Abstract
The enzyme HMG-CoA reductase (HMGR) has a key regulatory role in the mevalonate pathway for isoprenoid biosynthesis, critical not only for normal plant development, but also for the adaptation to demanding environmental conditions. Consistent with this notion, plant HMGR is modulated by many diverse endogenous signals and external stimuli. Protein phosphatase 2A (PP2A) is involved in auxin, abscisic acid, ethylene and brassinosteroid signaling and now emerges as a positive and negative multilevel regulator of plant HMGR, both during normal growth and in response to a variety of stress conditions. The interaction with HMGR is mediated by B″ regulatory subunits of PP2A, which are also calcium binding proteins. The new discoveries uncover the potential of PP2A to integrate developmental and calcium-mediated environmental signals in the control of plant HMGR.
Key words: HMG-CoA reductase, HMGR, mevalonate pathway, isoprenoid biosynthesis, protein phosphatase 2A, PP2A, salt stress
Plant 3-hydroxy-3-methylglutaryl-CoA reductase (HMG-CoA reductase or HMGR) was detected for the first time long ago1 and is modulated by many diverse endogenous signals and environmental factors. However, no protein factor interacting with and controlling plant HMGR in vivo had been described until recently.2 The finding that HMGR is under multilevel control by protein phosphatase 2A (PP2A)2 raises interesting hypotheses concerning the modulation of HMGR and, therefore, isoprenoid biosynthesis via the signal transduction network. In the present review we discuss these hypotheses, in connection the current knowledge on plant HMGR and PP2A. Other interesting reviews are available for a more detailed background on plant HMGR3,4 or PP2A.5–8
Plant HMG-CoA Reductase
The enzyme HMG-CoA reductase catalyzes the first committed step of the mevalonate (MVA) pathway for isoprenoid biosynthesis. In plants, this pathway provides precursors for a wide variety of isoprenoid products that are required for diverse functions including membrane biogenesis, respiration, control of growth and development, defense and protein prenylation and glycosylation.9,10 In addition, many isoprenoids have economical interest as drugs, nutraceuticals, flavors, fragrances, pigments, agrochemicals or desinfectants.11 Plant HMGR has a key regulatory role in the MVA pathway and is modulated by a myriad of endogenous signals and external stimuli such as phytohormones, calcium, calmodulin, light, chemical challenge, wounding, elicitor treatment and pathogen attack.3,4 It has been proposed that the major changes of HMGR activity would be determined at the transcriptional level, whereas post-translational mechanisms would allow a finer and faster adjustment.10 Transcriptional modulation of HMGR has been demonstrated in many plant systems, but evidence of post-translational control is still scarce.3 A. thaliana HMGR responds post-translationally to compensate the enhancement or depletion of the metabolic flux through the MVA pathway.12 Reversible phosphorylation at a conserved site of the catalytic domain,13 inhibition by calcium14 and protein degradation15 have been proposed as post-translational mechanisms regulating plant HMGR (Fig. 1).
Figure 1.
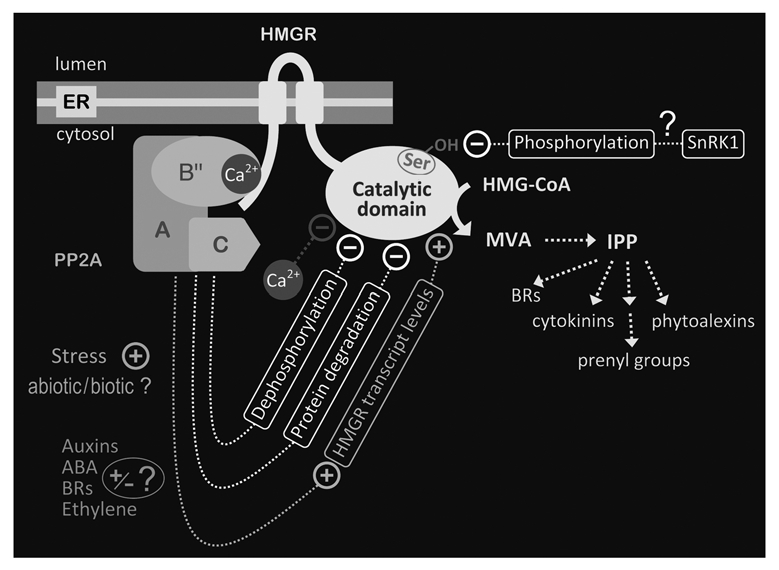
Working model on the modulation of plant HMGR by PP2A. Plant HMGR is a transmembrane protein of the ER, having the N-terminal region and catalytic domain in the cytosol. HMGR is a regulatory enzyme of the cytokinin, brassinosteroid (BR) and prenyl moiety biosynthetic pathway and can thus play a role in the signaling of these molecules. HMGR also regulates the synthesis of MVA-derived phytoalexins. PP2A is an heterotrimeric enzyme that binds the N-terminal region of HMGR through its B″ regulatory subunit, which is a calcium-binding protein. Calcium can inhibit plant HMGR post-translationally. In response to salt stress, HMGR is submitted to post-translational negative control by PP2A through dephosphorylation and protein degradation mechanisms. Dephosphorylation of HMGR by PP2A does not occur at the conserved serine residue of the catalytic domain previously proposed to be phosphorylated by the SNF1-related kinase (SnRK1). In the response to salt stress, PP2A also mediates the increase of HMGR transcript levels. Control of HMGR by PP2A could be modulated by environmental or developmental calcium- and phytohormone-mediated signals. Some of these signals could derive from the isoprenoid biosynthetic pathway, thus allowing feedback regulation on HMGR.
