn1/m1 and SaPIbov, respectively.31,60 SaPIs are highly mobile phage-related staphylococcal pathogenicity islands that can integrate into specific sites (known as the attC sites) in the chromosome determined by the specificity of SaPI-encoded integrases. SaPI particles are released by phage-induced lysis, and can infect and integrate into a new host with very high frequency.61 The sed gene is situated on a 27.6 kb penicillinase plasmid, pIB485, in S. aureus.24 Despite being encoded by different mobile genetic elements, the expression of seb, sec and sed genes is induced during the transition from the exponential to the stationary phase, an expression pattern characteristic of proteins encoded by genes regulated by the Agr regulatory system.24,58,62–65 Yet, the two se genes encoded by SaPIs, seb and sec, undergo a much more drastic induction than the plasmid-encoded sed.58
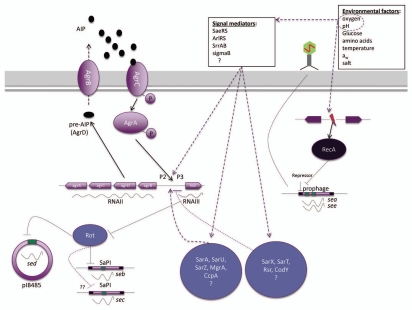
The Agr signal transduction system is a quorum sensing system that allows S. aureus to respond to cell density, as recently reviewed by Thoendel et al.66 Briefly, the agr locus generates two different transcripts, RNAII and RNAIII, driven by the promoters P2 and P3, respectively (Fig. 1). RNAII encodes the structural genes for the quorum sensing system agrB, agrD, agrC and agrA. AgrD and AgrB act to generate the quorum sensing molecule [autoinducing peptide (AIP)], which after reaching a threshold level stimulates activation of AgrC and AgrA, a two component regulatory system. Activated AgrA then upregulates the promoters P2 and P3, generating more RNAII and RNAIII transcripts. The P3 transcript, RNAIII, encodes delta-hemolysin but, more importantly, the RNAIII itself is the intracellular effector of gene regulation in the cell.66–68 As the cell grows the intracellular level of RNAIII increases due to the autoregulatory circuit of the Agr system, leading to increased transcription of secreted virulence factors such as enterotoxins, and reduced transcription of a subset of genes encoding cell wall proteins.69,70 Loss of the Agr signal transduction system is reported to result in substantial loss in the transcript level of seb, sec and sed and thus the corresponding SEB, SEC and SED production.24,63,65,71
Figure 1.
Regulation of se transcription in S. aureus. Black arrows represent identified direct regulation, while dashed purple arrows indicate potential regulatory pathways. The individual regulators and regulatory pathways are described in the text.66 AIP, autoinducing peptide.
The RNAIII-mediated impact on the transcription of seb and sed is indirect and is dependent on the presence of a functional Rot (repressor of toxins), which is a member of the Sar family of transcriptional factors of S. aureus. Rot binds to promoter regions, as shown for the seb promoter, thereby repressing the transcription of genes.72–74 When the Agr system is induced during post-exponential growth RNAIII basepairs with rot mRNA. This mediates translational repression of rot mRNA, and subsequently lowers the amount of cellular Rot.72 It is not known whether the activation of sec is regulated by the agr system via the RNAIII-Rot interaction, but it has been reported that SEC1 and SEC2 are produced during different growth phases, and the regulation of the enterotoxins of this biotype (C) may differ.64
Numerous transcriptional regulators affect the function of the agr system, and may in theory indirectly affect the levels of seb, sec and sed transcription (Fig. 1).66 These regulators respond to various environments and stresses also known to affect enterotoxin synthesis.66,75–78 The two-component system SrrAB and CodY are two of the interesting regulatory candidates known to directly control agr expression. SrrAB is involved in the adaptation to anaerobic growth, and inhibits RNAIII expression by binding to the P2 and P3 promoters. CodY is a transcriptional regulator, whose DNA binding ability is controlled by the cellular GTP pool, thus sensing nutrient availability.77–80 SarA, another member of the Sar family of homologs, positively affects agr transcription and its DNA-binding activity to P2 is dependent on cellular redox conditions and pH and has been demonstrated to positively control seb and sec.76,81,82 In accordance with the above observations, dissolved oxygen level, nutrient availability and pH have been shown to influence the formation of agr-regulated enterotoxins (Table 1). SaeRS, another two-component system, suggested to act downstream of agr, has been shown to positively regulate sec expression.83 It has been proposed that SaeRS responds to several environmental stimuli, including high glucose and salt levels, low aw and low pH.84
The enterotoxins B, C and D are, however, only partially upregulated by the Agr system and can be produced independently of agr.85,86 Although SarA is required for full agr loci transcription, SarA has also been shown to regulate seb transcription independently of RNAIII, and the alternative sigma factor, sigmaB, has been reported to reduce seb expression, possibly by repressing both the agr system and a second unidentified inducer.87,88 Notably, many of the environmental conditions known to repress seb transcription, such as high salt content and alkaline conditions, are also known activators of sigmaB.86,89
The non-classical enterotoxins (SElG-SElV).
Regarding regulation of the non-classical enterotoxins, results from a kinetic study indicate that the expression of the majority of the newly described se genes is not controlled by the agr system.58 Data from this study show that only the transcript level of seh, ser and sel increases in the post-exponential phase, which implies possible regulation by the Agr regulatory system. seh mRNA was found to undergo a much more drastic induction than ser and sel, and activation of seh took place earlier in the growth cycle than the classical agr-controlled seb and sed genes. This expression pattern is consistent with results reported by Sakai et al.90 showing that maximal SEH production takes place in the late exponential phase, while SEB is mainly produced in the stationary phase.58,70,90 The transcript level of other investigated se genes either remained unchanged during growth (sej, sek, seq, sep), or decreased slightly (seg, sei, sem, sen, seo, seu) after exponential growth. Most of the se genes with unchanged transcription are phage-encoded, and may therefore be regulated by the processes that govern lysogeny. In contrast, the se genes that showed a slight decrease in transcript level during growth are encoded by the egc operon and, notably, these enterotoxins could not be detected using two-dimensional gel electrophoresis.58,91 It is still unclear whether the non-classical enterotoxins are responsible for food poisoning, and so far SEH is the only non-classical enterotoxin detected in foods responsible for food poisoning.92,93
Impact of Environmental Factors on SE Production
SFP is often associated with growth in protein-rich food such as meat and dairy products.15,94 These products are highly complex matrices compared with broth, with respect to, e.g., microbial content, salt, pH, nutrient availability, oxygen availability and temperature.95 Generally, growth of S. aureus is necessary for enterotoxin production, although enterotoxin production does not always accompany growth, and in a few cases toxin production has been observed in non-replicating cell cultures, most recently by Wallin-Carlquist et al.8 in ham products.96–98 Studies have been performed to identify key parameters that prevent or stimulate enterotoxin production in laboratory media and in diverse food products, and a multifaceted network of environmental and genetic factors seems to regulate enterotoxin production.6,8,42 Some of the identified effects of environmental conditions on enterotoxin production are listed in Table 1.
Wallin-Carlquist et al.7,8 have recently studied SEA and SED formation in boiled and smoked ham. Notably, a prolonged sea expression and SEA formation were observed over the course of a week, instead of a short-term growth-associated sea expression8 and unexpectedly sed expression followed the same general pattern as the prophage-encoded SEA in both ham products, as the genes are regulated differently.7 The difference in the enterotoxin expression pattern observed for S. aureus on the ham products and in liquid culture is probably related to the different physiological states of the staphylococci. On the ham, S. aureus forms a biofilm, while in the culture the bacteria are planktonic. An active agr quorum sensing system is known to limit biofilm formation in S. aureus, and the activation of the agr system has been connected with the dispersal of S. aureus from an established biofilm upon glucose depletion.99 A second peak in sed expression observed in boiled and smoked ham could be due to glucose levels running low in the meat, activating the agr system, initiating the detachment of S. aureus from the biofilm, consequently inducing sed expression. Furthermore, recent studies show that mature biofilms are acidic environments.100–102 Low pH can cause prophage induction, leading to increased sea expression, explaining the prolonged sea expression observed on boiled and smoked ham. Enhanced transcription of phage-encoded virulence genes upon prophage induction has been demonstrated for Panton-Valentine leukocidin in S. aureus, streptococcal pyrogenic exotoxin C and a DNase in Streptococcus pyogenes, as well as the Shiga and Shiga-like toxins in Escherichia coli.57,103–106 Cretenet and colleagues6 recently used a transcriptomic approach to study virulence expression in a cheese matrix under the influence of Lactococcus lactis, showing that sea expression was slightly increased in this acidic environment. Another possible explanation of a second boost in sea expression may be that the temperate phage is activated by oxidative stress via RecA-mediated response.
