Abstract
Seed germination is an elaborate developmental process that is regulated through intricate signaling networks integrating diverse environmental cues into endogenous hormonal signaling pathways. Accumulating evidence in recent years supports the role of auxin in seed germination. Whereas the roles of gibberellic acid (GA) and abscisic acid (ABA) in the germination process have been studied extensively, how auxin modulates seed germination is largely unknown. We found that a membrane-bound NAC transcription factor NTM2 mediates the signaling crosstalk between auxin and salt stress via the IAA30 gene during seed germination in Arabidopsis. Germination of the NTM2-deficient ntm2-1 mutant seeds exhibited enhanced resistance to high salinity. However, the salt resistance was reduced in the ntm2-1 mutant overexpressing the IAA30 gene, which was induced by high salinity in a NTM2-dependent manner. Exogenous auxin treatment further suppressed the reduced germination rate of control seeds under high salinity. In contrast, the auxin effects disappeared in the ntm2-1 mutant. These observations indicate that NTM2 is a molecular link that incorporates auxin signal into salt stress signaling during seed germination, providing a role of auxin in modulating seed germination under high salinity.
Key words: Arabidopsis, auxin, IAA30, NTM2, high salinity, MTF, seed germination
Seed germination is the very first developmental process that is critical for plant establishment and propagation in nature. As sessile organisms, plants should stay throughout their life span at the place where their seeds germinate in most plant species. Therefore, seeds perceive diverse environmental cues, such as light intensity and quality, ambient temperature, water and oxygen availability and soil salinity, and incorporate these signals into endogenous developmental programs to determine when to germinate.1,2 It has been known that a variety of growth hormones, including GA, ABA and ethylene, plays a role in seed germination.3–5 These necessitate that identification and functional characterization of the molecular links that incorporate external cues into endogenous growth hormonal signaling are essential for understanding the germination process.
Auxin plays diverse roles in virtually all aspects of plant growth and developmental processes.6–8 It also plays a role in seed germination.9 Whereas roles of GA and ABA in seed germination have been extensively studied,10–13 those of auxin in seed germination is only poorly known. More direct evidence supporting the contribution of auxin to seed germination has been inferred from germination assays using transgenic plants overexpressing microRNA160 (miR160) or its target transcription factor gene Auxin Response Factor10 (ARF10).14 Seed germination of the miR160-overproducing transgenic plants is hyposensitive to ABA. In contrast, that of transgenic plants overexpressing a miR160-resistant ARF10 gene is relatively more sensitive to ABA. High soil salinity, which is likely to be mediated by ABA-dependent signaling pathways, also reduces seed germination.15 However, the relationship between auxin and high salinity in seed germination is unexplored yet.
We found that a NAM-ATAF1/2-CUC2 (NAC) transcription factor NTM2 comprises a salt signaling pathway functioning in seed germination. The NTM2 gene was induced by ABA to some degree. However, its role in seed germination under high salinity was essentially independent of ABA. The NTM2 gene was induced by high salinity primarily in the roots, a primary plant organ that perceives soil salinity. Germination of the ntm2-1 mutant seeds was less sensitive to high salinity, and accordingly the salt resistance response disappeared in the ntm2-1 mutant that was complemented with a wild-type NTM2 gene, confirming that the NTM2 gene plays a role in salt regulation of seed germination.
Notably, the NTM2-mediated salt signaling during seed germination was intimately linked with auxin signaling. Auxin does not affect the germination process under normal growth conditions. In contrast, it acts as a negative regulator of seed germination under high salinity. Intriguingly, germination of ntm2-1 seeds was influenced to a lesser degree by auxin, showing that auxin signals are interconnected with the NTM2-mediated salt signaling during seed germination under high salinity, in which the IAA30 gene plays a role. Whereas the transcript level of the IAA30 gene was elevated under high salinity, the salt induction of the IAA30 gene largely disappeared in the ntm2-1 mutant, indicating that the IAA30 gene is a component of the NTM2-mediated salt signaling. In addition, under high salinity, the germination percentage of the ntm2-1 mutant seeds overexpressing the IAA30 gene was lower than that of the parental ntm2-1 seeds but was comparable to that of control seeds, demonstrating that the effects of auxin on seed germination under high salinity is mediated at least in part by the IAA30 gene.