In all plant species studied so far, HMGR is encoded by a multigenic family composed of a variable number of genes. For instance, two HMGR-encoding genes (HMG1 and HMG2) are present in Arabidopsis thaliana,16,17 four in Solanum lycopersicum18 and at least seven in Medicago truncatula.19 Alternative splicing or transcription start could generate an even higher number of HMGR isoforms. In A. thaliana, HMGR1L and HMGR1S derive from the HMG1 gene and HMGR2 derives from HMG2.17,20 Gene expression and mutant analyses indicate a housekeeping role for HMGR1S and a more specialized function for HMGR1L and HMGR2.17,20–23 All known plant HMGR variants are targeted primarily to the endoplasmic reticulum (ER),24 but they may have specific locations in the endomembrane system.25 The diverged N-terminal region and the conserved catalytic domain of HMGR are located in the cytosol, whereas only a short stretch of amino acids connecting the two transmembrane segments is in the lumen (Fig. 1).24 This is consistent with the cytosol being the only site for MVA biosynthesis in plant cells. It was proposed that different HMGR variants might be integrated in specific arrays of enzymes or channels devoted to the synthesis of particular isoprenoid products.10 Each of these complexes could contain its own set of regulatory proteins.
Protein Phosphatase 2A
Protein phosphatase 2A (PP2A) is a structurally conserved heterotrimeric enzyme composed of a scaffolding A subunit, a regulatory B-type subunit and a catalytic C subunit (Fig. 1).6,26 Many studies in animals27–29 but still few in plants2,30 indicate that the B-type subunit determines the intracellular location and substrate specificity of the PP2A holoenzyme. So far, three distinct B-type protein families, named B, B′ and B″, have been confirmed as components of PP2A holoenzymes.5,6 PP2A is a major phosphatase that accounts for about 25% of the total protein phosphatase activity in crude homogenates from several plants.31 It is found in most plant tissues and in diverse subcellular locations including the nucleus, cytosol, membranes and insoluble fractions.8
PP2A has been proposed to modulate several metabolic processes in vascular plants, including sucrose synthesis,32 quinate metabolism,33 nitrogen assimilation,34 nocturnal CO2 fixation35 and more recently, isoprenoid biosynthesis (see below).2 Cumulative evidence indicates that PP2A participates in the signaling of diverse stress conditions.7 On the one hand, the transcript levels of particular PP2A subunits increase or decrease in response to biotic36–39 or abiotic39–47 challenges. On the other, PP2A activity or particular PP2A subunits participate as positive or negative transducers in the signaling of abscisic acid (ABA),48–50 and ethylene,51,52 in the response to cold53 or drought,45 or to ionic, osmotic and oxidative stress,54 and in the defense against biotic challenge.37 The broad distribution and functional diversity likely correspond to a multiplicity of PP2A holoenzymes (each formed by one A, one B-type and one C subunit), denoted at the genomic level. In A. thaliana, for instance, 3 genes code for A subunits, 17 for B-type subunits and 5 for C subunits, which together could theoretically form up to 255 different PP2A heterotrimers.55 The role of these A. thaliana PP2A subunits has been uncovered only in few cases: RCN1 (subunit