In the study by Cretenet and colleagues6 the expression of sec and two other genes that may be agr-controlled, seh and sel in S. aureus, isolated from a cheese matrix, was also followed. Using a transcriptomic approach, expression data revealed that both the dynamics and the levels of sec, seh and sel expression differed notably from those observed during growth in a chemically defined medium (CDM). The expression levels were significantly lower in the cheese matrix than in the CDM, and the post-exponential induction, characteristic of agr-regulated genes, was absent. Low water activity in the cheese matrix correlated with a reduction in saeRS level and was suggested to be responsible for, at least, the reduced sec expression in the cheese matrix.6 The expression of the se genes was also studied in the cheese matrix in the presence of L. lactis.107 In these mixed cultures, L. lactis has previously been reported to reduce the expression of sec, sel and seh, and for sec the reduction was suggested to be partly due to reduced activity of SarA and the agr system.107 The presence of L. lactis in the cheese matrix also downregulated the expression of RNAIII and sarA, while the rot level was increased. However, although the activity of the agr system was reduced in the presence of L. lactis and a decrease in sec, seh and sel levels had previously been observed in mixed cultures with L. lactis, both seh and sel were upregulated in the cheese matrix by the presence of L. lactis, while the sec level was not affected by its presence.6 These results illustrate that, despite the observation of similar expression patterns of sec, seh and sel in CDM and in mixed cultures in laboratory studies, the effects of environmental conditions in a food matrix are dependent on the type of enterotoxin, and that the regulatory organization is multifactorial.
Risk Assessment and Predicitive Microbiology to Control Enterotoxin-Producing S. aureus in Foods
In this section, predictive microbiology models and risk assessment of S. aureus are reviewed with the objectives of illustrating different approaches and highlighting challenges in relation to the findings presented in previous sections. The focus is on staphylococcal growth and enterotoxin production; other characteristics, such as antibiotic resistance, are not addressed.
Predictive microbiology models for staphylococcal growth and enterotoxin production.
One difficulty when estimating human exposure to a hazard or evaluating the safety of a production process is to quantify changes in the number of microorganisms or the amount of toxin in the food at the stage of food production or processing of interest. Predictive models are useful tools that can be used to estimate these changes, depending on the properties of the microorganism, and the nature of the food and the way it is handled, stored and processed.108 However, predictive models cannot be used as the sole determinant of product safety.109 If important decisions are to be made based on the results of a predictive model it must be validated in the food of interest.
Predictive models are based on data describing changes in numbers of microorganisms or levels of toxin fitted to primary models. Secondary models describe the effects of environmental factors, such as temperature and pH, on the parameters in primary models, e.g., the maximum specific growth rate. A summary of different models is provided by van Gerwen and Zwietering.110 Tertiary models are predictive models implemented in user-friendly software. Tertiary S. aureus models available free on the internet include the growth and survival models in the US FDA pathogen modeling program (PMP) and the growth model in the ComBase modeling toolbox, the ComBase Predictor.111 In addition, growth and inactivation curves from published studies are available in the ComBase database. There are also several kinetic models describing the physico-chemical effects of the food or the environment on S. aureus growth rates.112,113 Available kinetic tertiary models can be used to predict growth and lag time based on various input values, for example, temperature, pH, water activity, nitrite concentration and aerobic/anaerobic conditions (PMP) (ComBase Predictor) and temperature in various types of meat (THERM).114 An alternative approach is to describe the boundary between growth and no growth,95 or survival/death,115 the time before growth,116 or the probability of growth, as a function of environmental parameters. Such models can be useful in the design of food processes to prevent growth and enterotoxin production. For instance, Stewart et al.116 developed a time-to-growth model to study the effects of different humectants used to achieve shelf-stable intermediate-moisture foods. Similarly, Valero et al.95 developed a model for the probability of growth and found an abrupt transition of the interface between growth and no growth at low temperatures, where optimal levels of pH and water activity were required for growth. Obeso et al.115 described the effect of initial lytic phage titers and initial S. aureus contamination of pasteurized milk on the probability of S. aureus survival at different temperatures.
Food contamination is often incidental, and by few cells. Consequently, new approaches have emerged based on the study of individual cells to derive distributions of growth parameters for use in predictive models.117 For instance, Sado Kamdem et al.118 studied the effect of the concentration of fatty acids at two pH values on the distribution of division times among single cells of S. aureus. Considerable variation in division times was observed at the single cell level, which was masked when studies were performed at the population level. This emphasizes the difficulty in making predictions, especially under conditions of stress and at low levels of contamination. Variation has also been observed at strain level.119 The generation times of 34 S. aureus strains isolated from poultry and cultured in chicken broth at 17°C ranged from 2 to 17 h.120 These results emphasize the need to address biological variation and the usefulness of probabilistic approaches to predictive modeling using distributions of growth parameters instead of single fixed values. Vora et al.121 used a probabilistic simulation approach to evaluate the effect of contamination level of S. aureus on the survival/gradual decline in intermediate-moisture foods. They reported no effect of initial contamination levels but both simulations and observations indicated a wide variation in decline rates, including occasional increases in population. Interactions with other microorganisms, present in food or added, may have profound effects on S. aureus growth (as exploited by the use of starter cultures in fermented foods) and thus also on enterotoxin production.122 For example, Le Marc et al.123 developed a kinetic model that described the inhibitory effect of a starter culture of lactic acid bacteria on S. aureus growth in milk when the lactic acid bacteria had exceeded a critical density.
Although several studies of the kinetics of the production of different enterotoxins or expression of enterotoxin genes exist, very few predictive models of enterotoxin production are available. Fujikawa and Morozumi124 developed a model based on observations that SEA was detectable at levels greater than 6.5 log10 cfu ml−1, and increased linearly during the whole growth curve in a sterile milk medium. The rate of SEA production increased linearly with temperature from about 15 to 32°C, and was described by the following equation:
| (1) |
where p is the rate of SEA production (ng ml−1 h−1) and t is the temperature (°C). SEA was still produced at temperatures above 32°C, but the rate of increase with temperature leveled off. Thus, there is a lack of predictive staphylococcal enterotoxin models.
Overview of microbial risk assessments of S. aureus.
Risk assessments of S. aureus encompass a range of approaches from illustrative examples125 and partial risk assessments,126 to quantitative microbial risk assessments (QMRA) based on probabilistic modeling.127 Food products assessed include milk,128,129 skim milk,130 unripened raw-milk cheese,131 pork-based Korean food,127 kimbab,132,133 home-cooked foods126 and cream-filled baked goods.10 The results of risk assessments are equally varied. For model unripened raw milk cheese production Lindqvist et al.131 estimate that the probability for unsatisfactory concentrations of S. aureus (>6 log10 cfu g−1) is ∼4.5 × 10−2 in a high pH cheese and for on farm production of pasteurized drinking milk in the UK Barker et al.129 estimate that the filler tank contains significant levels of toxin with probability ∼2.9 × 10−5.
The hazard identified in risk assessments is S. aureus in general, or enterotoxigenic strains explicitly and/or the enterotoxin (SEA, or not specified). Although growth and subsequent toxin production can be prevented by storing “potentially hazardous” foods below 7°C and 10°C, respectively, poor personal hygiene and handling practices and inadequate refrigeration of foods have been identified as the main factors contributing to staphylococcal foodborne disease. As illustrated in the risk assessments, foods not stored below growth temperatures for sensory reasons (e.g., kimbab) or processing that includes steps under growth permissive conditions (e.g., cheese, milk) are also of concern.