A), involved in the targeting of auxin transport proteins,56,57 responsiveness to ABA,48 ethylene biosynthesis52 and seedling phototropism;58 B′α and B′β, involved in brassinosteroid (BR) signaling;30 B″α and B″β, involved in isoprenoid biosynthesis;2 and TONNEAU2 (subunit B″), involved in the organization of the cortical cytoskeleton.59 It was proposed that RCN1 might contribute to the crosstalk between auxin and ABA signaling pathways, which would be in turn modulated by accompanying regulatory and catalytic PP2A subunits.48 The identification of phosphorylated protein targets for PP2A in these processes is being pursued very actively at present. These studies confirm the involvement of PP2A in auxin signaling through the PIN auxin transport proteins as targets,57 seedling phototropism through the blue light sensors phot1 and phot2,58 brassinosteroid signaling through the BZR1 transcription factor,30 isoprenoid biosynthesis through the regulatory enzyme HMGR2 and ethylene biosynthesis through the regulatory enzyme 1-aminocyclopropane 1-carboxylate synthase (ACS).52
The B″ PP2A Subunit Family
The A. thaliana genome encodes five closely related B″ PP2A subunits (B″α, B″β, B″γ, B″δ and B″ε)2 and a diverged B″ variant named TONNEAU 2.59 In other plant species, the B″ PP2A subunit is also encoded by a multigenic family with two members in rice Oryza sativa (rapdb.dna.affrc.go.jp/), at least two members in Solanum lycopersicum (solgenomics.net/) and at least three members in Medicago truncatula (www.medicagohapmap.org/?genome). A. thaliana B″α and B″β where identified as proteins that interact specifically with HMGR1S and HMGR1L.2 They do not interact with HMGR2, the other member of the A. thaliana HMGR family.2 Whereas B″γ is nearly identical to B″β, with only two conservative changes in 536 amino acid residues, B″α and B″β are 67.4% identical (76.9% similar), i.e., nearly as divergent as the most distant members B″δ and B″ε (64.0% identical, 74.7% similar). Therefore, it would no be surprising that all five A. thaliana B″ isoforms bind HMGR1.2 The B″α (34% of the EST entries of A. thaliana B″ subunits), B″β (29%) and B″γ (27%) transcripts are much more abundant than those encoding B″δ (4%) and B″ε (6%). These observations indicate that the interaction with HMGR, contributed at least by B″α, B″β and B″γ, is a major role of the A. thaliana B″ protein family.
The ATH1 22k microarray data accessible at the Genevestigator facility (www.genevestigator.com/gv/index.jsp) suggest that the abundance of the B″α transcript increases in response to salt, ABA or mildew and decreases in response to potyvirus infection. Under salt stress, B″α is a negative regulator of root growth, whereas B″β does not affect this process.2 Interestingly, B″α and B″β are calcium binding proteins.2 They have a typical tandem of EF-hand calcium motifs and their binding to HMGR is stimulated by the cation.2 Calcium is a secondary messenger essential in the adaptation of plants to the environment. It mediates the signaling of many challenges such as heat- or cold-shock, slow cooling, UV radiation, red or blue light, oxidative stress, anoxia, drought/hyper-osmotic stress, salinity, hypo-osmotic stress, mechanical stimulation, heavy metals, pathogens and elicitors60,61 and might thus, be involved in the modulation of HMGR by PP2A under diverse stress conditions.