The hazard characterization step has focused on food poisoning symptoms, and has not addressed the issues of particularly susceptible populations or immunity. For toxigenic microorganisms dose-response relations are essentially that of a chemical toxin, i.e., a threshold model.134 Due to knowledge gaps various levels of enterotoxin have been used as the threshold. In several studies, levels of bacteria in the food were used as a proxy for potentially hazardous doses based on reported levels of S. aureus required for the detection of enterotoxin. Threshold levels for enterotoxin of 20,127 94128 and 20 or 100 ng130 per serving have been used. These levels are based on outbreak data. Threshold levels expressed as the number of S. aureus bacteria of 5 to 8 log CFU per g have been used.e.g.,131 Kim et al.127 used a constant relation between toxin production and cell numbers under the conditions they evaluated based on an equation developed from milk data by Soejima et al.130:
| (2) |
where Tox is the toxin production (log ng ml−1) and C is the number of cells (log cfu ml−1).
Exposure assessments have described initial contamination of the starting ingredients and their changes, mostly growth but also inactivation, due to cooking and during production, holding and storage. In contrast, consumer handling and consumption have not been described in any detail, and exposure is assessed per g or per serving. Exposure assessments based on measurements of enterotoxin in food and subsequent calculations have been reported in, for example, reference 130, but it is more common to rely on initial data on S. aureus levels and prevalence followed by modeling of the effect of processes on changes in S. aureus levels. Two studies have assessed enterotoxin production based on predicted numbers of S. aureus using either the model of Fujikawa and Morozumi124 and 15°C as the temperature limit for toxin production,128 or Equation 2.127
Risk characterization has been based on the number of S. aureus (CFU) or the concentration of enterotoxin (ng) per g or per serving. Sensitivity and scenario analyses in these studies have identified the initial contamination levels together with temperatures and storage/holding times127,130,131,133 and pH131 as having the greatest impact on the assessment endpoints. In one study, the assumption concerning the threshold level for the number of S. aureus cells required for hazardous levels of enterotoxin to be produced contributed most to the uncertainty in the risk estimate.131 This highlights the importance of filling the knowledge gap concerning the relationships between growth, survival and enterotoxin production in various foods and in dose-response relationships. This shortcoming is also reflected in the lack of predictive models for risk assessment and evaluation of process safety. Thus, safety is commonly evaluated based on predicted levels of S. aureus that have been associated with enterotoxin production, and more seldom in terms of the predicted enterotoxin level or the actual measured value. This may be a limitation in view of the dynamic and complex interplay between growth, gene expression, metabolism and enterotoxin levels and the potential uncoupling between cell numbers and the amount of enterotoxin produced, as reviewed in this manuscript. Potentially stronger models that describe the production of enterotoxin in conditions that correspond to food matrices could have the biggest impact on estimates of risk for SFP.
Advances in risk assessments.
It is clear that the amount of information and understanding relating to the biology of S. aureus and to SFP, is increasing rapidly. Novel molecular techniques, in particular, provide improved understanding of virulence and survival mechanisms, etc., as well as providing opportunities for improved detection and improved typing of S. aureus and SEs. However, this increase in available information is not fully reflected by developments in risk assessment and several challenges remain.
Established risk assessments focus on enterotoxigenic strains of S. aureus in order to estimate hazard strength but, in general, they do not discriminate other (sub)types; for example, types based on serology, on toxin genes or on the molecular markers that are commonly used in epidemiology.e.g.135 For risk assessments type discrimination establishes a heterogeneous population that requires an assignment of (exponentially) increasing numbers of parameters to give a complete quantitative description. It seems inevitable that improved risk assessments will use clustering of types, based on hazard potential, but appropriate clusters are currently unknown. Toxin type appears most appropriate for clustering of S. aureus hazards and, in particular, the toxin gene regulation mechanisms are crucial,e.g.136 however, quantitative models for inclusion in risk assessments have not been established. For some hazards, such as those associated with dairy products, there is an indication that small groups of toxin types, e.g., A, G and I, could identify populations that can be treated as homogeneous for risk assessments.137
Similar considerations, related to a reduction in complexity, surround assessments for hazards, such as those in dairy products, where S. aureus populations co-exist with other bacteria. Predictive models for population kinetics do not routinely account for complex competing populations and, in many cases, the precise mechanism of competition remains unclear. For hazards associated with S. aureus, improved risk assessments will include the role of coexisting bacteria but, in order to avoid overwhelming complexities, will distill the relevant interaction properties rather than describe detailed coupled dynamics. This approach is currently being used to establish the role of starter cultures in virulence expression of S. aureus and hence on hazards associated with S. aureus in cheese.e.g.,107
Current quantitative risk assessments for staphylococcal food poisoning include very generic information concerning the production of toxins, whereas molecular approaches highlight an increasing diversity of toxin types and regulation mechanisms. Sensitivity analysis indicates that, for endpoint measures related to enterotoxin, a population threshold for the initiation of toxin production is more significant than a temperature threshold or rate parameters.129 This indicates that quantitative details for the population sensing mechanisms, and its relationship with the environment and with toxin types, will be necessary to inform improved risk assessments. Molecular methods have been used to identify differential expression of the staphylococcal enterotoxin genes during cell growth58 and at explicit points during cheese making;138 the details of transcription for SE genes can potentially inform improved models for toxin production and, hence, improved risk assessments.
Generally, risk assessments of the hazards associated with S. aureus have concentrated on the dynamics of cell populations and toxin production, whereas during the past decade research has been dominated by the association between virulence factors and accessory genetic elements, such as plasmids, prophages and pathogenicity islands.e.g.,139 Currently the mechanism for including mobile genetics and horizontal transfer, into risk assessment methodology is uncertain but it is clear that this will be crucial for improved quantitative understanding of hazards associated with S. aureus. The association between toxin genes and mobile elements that supply antibiotic resistance, which are more widely studied, will contribute to this development.140
Future Perspectives
Foodborne diseases caused by bacteria present a constantly evolving challenge, and although a great deal is known about these bacteria, we are still not able to control them. Consumer trends and demands for fresh, minimally processed food are the driving force for the development of new innovative methods of food processing and preservation. Knowledge concerning pathogenic virulence may improve our understanding of foodborne diseases, allowing new solutions to the problem to be developed. This review has described how different food parameters influence SE formation and the importance of in situ studies. The traditional view is that SE production is correlated with bacterial growth, i.e., the more cells, the more toxin and the number of bacteria is usually counted to determine whether a food product is safe for human consumption. However, bacterial growth and SE production may be decoupled, and the behavior of S. aureus in food environments may be very different from that of pure bacteria in liquid cultures. This highlights the importance of performing studies in food matrices if the results are to be applied to real food products. A better understanding of how bacterial growth and virulence expression are related and regulated by environmental factors and food preservatives will provide safer food products and give rise to new approaches to disease prevention and control in the future, through the improvement of quantitative risk assessments.
Acknowledgments
We would like to acknowledge the Swedish research council for environment, agricultural sciences and spatial planning (FORMAS) for financial support. G.C.B. acknowledges financial support from a competitive strategic grant from the BBSRC, United Kingdom.
Abbreviations
- SFP
staphylococcal food poisoning
- SE
staphylococcal enterotoxin
- SaPI
Staphylococcus aureus pathogenicity island
References
- 1.Zink DL. The impact of consumer demands and trends on food processing. Emerg Infect Dis. 1997;3:467–469. doi: 10.3201/eid0304.970408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hedberg CW, MacDonald KL, Osterholm MT. Changing epidemiology of food-borne disease: a Minnesota perspective. Clin Infect Dis. 1994;18:671–682. doi: 10.1093/clinids/18.5.671. [DOI] [PubMed] [Google Scholar]
- 3.WHO, author. Initiative to estimate the Global Burden of Foodborne Diseases; Foodborne Diseases—a Growing Risk. World Health Organization; 2010. [DOI] [PubMed] [Google Scholar]
- 4.Adams MR, Moss MO. Bacterial agents of foodborne illness—Staphylococcus aureus. Food Microbiology. Cambridge: Royal Society of Chemistry; 2008. [Google Scholar]
- 5.EFSA, author. The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and food-borne outbreaks in 2009. EFSA J. 2011:2090. doi: 10.2903/j.efsa.2018.5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cretenet M, Nouaille S, Thouin J, Rault L, Stenz L, Francois P, et al. Staphylococcus aureus virulence and metabolism are dramatically affected by Lactococcus lactis in cheese matrix. Environ Microbiol Rep. 2011;3:340–351. doi: 10.1111/j.1758-2229.2010.00230.x. [DOI] [PubMed] [Google Scholar]
- 7.Márta D, Wallin-Carlquist N, Schelin J, Borch E, Radstrom P. Extended staphylococcal enterotoxin D expression in ham products. Food Microbiol. 2011;28:617–620. doi: 10.1016/j.fm.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 8.Wallin-Carlquist N, Marta D, Borch E, Radstrom P. Prolonged expression and production of Staphylococcus aureus enterotoxin A in processed pork meat. Int J Food Microbiol. 2010;141:69–74. doi: 10.1016/j.ijfoodmicro.2010.03.028. [DOI] [PubMed] [Google Scholar]
- 9.Havelaar AH, Brul S, de Jong A, de Jonge R, Zwietering MH, Ter Kuile BH. Future challenges to microbial food safety. Int J Food Microbiol. 2010;139:79–94. doi: 10.1016/j.ijfoodmicro.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 10.Stewart CM, Cole MB, Schaffner DW. Managing the risk of staphylococcal food poisoning from cream-filled baked goods to meet a food safety objective. J Food Prot. 2003;66:1310–1325. doi: 10.4315/0362-028x-66.7.1310. [DOI] [PubMed] [Google Scholar]
- 11.Principles and guidelines for the conduct of microbiological risk management (MRM) Guidance document. 2007:1–19. Anonymous. [Google Scholar]
- 12.Havelaar AH, Nauta MJ, Jansen JT. Fine-tuning Food Safety Objectives and risk assessment. Int J Food Microbiol. 2004;93:11–29. doi: 10.1016/j.ijfoodmicro.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Murray PR, Rosenthal KS, Kobayashi GS, Pfaller MA. Medical Microbiology. Mosby: St. Louis; 2002. [Google Scholar]
- 14.Prescott LM, Harley JP, Klein DA. Microbiology. 2005. [Google Scholar]
- 15.Le Loir Y, Baron F, Gautier M. Staphylococcus aureus and food poisoning. Genet Mol Res. 2003;2:63–76. [PubMed] [Google Scholar]
- 16.Smyth CJ, Smyth DS, Kennedy J, Twohig J, Bolton DJ. Staphylococcus aureus: from man or animal—an enterotoxin iceberg? In: Maunsell B, Sheridan J, Bolton DJ, editors. EU-RAIN. Padua, Italy: Teagasc—The National Food Centre; 2004. [Google Scholar]
- 17.Borch E, Nesbakken T, Christensen H. Hazard identification in swine slaughter with respect to foodborne bacteria. Int J Food Microbiol. 1996;30:9–25. doi: 10.1016/0168-1605(96)00988-9. [DOI] [PubMed] [Google Scholar]
- 18.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis. 2011;17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asao T, Kumeda Y, Kawai T, Shibata T, Oda H, Haruki K, et al. An extensive outbreak of staphylococcal food poisoning due to low-fat milk in Japan: estimation of enterotoxin A in the incriminated milk and powdered skim milk. Epidemiol Infect. 2003;130:33–40. doi: 10.1017/S0950268802007951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlievert PM, Case LC. Molecular analysis of staphylococcal superantigens. Methods Mol Biol. 2007;391:113–126. doi: 10.1007/978-1-59745-468-1_9. [DOI] [PubMed] [Google Scholar]
- 21.Thomas DY, Jarraud S, Lemercier B, Cozon G, Echasserieau K, Etienne J, et al. Staphylococcal enterotoxin-like toxins U2 and V, two new staphylococcal superantigens arising from recombination within the enterotoxin gene cluster. Infect Immun. 2006;74:4724–4734. doi: 10.1128/IAI.00132-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lina G, Bohach GA, Nair SP, Hiramatsu K, Jouvin-Marche E, Mariuzza R. Standard nomenclature for the superantigens expressed by Staphylococcus. J Infect Dis. 2004;189:2334–2336. doi: 10.1086/420852. [DOI] [PubMed] [Google Scholar]
- 23.Altboum Z, Hertman I, Sarid S. Penicillinase plasmidlinked genetic determinants for enterotoxins B and C1 production in Staphylococcus aureus. Infect Immun. 1985;47:514–521. doi: 10.1128/iai.47.2.514-521.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayles KW, Iandolo JJ. Genetic and molecular analyses of the gene encoding staphylococcal enterotoxin D. J Bacteriol. 1989;171:4799–4806. doi: 10.1128/jb.171.9.4799-4806.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betley MJ, Mekalanos JJ. Staphylococcal enterotoxin A is encoded by phage. Science. 1985;229:185–187. doi: 10.1126/science.3160112. [DOI] [PubMed] [Google Scholar]
- 26.Borst DW, Betley MJ. Phage-associated differences in staphylococcal enterotoxin A gene (sea) expression correlate with sea allele class. Infect Immun. 1994;62:113–118. doi: 10.1128/iai.62.1.113-118.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Couch JL, Soltis MT, Betley MJ. Cloning and nucleotide sequence of the type E staphylococcal enterotoxin gene. J Bacteriol. 1988;170:2954–2960. doi: 10.1128/jb.170.7.2954-2960.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fitzgerald JR, Monday SR, Foster TJ, Bohach GA, Hartigan PJ, Meaney WJ, et al. Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens. J Bacteriol. 2001;183:63–70. doi: 10.1128/JB.183.1.63-70.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jarraud S, Peyrat MA, Lim A, Tristan A, Bes M, Mougel C, et al. egc, a highly prevalent operon of enterotoxin gene, forms a putative nursery of superantigens in Staphylococcus aureus. J Immunol. 2001;166:669–677. doi: 10.4049/jimmunol.166.1.669. [DOI] [PubMed] [Google Scholar]
- 30.Khan SA, Novick RP. Structural analysis of plasmid pSN2 in Staphylococcus aureus: no involvement in enterotoxin B production. J Bacteriol. 1982;149:642–649. doi: 10.1128/jb.149.2.642-649.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, et al. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225–1240. doi: 10.1016/S0140-6736(00)04403-2. [DOI] [PubMed] [Google Scholar]
- 32.Letertre C, Perelle S, Dilasser F, Fach P. Identification of a new putative enterotoxin SEU encoded by the egc cluster of Staphylococcus aureus. J Appl Microbiol. 2003;95:38–43. doi: 10.1046/j.1365-2672.2003.01957.x. [DOI] [PubMed] [Google Scholar]