Control of HMG-CoA Reductase by PP2A
It was recently shown that PP2A exerts a multilevel control on HMGR through the B″ protein family in A. thaliana (Fig. 1).2 PP2A is not only a post-translational negative regulator of HMGR activity and protein levels, but also a positive regulator of HMG1 transcript levels. The post-translational control by PP2A involves HMGR dephosphorylation and degradation.2 Dephosphorylation of HMGR by PP2A should occur at a position different to the conserved phosphorylation site of the catalytic domain.2 Whereas B″β plays a role in the post-transcriptional repression of HMGR in unchallenged seedlings, B″α modulates HMGR transcript, protein and activity levels in response to salt challenge. The knock-out of the A. thaliana B″α gene completely abolished the modulation of HMGR by salt stress, indicating that the B″ PP2A subunits have specific roles and can not substitute for each other in the response to this challenge.2 When seedlings were transferred to salt-containing medium, B″α and PP2A mediated the decrease and subsequent increase of HMGR activity, which resulted from a steady rise of HMGR1-encoding transcript levels and an initial sharper reduction of HMGR protein level.2 A similar biphasic profile of HMGR activity was previously observed when BY-2 tobacco cells were subjected to a block of isoprenoid biosynthesis with mevinolin62 or when potato tubers were subjected to wounding,63 suggesting that PP2A and B″ subunits might also participate in these responses. HMGR transcript, protein and activity levels were also altered by the mere transfer of the seedlings to fresh plates that did not contain additional salt.2 PP2A and B″α were required for this HMGR response, likely induced by short-time desiccation or soft mechanical stimulus.2 Altogether, the data indicate that PP2A controls HMGR transcript and activity levels in different plant systems, during normal development and in response to a variety of challenging conditions. Since PP2A is involved ABA and auxin signaling, it is tempting to speculate that the effect of these phytohormones on HMGR activity observed in the past64,65 could be mediated by PP2A complexes.2
HMGR is a key regulatory enzyme of the isoprenoid biosynthetic pathway and its modulation by PP2A may be considered as an end point of the signal transduction network. PP2A might be at the heart of the control of HMGR, integrating phytohormone signals and environmental calcium-mediated stimuli (Fig. 1). The complexity of the PP2A-HMGR system and, in particular, the fact that HMGR is subjected to both positive and negative control by the same regulatory factor may account for the lengthy failure in identifying HMGR regulatory partners. Interestingly, positive and negative control by PP2A on the same target is not exclusive of HMGR. It was recently shown that A. thaliana ACS is subjected both to positive and negative regulation by PP2A.52 PP2A mediates the degradation of Type I ACS isoforms (ACS2 and ACS6) by the 26S proteasome and the stabilization of Type II ACS isoform (ACS5).52 Both HMGR and ACS catalyze key regulatory steps of biosynthetic pathways that generate phytohormone or prenyl moiety signals (cytokinins, brassinosteroids and prenyl groups for the former and ethylene for the later). PP2A connects these key nodes of metabolism with the signal transduction network, allowing modulation by environmental stimuli and, maybe, feedback regulation by the phytohormone or prenyl moiety products.
Concluding Remarks and Future Perspectives
Plant HMGR is a key regulatory enzyme of the MVA pathway and is modulated by many diverse endogenous signals and environmental factors, but the mechanisms behind this control have remained largely unknown. The recent finding of a complex HMGR control by PP2A under particular stress conditions opens new perspectives in the research of both HMGR and PP2A. It will be interesting to analyze whether PP2A is involved in the response of HMGR to other environmental challenges and to define the role of phytohormone and calcium signals to this respect. The study could uncover additional connections of HMGR with the signal transduction network. The specialization of HMGR isoforms in the synthesis of particular defense products will be relevant to this respect. The observation that PP2A can exert both positive and negative control on HMGR is intriguing. From a conceptual point of view, it will be appealing to determine whether such a behavior may result from the integration of different signals in the PP2A regulatory center.
Acknowledgments
This work was supported by grants of the Spanish Ministerio de Ciencia e Innovación (BMC2003-03450 and BFU200614655 to N.C., BFU2006-00544 and BIO2009-06984 to A.F. and BIO2009-09523 to A.B., all of them including FEDER funds), the Spanish Consolider-Ingenio 2010 Program (CSD2007-00036 Centre for Research in Agrigenomics) and the Generalitat de Catalunya (2009SGR0026). M.A. and P.L. were recipients of fellowships, respectively, from the Spanish Ministerio de Educación Cultura y Deporte and the Comissió Interdepartamental de Recerca i Innovació Tecnològica.