- 33.Munson SH, Tremaine MT, Betley MJ, Welch RA. Identification and characterization of staphylococcal enterotoxin types G and I from Staphylococcus aureus. Infect Immun. 1998;66:3337–3348. doi: 10.1128/iai.66.7.3337-3348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Omoe K, Hu DL, Takahashi-Omoe H, Nakane A, Shinagawa K. Identification and characterization of a new staphylococcal enterotoxin-related putative toxin encoded by two kinds of plasmids. Infect Immun. 2003;71:6088–6094. doi: 10.1128/IAI.71.10.6088-94.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orwin PM, Leung DY, Donahue HL, Novick RP, Schlievert PM. Biochemical and biological properties of Staphylococcal enterotoxin K. Infect Immun. 2001;69:360–366. doi: 10.1128/IAI.69.1.360-6.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shafer WM, Iandolo JJ. Chromosomal locus for staphylococcal enterotoxin B. Infect Immun. 1978;20:273–278. doi: 10.1128/iai.20.1.273-278.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shalita Z, Hertman I, Sarid S. Isolation and characterization of a plasmid involved with enterotoxin B production in Staphylococcus aureus. J Bacteriol. 1977;129:317–325. doi: 10.1128/jb.129.1.317-325.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yarwood JM, McCormick JK, Paustian ML, Orwin PM, Kapur V, Schlievert PM. Characterization and expression analysis of Staphylococcus aureus pathogenicity island 3. Implications for the evolution of staphylococcal pathogenicity islands. J Biol Chem. 2002;277:13138–13147. doi: 10.1074/jbc.M111661200. [DOI] [PubMed] [Google Scholar]
- 39.Zhang S, Iandolo JJ, Stewart GC. The enterotoxin D plasmid of Staphylococcus aureus encodes a second enterotoxin determinant (sej) FEMS Microbiol Lett. 1998;168:227–233. doi: 10.1111/j.1574-6968.1998.tb13278.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang S, Stewart GC. Staphylococcal enterotoxins. In: Honeyman A, Friedman H, Bendinelli M, editors. Staphylococcus aureus infection and disease. New York: Plenum Publishing Corporation; 2001. pp. 117–136. [Google Scholar]
- 41.Denny CB, Tan PL, Bohrer CW. Heat inactivation of staphylococcal enterotoxin A. J Food Sci. 1966;31:762–767. doi: 10.1111/j.1365-2621.1966.tb01938.x. [DOI] [Google Scholar]
- 42.Genigeorgis CA. Present state of knowledge on staphylococcal intoxication. Int J Food Microbiol. 1989;9:327–360. doi: 10.1016/0168-1605(89)90100-1. [DOI] [PubMed] [Google Scholar]
- 43.Daoud SM, Debevere JM. The effect of Bacillus subtilis and Streptococcus faecalis var. liquefaciens on staphylococcal enterotoxin A activity. Int J Food Microbiol. 1985;2:211–218. doi: 10.1016/0168-1605(85)90011-X. [DOI] [Google Scholar]
- 44.Donnelly CB, Leslie JE, Black LA. Production of enterotoxin A in milk. Appl Microbiol. 1968;16:917–924. doi: 10.1128/am.16.6.917-924.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chordash RA, Potter NN. Stability of staphylococcal enterotoxin A in selected conditions encountered in foods. J Food Sci. 1976;41:906–909. doi: 10.1111/j.1365-2621.1976.tb00750_41_4.x. [DOI] [Google Scholar]
- 46.Hallis BA, Thurston CF, Mason JR. Glucose control of staphylococcal enterotoxin A synthesis and location is mediated by cyclic AMP. FEMS Microbiol Lett. 1991;80:247–251. doi: 10.1111/j.1574-6968.1991.tb04670.x. [DOI] [PubMed] [Google Scholar]
- 47.Bore E, Langsrud S, Langsrud O, Rode TM, Holck A. Acid-shock responses in Staphylococcus aureus investigated by global gene expression analysis. Microbiology. 2007;153:2289–2303. doi: 10.1099/mic.0.2007/005942-0. [DOI] [PubMed] [Google Scholar]
- 48.Fraser JD, Proft T. The bacterial superantigen and superantigen-like proteins. Immunol Rev. 2008;225:226–243. doi: 10.1111/j.1600-065X.2008.00681.x. [DOI] [PubMed] [Google Scholar]
- 49.Wieneke AA, Roberts D, Gilbert RJ. Staphylococcal food poisoning in the United Kingdom 1969–90. Epidemiol Infect. 1993;110:519–531. doi: 10.1017/S0950268800050949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kérouanton A, Hennekinne JA, Letertre C, Petit L, Chesneau O, Brisabois A, et al. Characterization of Staphylococcus aureus strains associated with food poisoning outbreaks in France. Int J Food Microbiol. 2007;115:369–375. doi: 10.1016/j.ijfoodmicro.2006.10.050. [DOI] [PubMed] [Google Scholar]
- 51.Cha JO, Lee JK, Jung YH, Yoo JI, Park YK, Kim BS, et al. Molecular analysis of Staphylococcus aureus isolates associated with staphylococcal food poisoning in South Korea. J Appl Microbiol. 2006;101:864–871. doi: 10.1111/j.1365-2672.2006.02957.x. [DOI] [PubMed] [Google Scholar]
- 52.Wallin-Carlquist N, Cao R, Marta D, da Silva AS, Schelin J, Radstrom P. Acetic acid increases the phage-encoded enterotoxin A expression in Staphylococcus aureus. BMC Microbiol. 2010;10:147. doi: 10.1186/1471-2180-10-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baba T, Bae T, Schneewind O, Takeuchi F, Hiramatsu K. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J Bacteriol. 2008;190:300–310. doi: 10.1128/JB.01000-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K, Oguchi A, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/S0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 55.Holden MT, Feil EJ, Lindsay JA, Peacock SJ, Day NP, Enright MC, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci USA. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, Grumann D, et al. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J Bacteriol. 2009;191:3462–3468. doi: 10.1128/JB.01804-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sumby P, Waldor MK. Transcription of the toxin genes present within the Staphylococcal phage phiSa3ms is intimately linked with the phage's life cycle. J Bacteriol. 2003;185:6841–6851. doi: 10.1128/JB.185.23.6841-51.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Derzelle S, Dilasser F, Duquenne M, Deperrois V. Differential temporal expression of the staphylococcal enterotoxins genes during cell growth. Food Microbiol. 2009;26:896–904. doi: 10.1016/j.fm.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 59.Novick RP, Schlievert P, Ruzin A. Pathogenicity and resistance islands of staphylococci. Microbes Infect. 2001;3:585–594. doi: 10.1016/S1286-4579(01)01414-9. [DOI] [PubMed] [Google Scholar]
- 60.Novick RP. Mobile genetic elements and bacterial toxinoses: the superantigen-encoding pathogenicity islands of Staphylococcus aureus. Plasmid. 2003;49:93–105. doi: 10.1016/S0147-619X(02)00157-9. [DOI] [PubMed] [Google Scholar]
- 61.Novick RP, Christie GE, Penades JR. The phage-related chromosomal islands of Gram-positive bacteria. Nat Rev Microbiol. 2010;8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Czop JK, Bergdoll MS. Staphylococcal enterotoxin synthesis during the exponential, transitional and stationary growth phases. Infect Immun. 1974;9:229–235. doi: 10.1128/iai.9.2.229-235.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gaskill ME, Khan SA. Regulation of the enterotoxin B gene in Staphylococcus aureus. J Biol Chem. 1988;263:6276–6280. [PubMed] [Google Scholar]
- 64.Otero A, Garcia ML, Garcia MC, Moreno B, Bergdoll MS. Production of staphylococcal enterotoxins C1 and C2 and thermonuclease throughout the growth cycle. Appl Environ Microbiol. 1990;56:555–559. doi: 10.1128/aem.56.2.555-559.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Regassa LB, Couch JL, Betley MJ. Steady-state staphylococcal enterotoxin type C mRNA is affected by a product of the accessory gene regulator (agr) and by glucose. Infect Immun. 1991;59:955–962. doi: 10.1128/iai.59.3.955-962.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the staphylococci. Chem Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Novick RP. Gram Positive Pathogens. Washington DC: ASM press; 2006. Staphylococcal Pathogenesis and Pathogenicity Factors: Genetics and Regulation; pp. 486–516. [Google Scholar]
- 68.Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–564. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 69.Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, et al. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 2001;183:7341–7353. doi: 10.1128/JB.183.24.7341-53.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morfeldt E, Tegmark K, Arvidson S. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol Microbiol. 1996;21:1227–1237. doi: 10.1046/j.1365-2958.1996.751447.x. [DOI] [PubMed] [Google Scholar]
- 71.Compagnone-Post P, Malyankar U, Khan SA. Role of host factors in the regulation of the enterotoxin B gene. J Bacteriol. 1991;173:1827–1830. doi: 10.1128/jb.173.5.1827-1830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tseng CW, Stewart GC. Rot repression of enterotoxin B expression in Staphylococcus aureus. J Bacteriol. 2005;187:5301–5309. doi: 10.1128/JB.187.15.5301-9.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tseng CW, Zhang S, Stewart GC. Accessory gene regulator control of staphyloccoccal enterotoxin D gene expression. J Bacteriol. 2004;186:1793–1801. doi: 10.1128/JB.186.6.1793-801.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barber LE, Deibel RH. Effect of pH and oxygen tension on staphylococcal growth and enterotoxin formation in fermented sausage. Appl Microbiol. 1972;24:891–898. doi: 10.1128/am.24.6.891-898.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Fujimoto DF, Higginbotham RH, Sterba KM, Maleki SJ, Segall AM, Smeltzer MS, et al. Staphylococcus aureus SarA is a regulatory protein responsive to redox and pH that can support bacteriophage lambda integrasemediated excision/recombination. Mol Microbiol. 2009;74:1445–1458. doi: 10.1111/j.1365-2958.2009.06942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pragman AA, Yarwood JM, Tripp TJ, Schlievert PM. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J Bacteriol. 2004;186:2430–2438. doi: 10.1128/JB.186.8.2430-8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yarwood JM, McCormick JK, Schlievert PM. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J Bacteriol. 2001;183:1113–1123. doi: 10.1128/JB.183.4.1113-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. Staphylococcus aureus CodY negatively regulates virulence gene expression. J Bacteriol. 2008;190:2257–2265. doi: 10.1128/JB.01545-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shivers RP, Sonenshein AL. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Microbiol. 2004;53:599–611. doi: 10.1111/j.1365-2958.2004.04135.x. [DOI] [PubMed] [Google Scholar]
- 81.Bronner S, Monteil H, Prevost G. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol Rev. 2004;28:183–200. doi: 10.1016/j.femsre.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 82.Heinrichs JH, Bayer MG, Cheung AL. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J Bacteriol. 1996;178:418–423. doi: 10.1128/jb.178.2.418-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Voyich JM, Vuong C, DeWald M, Nygaard TK, Kocianova S, Griffith S, et al. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J Infect Dis. 2009;199:1698–1706. doi: 10.1086/598967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 85.Regassa LB, Betley MJ. High sodium chloride concentrations inhibit staphylococcal enterotoxin C gene (sec) expression at the level of sec mRNA. Infect Immun. 1993;61:1581–1585. doi: 10.1128/iai.61.4.1581-1585.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yarwood JM, Schlievert PM. Quorum sensing in Staphylococcus infections. J Clin Invest. 2003;112:1620–1625. doi: 10.1172/JCI20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chan PF, Foster SJ. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J Bacteriol. 1998;180:6232–6241. doi: 10.1128/jb.180.23.6232-6241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schmidt KA, Donegan NP, Kwan WA, Jr, Cheung A. Influences of sigmaB and agr on expression of staphylococcal enterotoxin B (seb) in Staphylococcus aureus. Can J Microbiol. 2004;50:351–360. doi: 10.1139/w04-017. [DOI] [PubMed] [Google Scholar]
- 89.Kullik I, Giachino P, Fuchs T. Deletion of the alternative sigma factor sigmaB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J Bacteriol. 1998;180:4814–4820. doi: 10.1128/jb.180.18.4814-4820.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sakai F, Ihara H, Aoyama K, Igarashi H, Yanahira S, Ohkubo T, et al. Characteristics of enterotoxin H-producing Staphylococcus aureus isolated from clinical cases and properties of the enterotoxin productivity. J Food Prot. 2008;71:1855–1860. doi: 10.4315/0362-028x-71.9.1855. [DOI] [PubMed] [Google Scholar]
- 91.Pocsfalvi G, Cacace G, Cuccurullo M, Serluca G, Sorrentino A, Schlosser G, et al. Proteomic analysis of exoproteins expressed by enterotoxigenic Staphylococcus aureus strains. Proteomics. 2008;8:2462–2476. doi: 10.1002/pmic.200700965. [DOI] [PubMed] [Google Scholar]
- 92.Ikeda T, Tamate N, Yamaguchi K, Makino S. Mass outbreak of food poisoning disease caused by small amounts of staphylococcal enterotoxins A and H. Appl Environ Microbiol. 2005;71:2793–2795. doi: 10.1128/AEM.71.5.2793-5.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jørgensen HJ, Mathisen T, Lovseth A, Omoe K, Qvale KS, Loncarevic S. An outbreak of staphylococcal food poisoning caused by enterotoxin H in mashed potato made with raw milk. FEMS Microbiol Lett. 2005;252:267–272. doi: 10.1016/j.femsle.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 94.De Buyser ML, Dufour B, Maire M, Lafarge V. Implication of milk and milk products in food-borne diseases in France and in different industrialised countries. Int J Food Microbiol. 2001;67:1–17. doi: 10.1016/S0168-1605(01)00443-3. [DOI] [PubMed] [Google Scholar]
- 95.Valero A, Perez-Rodriguez F, Carrasco E, Fuentes-Alventosa JM, Garcia-Gimeno RM, Zurera G. Modelling the growth boundaries of Staphylococcus aureus: Effect of temperature, pH and water activity. Int J Food Microbiol. 2009;133:186–194. doi: 10.1016/j.ijfoodmicro.2009.05.023. [DOI] [PubMed] [Google Scholar]
- 96.Bang W, Hanson DJ, Drake MA. Effect of salt and sodium nitrite on growth and enterotoxin production of Staphylococcus aureus during the production of air-dried fresh pork sausage. J Food Prot. 2008;71:191–195. doi: 10.4315/0362-028x-71.1.191. [DOI] [PubMed] [Google Scholar]
- 97.Jarvis AW, Lawrence RC, Pritchard GG. Glucose repression of enterotoxins A, B and C and other extracellular proteins in staphlyococci in batch and continuous culture. J Gen Microbiol. 1975;86:75–87. doi: 10.1099/00221287-86-1-75. [DOI] [PubMed] [Google Scholar]
- 98.Markus Z, Silverman GJ. Enterotoxin B synthesis by replicating and nonreplicating cells of Staphylococcus aureus. J Bacteriol. 1969;97:506–512. doi: 10.1128/jb.97.2.506-512.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Boles BR, Horswill AR. agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4:1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weinrick B, Dunman PM, McAleese F, Murphy E, Projan SJ, Fang Y, et al. Effect of mild acid on gene expression in Staphylococcus aureus. J Bacteriol. 2004;186:8407–8423. doi: 10.1128/JB.186.24.8407-23.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 102.Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, et al. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:4665–4684. doi: 10.1128/JB.186.14.4665-84.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wagner PL, Neely MN, Zhang X, Acheson DW, Waldor MK, Friedman DI. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J Bacteriol. 2001;183:2081–2085. doi: 10.1128/JB.183.6.2081-5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Broudy TB, Pancholi V, Fischetti VA. Induction of lysogenic bacteriophage and phage-associated toxin from group A streptococci during coculture with human pharyngeal cells. Infect Immun. 2001;69:1440–1443. doi: 10.1128/IAI.69.3.1440-3.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mühldorfer I, Hacker J, Keusch GT, Acheson DW, Tschape H, Kane AV, et al. Regulation of the Shiga-like toxin II operon in Escherichia coli. Infect Immun. 1996;64:495–502. doi: 10.1128/iai.64.2.495-502.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wirtz C, Witte W, Wolz C, Goerke C. Transcription of the phage-encoded Panton-Valentine leukocidin of Staphylococcus aureus is dependent on the phage life-cycle and on the host background. Microbiology. 2009;155:3491–3499. doi: 10.1099/mic.0.032466-0. [DOI] [PubMed] [Google Scholar]