Abbreviations
- ABA
abscisic acid
- ACS
1-aminocyclopropane 1-carboxylate synthase
- BR
brassinosteroid
- ER
endoplasmic reticulum
- HMG-CoA
3-hydroxy-3-methylglutaryl-CoA
- HMGR
3-hydroxy-3-methylglutaryl-CoA reductase
- MVA
mevalonate
- PP2A
protein phosphatase 2A
- SnRK1
SNF1-related kinase
References
- 1.Brooker JD, Russell DW. Some properties of 3-hydroxy-3-methylglutaryl coenzyme A reductase from Pisum sativum. Roy Soc New Zealand Bull. 1974;12:365–370. [Google Scholar]
- 2.Leivar P, Antolín-Llovera M, Ferrero S, Closa M, Arró M, Ferrer A, et al. Multilevel control of Arabidopsis 3-hydroxy-3-methylglutaryl coenzyme A reductase by protein phosphatase 2A. Plant Cell. 2011;23:1494–1511. doi: 10.1105/tpc.110.074278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodríguez-Concepción M, Campos N, Ferrer A, Boronat A. Biosynthesis of isoprenoid precursors in Arabidopsis. In: Bach TJ, Rohmer M, Gershenzon J, editors. Isoprenoid Synthesis and Function in Plants and Microorganisms: New Concepts and Experimental Approaches. New York: Springer; 2011. In press. [Google Scholar]
- 4.Stermer BA, Bianchini GM, Korth KL. Regulation of HMG-CoA reductase activity in plants. J Lipid Res. 1994;35:1133–1140. [PubMed] [Google Scholar]
- 5.DeLong A. Switching the flip: protein phosphatase roles in signaling pathways. Curr Opin Plant Biol. 2006;9:470–477. doi: 10.1016/j.pbi.2006.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Luan S. Protein phosphatases in plants. Annu Rev Plant Biol. 2003;54:63–92. doi: 10.1146/annurev.arplant.54.031902.134743. [DOI] [PubMed] [Google Scholar]
- 7.País SM, Tellez-Iñón MT, Capiati DA. Serine/threonine protein phosphatases type 2A and their roles in stress signaling. Plant Signal Behav. 2009;4:1013–1015. doi: 10.4161/psb.4.11.9783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith RD, Walker JC. Plant protein phosphatases. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:101–125. doi: 10.1146/annurev.arplant.47.1.101. [DOI] [PubMed] [Google Scholar]
- 9.Bach TJ. Some new aspects of isoprenoid biosynthesis in plants—A review. Lipids. 1995;30:191–202. doi: 10.1007/BF02537822. [DOI] [PubMed] [Google Scholar]
- 10.Chappell J. Biochemistry and molecular biology of the isoprenoid biosynthetic pathway in plants. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:521–547. [Google Scholar]
- 11.Bohlmann J, Keeling CI. Terpenoid biomaterials. Plant J. 2008;54:656–669. doi: 10.1111/j.1365-313X.2008.03449.x. [DOI] [PubMed] [Google Scholar]
- 12.Nieto B, Forés O, Arró M, Ferrer A. Arabidopsis 3-hydroxy-3-methylglutaryl-CoA reductase is regulated at the post-translational level in response to alterations of the sphingolipid and the sterol biosynthetic pathways. Phytochemistry. 2009;70:53–59. doi: 10.1016/j.phytochem.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 13.Dale S, Arró M, Becerra B, Morrice NG, Boronat A, Hardie DG, et al. Bacterial expression of the catalytic domain of 3-hydroxy-3-methylglutaryl-CoA reductase (isoform HMGR1) from Arabidopsis thaliana, and its inactivation by phosphorylation at Ser577 by Brassica oleracea 3-hydroxy-3-methylglutaryl-CoA reductase kinase. Eur J Biochem. 1995;233:506–513. doi: 10.1111/j.1432-1033.1995.506_2.x. [DOI] [PubMed] [Google Scholar]
- 14.Russell DW, Knight JS, Wilson TM. Pea seedling HMG-CoA reductases: regulation of activity in vitro by phosphorylation and Ca2+, and posttranslational control in vivo by phytochrome and isoprenoid hormones. In: Randall DD, Blevins DG, Larson RL, Kagawa T, editors. Current Topics in Plant Biochemistry and Physiology. Columbia: University of Missouri-Columbia; 1985. pp. 191–206. [Google Scholar]
- 15.Korth KL, Jaggard DAW, Dixon RA. Developmental and light-regulated post-translational control of 3-hydroxy-3-methylglutaryl-CoA reductase levels in potato. Plant J. 2000;23:507–516. doi: 10.1046/j.1365-313x.2000.00821.x. [DOI] [PubMed] [Google Scholar]
- 16.Caelles C, Ferrer A, Balcells L, Hegardt FG, Boronat A. Isolation and structural characterization of a cDNA encoding Arabidopsis thaliana 3-hydroxy-3-methylglutaryl coenzyme-A reductase. Plant MolBiol. 1989;13:627–638. doi: 10.1007/BF00016018. [DOI] [PubMed] [Google Scholar]