- 107.Even S, Charlier C, Nouaille S, Ben Zakour NL, Cretenet M, Cousin FJ, et al. Staphylococcus aureus virulence expression is impaired by Lactococcus lactis in mixed cultures. Appl Environ Microbiol. 2009;75:4459–4472. doi: 10.1128/AEM.02388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ross T, McMeekin TA. Modeling microbial growth within food safety risk assessments. Risk Anal. 2003;23:179–197. doi: 10.1111/1539-6924.00299. [DOI] [PubMed] [Google Scholar]
- 109.Castillejo-Rodriguez AM, Gimeno RM, Cosano GZ, Alcala EB, Perez MR. Assessment of mathematical models for predicting Staphylococcus aureus growth in cooked meat products. J Food Prot. 2002;65:659–665. doi: 10.4315/0362-028x-65.4.659. [DOI] [PubMed] [Google Scholar]
- 110.van Gerwen SJ, Zwietering MH. Growth and inactivation models to be used in quantitative risk assessments. J Food Prot. 1998;61:1541–1549. doi: 10.4315/0362-028x-61.11.1541. [DOI] [PubMed] [Google Scholar]
- 111.McMeekin TA, Baranyi J, Bowman J, Dalgaard P, Kirk M, Ross T, et al. Information systems in food safety management. Int J Food Microbiol. 2006;112:181–194. doi: 10.1016/j.ijfoodmicro.2006.04.048. [DOI] [PubMed] [Google Scholar]
- 112.Seo KY, Heo SK, Lee C, Chung DH, Kim MG, Lee KH, et al. Development of predictive mathematical model for the growth kinetics of Staphylococcus aureus by response surface model. J Microbiol Biotechnol. 2007;17:1437–1444. [PubMed] [Google Scholar]
- 113.Belay N, Rasooly A. Staphylococcus aureus growth and enterotoxin A production in an anaerobic environment. J Food Prot. 2002;65:199–204. doi: 10.4315/0362-028x-65.1.199. [DOI] [PubMed] [Google Scholar]
- 114.Borneman DL, Ingham SC, Ane C. Mathematical approaches to estimating lag-phase duration and growth rate for predicting growth of Salmonella serovars, Escherichia coli O157:H7 and Staphylococcus aureus in raw beef, bratwurst and poultry. J Food Prot. 2009;72:1190–1200. doi: 10.4315/0362-028x-72.6.1190. [DOI] [PubMed] [Google Scholar]
- 115.Obeso JM, Garcia P, Martinez B, Arroyo-Lopez FN, Garrido-Fernandez A, Rodriguez A. Use of logistic regression for prediction of the fate of Staphylococcus aureus in pasteurized milk in the presence of two lytic phages. Appl Environ Microbiol. 2010;76:6038–6046. doi: 10.1128/AEM.00613-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stewart CM, Cole MB, Legan JD, Slade L, Vandeven MH, Schaffner DW. Staphylococcus aureus growth boundaries: moving towards mechanistic predictive models based on solute-specific effects. Appl Environ Microbiol. 2002;68:1864–1871. doi: 10.1128/AEM.68.4.1864-71.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Métris A, Le Marc Y, Elfwing A, Ballagi A, Baranyi J. Modelling the variability of lag times and the first generation times of single cells of E. coli. Int J Food Microbiol. 2005;100:13–19. doi: 10.1016/j.ijfoodmicro.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 118.Sado Kamdem S, Guerzoni ME, Baranyi J, Pin C. Effect of capric, lauric and alpha-linolenic acids on the division time distributions of single cells of Staphylococcus aureus. Int J Food Microbiol. 2008;128:122–128. doi: 10.1016/j.ijfoodmicro.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 119.Dengremont E, Membre JM. Statistical approach for comparison of the growth rates of five strains of Staphylococcus aureus. Appl Environ Microbiol. 1995;61:4389–4395. doi: 10.1128/aem.61.12.4389-4395.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lindqvist R. Estimation of Staphylococcus aureus growth parameters from turbidity data: characterization of strain variation and comparison of methods. Appl Environ Microbiol. 2006;72:4862–4870. doi: 10.1128/AEM.00251-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vora P, Senecal A, Schaffner DW. Survival of Staphylococcus aureus ATCC 13565 in intermediate moisture foods is highly variable. Risk Anal. 2003;23:229–236. doi: 10.1111/1539-6924.00302. [DOI] [PubMed] [Google Scholar]
- 122.Charlier C, Cretenet M, Even S, Le Loir Y. Interactions between Staphylococcus aureus and lactic acid bacteria: an old story with new perspectives. Int J Food Microbiol. 2009;131:30–39. doi: 10.1016/j.ijfoodmicro.2008.06.032. [DOI] [PubMed] [Google Scholar]
- 123.Le Marc Y, Valik L, Medvedova A. Modelling the effect of the starter culture on the growth of Staphylococcus aureus in milk. Int J Food Microbiol. 2009;129:306–311. doi: 10.1016/j.ijfoodmicro.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 124.Fujikawa H, Morozumi S. Modeling Staphylococcus aureus growth and enterotoxin production in milk. Food Microbiol. 2006;23:260–267. doi: 10.1016/j.fm.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 125.Walls I, Scott VN. Use of predictive microbiology in microbial food safety risk assessment. Int J Food Microbiol. 1997;36:97–102. doi: 10.1016/S0168-1605(97)01260-9. [DOI] [PubMed] [Google Scholar]
- 126.Taulo S, Wetlesen A, Abrahamsen R, Kululanga G, Mkakosya R, Grimason A. Microbiological hazard identification and exposure assessment of food prepared and served in rural households of Lungwena, Malawi. Int J Food Microbiol. 2008;125:111–116. doi: 10.1016/j.ijfoodmicro.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 127.Kim HJ, Griffiths MW, Fazil AM, Lammerding AM. Probabilistic risk model for staphylococcal intoxication from pork-based food dishes prepared in food service establishments in Korea. J Food Prot. 2009;72:1897–1908. doi: 10.4315/0362-028x-72.9.1897. [DOI] [PubMed] [Google Scholar]
- 128.Heidinger JC, Winter CK, Cullor JS. Quantitative microbial risk assessment for Staphylococcus aureus and Staphylococcus enterotoxin A in raw milk. J Food Prot. 2009;72:1641–1653. doi: 10.4315/0362-028x-72.8.1641. [DOI] [PubMed] [Google Scholar]
- 129.Barker GC, Goméz-Tomé N. A risk assessment model for enterotoxigenic Staphylococcus aureus in pasteurized milk; a potential route to source level inference. Risk Anal. 2011 doi: 10.1111/j.1539-6924.2011.01667.x. In press. [DOI] [PubMed] [Google Scholar]
- 130.Soejima T, Nagao E, Yano Y, Yamagata H, Kagi H, Shinagawa K. Risk evaluation for staphylococcal food poisoning in processed milk produced with skim milk powder. Int J Food Microbiol. 2007;115:29–34. doi: 10.1016/j.ijfoodmicro.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 131.Lindqvist R, Sylven S, Vagsholm I. Quantitative microbial risk assessment exemplified by Staphylococcus aureus in unripened cheese made from raw milk. Int J Food Microbiol. 2002;78:155–170. doi: 10.1016/S0168-1605(02)00237-4. [DOI] [PubMed] [Google Scholar]
- 132.Rho MJ, Schaffner DW. Microbial risk assessment of staphylococcal food poisoning in Korean kimbab. Int J Food Microbiol. 2007;116:332–338. doi: 10.1016/j.ijfoodmicro.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 133.Bahk GJ, Hong CH, Oh DH, Ha SD, Park KH, Todd EC. Modeling the level of contamination of Staphylococcus aureus in ready-to-eat kimbab in Korea. J Food Prot. 2006;69:1340–1346. doi: 10.4315/0362-028x-69.6.1340. [DOI] [PubMed] [Google Scholar]
- 134.Buchanan RL, Smith JL, Long W. Microbial risk assessment: dose-response relations and risk characterization. Int J Food Microbiol. 2000;58:159–172. doi: 10.1016/S0168-1605(00)00270-1. [DOI] [PubMed] [Google Scholar]
- 135.Melles DC, van Leeuwen WB, Snijders SV, Horst-Kreft D, Peeters JK, Verbrugh HA, et al. Comparison of multilocus sequence typing (MLST), pulsed-field gel electrophoresis (PFGE) and amplified fragment length polymorphism (AFLP) for genetic typing of Staphylococcus aureus. J Microbiol Methods. 2007;69:371–375. doi: 10.1016/j.mimet.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 136.George EA, Muir TW. Molecular mechanisms of agr quorum sensing in virulent staphylococci. ChemBioChem. 2007;8:847–855. doi: 10.1002/cbic.200700023. [DOI] [PubMed] [Google Scholar]
- 137.Rosengren A, Fabricius A, Guss B, Sylven S, Lindqvist R. Occurrence of foodborne pathogens and characterization of Staphylococcus aureus in cheese produced on farm-dairies. Int J Food Microbiol. 2010;144:263–269. doi: 10.1016/j.ijfoodmicro.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 138.Duquenne M, Fleurot I, Aigle M, Darrigo C, Borezée-Durant E, Derzelle S, et al. Tool for quantification of staphylococcal enterotoxin gene expression in cheese. Appl Environ Microbiol. 2010;76:1367–1374. doi: 10.1128/AEM.01736-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Maiques E, Ubeda C, Tormo MA, Ferrer MD, Lasa I, Novick RP, et al. Role of staphylococcal phage and SaPI integrase in intra- and interspecies SaPI transfer. J Bacteriol. 2007;189:5608–5616. doi: 10.1128/JB.00619-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Otto M. Staphylococcus aureus toxin gene hitchhikes on a transferable antibiotic resistance element. Virulence. 2010;1:49–51. doi: 10.4161/viru.1.1.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang SE, Yu RC, Chou CC. Influence of holding temperature on the growth and survival of Salmonella spp and Staphylococcus aureus and the production of staphylococcal enterotoxin in egg products. Int J Food Microbiol. 2001;63:99–107. doi: 10.1016/S0168-1605(00)00416-5. [DOI] [PubMed] [Google Scholar]
- 142.Tatini S. Influence of food environments on growth of Staphylococcus aureus and production of various enterotoxins. J Milk Food Technol. 1973;36:559–563. [Google Scholar]
- 143.Scheusner DL, Hood LL, Harmon LG. Effect of temperature and pH on growth and enterotoxin productin by Staphylococcus aureus. J Milk Food Technol. 1973;36:249–252. [Google Scholar]
- 144.Notermans S, Heuvelman CJ. Combined effect of water activity, pH and sub-optimal temperature on growth and enterotoxin production of Staphylococcus aureus. J Food Sci. 1983;48:1832–1835. doi: 10.1111/j.1365-2621.1983.tb05096.x. [DOI] [Google Scholar]
- 145.Regassa LB, Betley MJ. Alkaline pH decreases expression of the accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1992;174:5095–5100. doi: 10.1128/jb.174.15.5095-5100.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Domenech A, Hernandez FJ, Orden JA, Goyache J, Lopez B, Suarez G, et al. Effect of six organic acids on staphylococcal growth and enterotoxin production. Z Lebensm Unters Forsch. 1992;194:124–128. doi: 10.1007/BF01190181. [DOI] [PubMed] [Google Scholar]
- 147.Genigeorgis CA. Quality control for fermented meats. J Am Vet Med Assoc. 1976;169:1220–1228. [PubMed] [Google Scholar]
- 148.Onoue Y, Mori M. Amino acid requirements for the growth and enterotoxin production by Staphylococcus aureus in chemically defined media. Int J Food Microbiol. 1997;36:77–82. doi: 10.1016/S0168-1605(97)01250-6. [DOI] [PubMed] [Google Scholar]
- 149.Qi Y, Miller KJ. Effect of low water activity on staphylococcal enterotoxin A and B biosynthesis. J Food Prot. 2000;63:473–478. doi: 10.4315/0362-028x-63.4.473. [DOI] [PubMed] [Google Scholar]
- 150.Troller JA. Effect of water activity on enterotoxin B production and growth of Staphylococcus aureus. Appl Microbiol. 1971;21:435–439. doi: 10.1128/am.21.3.435-439.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Troller JA. Effect of water activity on enterotoxin A production and growth of Staphylococcus aureus. Appl Microbiol. 1972;24:440–443. doi: 10.1128/am.24.3.440-443.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lee RY, Silverman GJ, Munsey ST. Growth and enterotoxin A production by Staphylococcus aureus in precooked bacon in the intermediate moisture range. J Food Sci. 1981;46:1687–1692. doi: 10.1111/j.1365-2621.1981.tb04464.x. [DOI] [Google Scholar]
- 153.Genigeorgis C, Sadler WW. Effect of sodium chloride and pH on enterotoxin B production. J Bacteriol. 1966;92:1383–1387. doi: 10.1128/jb.92.5.1383-1387.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Genigeorgis C, Foda MS, Mantis A, Sadler WW. Effect of sodium chloride and pH on enterotoxin C production. Appl Microbiol. 1971;21:862–866. doi: 10.1128/am.21.5.862-866.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hughes A, Hurst A. The effect of NaCl on the upper temperature limit for growth of and enterotoxin synthesis by Staphylococcus aureus. Can J Microbiol. 1980;26:507–510. doi: 10.1139/m80-085. [DOI] [PubMed] [Google Scholar]
- 156.McLean RA, Lilly HD, Alford JA. Effects of meat-curing salts and temperature on production of staphylococcal enterotoxin B. J Bacteriol. 1968;95:1207–1211. doi: 10.1128/jb.95.4.1207-1211.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Baird-Parker AC. Symposium on microbial changes in foods. Factors affecting the production of bacterial food poisoning toxins. J Appl Bacteriol. 1971;34:181–197. doi: 10.1111/j.1365-2672.1971.tb02277.x. [DOI] [PubMed] [Google Scholar]
- 158.Dietrich GG, Watson RJ, Silverman GJ. Effect of shaking speed on the secretion of enterotoxin B by Staphylococcus aureus. Appl Microbiol. 1972;24:561–566. doi: 10.1128/am.24.4.561-566.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Carpenter DF, Silverman GJ. Staphylococcal enterotoxin B and nuclease production under controlled dissolved oxygen conditions. Appl Microbiol. 1974;28:628–637. doi: 10.1128/am.28.4.628-637.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Carpenter DF, Silverman GJ. Synthesis of staphylococcal enterotoxin A and nuclease under controlled fermentor conditions. Appl Environ Microbiol. 1976;31:243–248. doi: 10.1128/aem.31.2.243-248.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Woodburn M, Morita TN, Venn SZ. Production of staphylococcal enterotoxins A, B and C in colloidal dispersions. Appl Microbiol. 1973;25:825–833. doi: 10.1128/am.25.5.825-833.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gómez-Lucía E, Goyache J, Orden JA, Domenech A, Javier Hernandez F, Ruiz-Santa Quiteria JA, et al. Growth of Staphylococcus aureus and synthesis of enterotoxin during ripening of experimental Manchego-type cheese. J Dairy Sci. 1992;75:19–26. doi: 10.3168/jds.S0022-0302(92)77733-9. [DOI] [PubMed] [Google Scholar]
- 163.Bhatia A, Zahoor S. Staphylococcus aureus enterotoxins: a review. J Clin Diag Res. 2007;1:188–197. [Google Scholar]
- 164.Marr JC, Lyon JD, Roberson JR, Lupher M, Davis WC, Bohach GA. Characterization of novel type C staphylococcal enterotoxins: biological and evolutionary implications. Infect Immun. 1993;61:4254–4262. doi: 10.1128/iai.61.10.4254-4262.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Abe J, Ito Y, Onimaru M, Kohsaka T, Takeda T. Characterization and distribution of a new enterotoxin-related superantigen produced by Staphylococcus aureus. Microbiol Immunol. 2000;44:79–88. doi: 10.1111/j.1348-0421.2000.tb01250.x. [DOI] [PubMed] [Google Scholar]
- 166.Orwin PM, Leung DY, Donahue HL, Novick RP, Schlievert PM. Biochemical and biological properties of Staphylococcal enterotoxin K. Infect Immun. 2001;69:360–366. doi: 10.1128/IAI.69.1.360-6.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Omoe K, Imanishi K, Hu DL, Kato H, Fugane Y, Abe Y, et al. Characterization of novel staphylococcal enterotoxin-like toxin type P. Infect Immun. 2005;73:5540–5546. doi: 10.1128/IAI.73.9.5540-6.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ono HK, Omoe K, Imanishi K, Iwakabe Y, Hu DL, Kato H, et al. Identification and characterization of two novel staphylococcal enterotoxins, types S and T. Infect Immun. 2008;76:4999–5005. doi: 10.1128/IAI.00045-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Genigeorgis C, Riemann H, Sadler WW. Production of Enterotoxin-B in Cured Meats. J Food Sci. 1969;34:63–68. doi: 10.1111/j.1365-2621.1969.tb14363.x. [DOI] [Google Scholar]