- 17.Enjuto M, Balcells L, Campos N, Caelles C, Arró M, Boronat A. Arabidopsis thaliana contains two differentially expressed 3-hydroxy-3-methylglutaryl-CoA reductase genes, which encode microsomal forms of the enzyme. Proc Natl Acad Sci USA. 1994;91:927–931. doi: 10.1073/pnas.91.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cramer CL, Weissenborn D, Cottingham CK, Denbow CJ, Eisenback JD, Radin DN, et al. Regulation of defense-related gene expression during plant-pathogen interactions. J Nematol. 1993;25:507–518. [PMC free article] [PubMed] [Google Scholar]
- 19.Kevei Z, Lougnon G, Mergaert P, Horváth GV, Kereszt A, Jayaraman D, et al. 3-Hydroxy-3-methylglutaryl coenzyme a reductase 1 interacts with NORK and is crucial for nodulation in Medicago truncatula. Plant Cell. 2007;19:3974–3989. doi: 10.1105/tpc.107.053975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lumbreras V, Campos N, Boronat A. The use of an alternative promoter in the Arabidopsis thaliana HMG1 gene generates an mRNA that encodes a novel 3-hydroxy-3-methylglutaryl coenzyme A reductase isoform with an extended N-terminal region. Plant J. 1995;8:541–549. doi: 10.1046/j.1365-313x.1995.8040541.x. [DOI] [PubMed] [Google Scholar]
- 21.Enjuto M, Lumbreras V, Marín C, Boronat A. Expression of the Arabidopsis HMG2 gene, encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase, is restricted to meristematic and floral tissues. Plant Cell. 1995;7:517–527. doi: 10.1105/tpc.7.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohyama K, Suzuki M, Masuda K, Yoshida S, Muranaka T. Chemical phenotypes of the hmg1 and hmg2 mutants of Arabidopsis demonstrate the In-planta role of HMG-CoA reductase in triterpene biosynthesis. Chem Pharm Bull. 2007;55:1518–1521. doi: 10.1248/cpb.55.1518. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki M, Kamide Y, Nagata N, Seki H, Ohyama K, Kato H, et al. Loss of function of 3-hydroxy-3-methylglutaryl coenzyme A reductase 1 (HMG1) in Arabidopsis leads to dwarfing, early senescence and male sterility, and reduced sterol levels. Plant J. 2004;37:750–761. doi: 10.1111/j.1365-313x.2004.02003.x. [DOI] [PubMed] [Google Scholar]
- 24.Campos N, Boronat A. Targeting and topology in the membrane of plant 3-hydroxy-3-methylglutaryl coenzyme A reductase. Plant Cell. 1995;7:2163–2174. doi: 10.1105/tpc.7.12.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leivar P, González VM, Castel S, Trelease RN, López-Iglesias C, Arró M, et al. Subcellular localization of Arabidopsis 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Plant Physiol. 2005;137:57–69. doi: 10.1104/pp.104.050245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer-Jaekel RE, Hemmings BA. Protein phosphatase 2A: a “menage a trois”. Trends Cell Biol. 1994;4:287–291. doi: 10.1016/0962-8924(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 27.Janssens V, Goris J. Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell-growth and signalling. Biochem J. 2001;353:417–439. doi: 10.1042/0264-6021:3530417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sontag E. Protein phosphatase 2A: the trojan horse of cellular signaling. Cell Signal. 2001;13:7–16. doi: 10.1016/s0898-6568(00)00123-6. [DOI] [PubMed] [Google Scholar]
- 29.Virshup DM. Protein phosphatase 2A: a panoply of enzymes. Curr Opin Cell Biol. 2000;12:180–185. doi: 10.1016/s0955-0674(99)00074-5. [DOI] [PubMed] [Google Scholar]
- 30.Tang W, Yuan M, Wang R, Yang Y, Wang C, Oses-Prieto JA, et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat Cell Biol. 2011;13:124–131. doi: 10.1038/ncb2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacKintosh C, Cohen P. Identification of high-levels of type 1 and type 2A protein phosphatases in higher plants. Biochem J. 1989;262:335–339. doi: 10.1042/bj2620335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siegl G, Mackintosh C, Stitt M. Sucrose-phosphate synthase is dephosphorylated by protein phosphatase 2A in spinach leaves. Evidence from the effects of okadaic acid and microcystin. FEBS Lett. 1990;270:198–202. doi: 10.1016/0014-5793(90)81267-r. [DOI] [PubMed] [Google Scholar]
- 33.MacKintosh C, Coggins J, Cohen P. Plant protein phosphatases. Subcellular distribution, detection of protein phosphatase 2C and identification of protein phosphatase 2A as the major quinate dehydrogenase phosphatase. Biochem J. 1991;273:733–738. doi: 10.1042/bj2730733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacKintosh C. Regulation of spinach leaf nitrate reductase by reversible phosphorylation. Biochim Biophys Acta. 1992;1137:121–126. doi: 10.1016/0167-4889(92)90109-o. [DOI] [PubMed] [Google Scholar]
- 35.Dong LY, Ermolova NV, Chollet R. Partial purification and biochemical characterization of a heteromeric protein phosphatase 2A holoenzyme from maize (Zea mays L.) leaves that dephosphorylates C4 phosphoenolpyruvate carboxylase. Planta. 2001;213:379–389. doi: 10.1007/s004250100604. [DOI] [PubMed] [Google Scholar]
- 36.Alignan M, Hewezi T, Petitprez M, Dechamp-Guillaume G, Gentzbittel L. A cDNA microarray approach to decipher sunflower (Helianthus annuus) responses to the necrotrophic fungus Phoma macdonaldii. New Phytol. 2006;170:523–536. doi: 10.1111/j.1469-8137.2006.01696.x. [DOI] [PubMed] [Google Scholar]
- 37.He XH, Anderson JC, del Pozo O, Gu YQ, Tang XY, Martin GB. Silencing of subfamily I of protein phosphatase 2A catalytic subunits results in activation of plant defense responses and localized cell death. Plant J. 2004;38:563–577. doi: 10.1111/j.1365-313X.2004.02073.x. [DOI] [PubMed] [Google Scholar]
- 38.Mysore KS, Crasta OR, Tuori RP, Folkerts O, Swirsky PB, Martin GB. Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 2002;32:299–315. doi: 10.1046/j.1365-313x.2002.01424.x. [DOI] [PubMed] [Google Scholar]
- 39.País SM, González MA, Téllez-Iñón MT, Capiati DA. Characterization of potato (Solanum tuberosum) and tomato (Solanum lycopersicum) protein phosphatases type 2A catalytic subunits and their involvement in stress responses. Planta. 2009;230:13–25. doi: 10.1007/s00425-009-0923-5. [DOI] [PubMed] [Google Scholar]
- 40.Binh LT, Oono K. Molecular cloning and characterization of genes related to chilling tolerance in rice. Plant Physiol. 1992;99:1146–1150. doi: 10.1104/pp.99.3.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris DM, Myrick TL, Rundle SJ. The Arabidopsis homolog of yeast TAP42 and mammalian α4 binds to the catalytic subunit of protein phosphatase 2A and is induced by chilling. Plant Physiol. 1999;121:609–617. doi: 10.1104/pp.121.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haynes JG, Hartung AJ, Hendershot JD, III, Passingham RS, Rundle SJ. Molecular characterization of the B′ regulatory subunit gene family of Arabidopsis protein phosphatase 2A. Eur J Biochem. 1999;260:127–136. doi: 10.1046/j.1432-1327.1999.00154.x. [DOI] [PubMed] [Google Scholar]
- 43.Latorre KA, Harris DM, Rundle SJ. Differential expression of three Arabidopsis genes encoding the B' regulatory subunit of protein phosphatase 2A. Eur J Biochem. 1997;245:156–163. doi: 10.1111/j.1432-1033.1997.00156.x. [DOI] [PubMed] [Google Scholar]
- 44.Tóth EC, Vissi E, Kovács I, Szöke A, Ariño J, Gergely P, et al. Protein phosphatase 2A holoenzyme and its subunits from Medicago sativa. Plant MolBiol. 2000;43:527–536. doi: 10.1023/a:1006436925253. [DOI] [PubMed] [Google Scholar]
- 45.Xu C, Jing R, Mao X, Jia X, Chang X. A wheat (Triticum aestivum) protein phosphatase 2A catalytic subunit gene provides enhanced drought tolerance in tobacco. Ann Bot. 2007;99:439–450. doi: 10.1093/aob/mcl285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu RM, Wong MM, Jack RW, Kong RY. Structure, evolution and expression of a second subfamily of protein phosphatase 2A catalytic subunit genes in the rice plant (Oryza sativa L.) Planta. 2005;222:757–768. doi: 10.1007/s00425-005-0018-x. [DOI] [PubMed] [Google Scholar]
- 47.Yu RMK, Zhou Y, Xu ZF, Chye ML, Kong RYC. Two genes encoding protein phosphatase 2A catalytic subunits are differentially expressed in rice. Plant MolBiol. 2003;51:295–311. doi: 10.1023/a:1022006023273. [DOI] [PubMed] [Google Scholar]
- 48.Kwak JM, Moon JH, Murata Y, Kuchitsu K, Leonhardt N, Delong A, et al. Disruption of a guard cell-expressed protein phosphatase 2A regulatory subunit, RCN1, confers abscisic-acid insensitivity in Arabidopsis. Plant Cell. 2002;14:2849–2861. doi: 10.1105/tpc.003335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo J, Shen G, Yan J, He C, Zhang H. AtCHIP functions as an E3 ubiquitin ligase of protein phosphatase 2A subunits and alters plant response to abscisic acid treatment. Plant J. 2006;46:649–657. doi: 10.1111/j.1365-313X.2006.02730.x. [DOI] [PubMed] [Google Scholar]
- 50.Pernas M, Garcia-Casado G, Rojo E, Solano R, Sanchez-Serrano JJ. A protein phosphatase 2A catalytic subunit is a negative regulator of abscisic acid signalling. Plant J. 2007;51:763–778. doi: 10.1111/j.1365-313X.2007.03179.x. [DOI] [PubMed] [Google Scholar]
- 51.Larsen PB, Cancel JD. Enhanced ethylene responsiveness in the Arabidopsis eer1 mutant results from a loss-of-function mutation in the protein phosphatase 2A A regulatory subunit, RCN1. Plant J. 2003;34:709–718. doi: 10.1046/j.1365-313x.2003.01762.x. [DOI] [PubMed] [Google Scholar]
- 52.Skottke KR, Yoon GM, Kieber JJ, Delong A. Protein phosphatase 2A controls ethylene biosynthesis by differentially regulating the turnover of ACC synthase isoforms. PLoS Genet. 2011;7:1001370. doi: 10.1371/journal.pgen.1001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monroy AF, Sangwan V, Dhindsa RS. Low-temperature signal transduction during cold acclimation: protein phosphatase 2A as an early target for cold-inactivation. Plant J. 1998;13:653–660. [Google Scholar]
- 54.Blakeslee JJ, Zhou HW, Heath JT, Skottke KR, Rodríguez-Barrios JA, Liu SY, et al. Specificity of RCN1-mediated protein phosphatase 2A regulation in meristem organization and stress response in roots. Plant Physiol. 2008;146:539–553. doi: 10.1104/pp.107.112995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou HW, Nussbaumer C, Chao Y, DeLong A. Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell. 2004;16:709–722. doi: 10.1105/tpc.018994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garbers C, DeLong A, Deruère J, Bernasconi P, Söll D. A mutation in protein phosphatase 2A regulatory subunit A affects auxin transport in Arabidopsis. EMBO J. 1996;15:2115–2124. [PMC free article] [PubMed] [Google Scholar]
- 57.Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, et al. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130:1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 58.Tseng TS, Briggs WR. The Arabidopsis rcn1-1 mutation impairs dephosphorylation of phot2, resulting in enhanced blue light responses. Plant Cell. 2010;22:392–402. doi: 10.1105/tpc.109.066423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Camilleri C, Azimzadeh J, Pastuglia M, Bellini C, Grandjean O, Bouchez D. The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell. 2002;14:833–845. doi: 10.1105/tpc.010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lecourieux D, Raneva R, Pugin A. Calcium in plant defence-signalling pathways. New Phytol. 2006;171:249–269. doi: 10.1111/j.1469-8137.2006.01777.x. [DOI] [PubMed] [Google Scholar]
- 61.White PJ, Broadley MR. Calcium in plants. Ann Bot. 2003;92:487–511. doi: 10.1093/aob/mcg164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hemmerlin A, Hoeffler JF, Meyer O, Tritsch D, Kagan IA, Grosdemange-Billiard C, et al. Cross-talk between the cytosolic mevalonate and the plastidial methylerythritol phosphate pathways in tobacco Bright Yellow-2 cells. J Biol Chem. 2003;278:26666–26676. doi: 10.1074/jbc.M302526200. [DOI] [PubMed] [Google Scholar]
- 63.Yang ZB, Park HS, Lacy GH, Cramer CL. Differential activation of potato 3-hydroxy-3-methylglutaryl coenzyme A reductase genes by wounding and pathogen challenge. Plant Cell. 1991;3:397–405. doi: 10.1105/tpc.3.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nishi A, Tsuritani I. Effect of auxin on the metabolism of mevalonic acid in suspension-cultured carrot cells. Phytochemistry. 1983;22:399–401. [Google Scholar]
- 65.Russell DW, Davidson H. Regulation of cytosolic HMG-CoA reductase activity in pea seedlings: contrasting responses to different hormones and hormone-product interaction, suggest hormonal modulation of activity. Biochem Biophys Res Commun. 1982;104:1537–1543. doi: 10.1016/0006-291x(82)91426-7. [DOI] [PubMed] [Google Scholar]