Our electrophoretic mobility shift assays (EMSA) and transcriptional activation activity assays in Arabidopsis protoplasts revealed that the NTM2 protein acts as a transcriptional activator and binds directly to the IAA30 gene promoter, providing a direct molecular evidence that the salt-responsive NTM2 transcription factor is a component of IAA30-mediated auxin signaling functioning during seed germination under high salinity.
Our data show that seed germination is suppressed by auxin under high salinity. To verify the notion that auxin plays a negative regulatory role in seed germination under high salinity, we generated transgenic plants overexpressing the YUCCA3 (YUC3) gene, which encodes an auxin biosynthetic enzyme.16 It was observed that seed germination of the YUC3-overexpressing plants was more sensitive to high salinity. Histochemical assays using the DR5-GUS reporter system also showed that elevation of GUS activities in the emerging radicles of germinating seeds is correlated with delayed seed germination. We also observed that GUS activity is elevated by high salinity as well as by auxin in the emerging radicles. The GUS activity was further elevated in the presence of both auxin and salt, the combination of which significantly delays seed germination.
Altogether, our observations demonstrate that auxin signals are incorporated into the NTM2-mediated salt signal transduction pathway in modulating seed germination under high salinity (Fig. 1). In this signaling crosstalk, the IAA30 gene incorporates auxin and salt signals into the germination process. Our data also support that NTM2 serves as a molecular link that interconnects a developmental feedback loop of auxin signaling with a salt signal transduction pathway during seed germination by direct regulation of the IAA30 gene.
Figure 1.
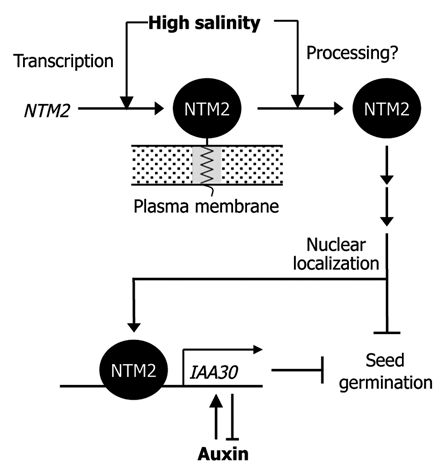
Figure 1. Schematic working model for NTM2 function in seed germination. High salinity induces NTM2 gene transcription and NTM2 protein release from the plasma membranes. The transcriptionally active NTM 2 transcription factor binds directly to the IAA30 gene promoter. In this signaling crosstalk, the NTM2 transcription factor links salt signals into an auxin signaling pathway functioning in seed germination.
A question to be answered is how the plasma membrane-localized NTM2 protein is released from the membranes and transported into the nucleus.12,17,18 We observed in subcellular localization assays that more NTM2 proteins are localized into the nucleus of Arabidopsis protoplasts after salt treatments, showing that NTM2 processing step is also influenced by high salinity. It has been known in both animals and plants that membrane-bound transcription factors (MTFs) are released proteolytically from the cellular membranes by either intramembrane proteolysis or ubiquitin/proteasome-dependent processing.19 It will be interesting to examine whether NTM2 processing by high salinity is mediated by a specific membrane-bound protease or by ubiquitin-mediated processing and how this activation process is related with regulation of seed germination by high salinity.
Acknowledgments
This work was supported by the Leaping Research Program (20100014373) provided by the National Research Foundation of Korea and by grants from the Plant Signaling Network Research Center (20100001457), the National Research Foundation of Korea (20100028147), and from the Agricultural R & D Promotion Center (309017-05-2-HD130), Korea Ministry for Food, Agriculture, Forestry and Fisheries.
Abbreviations
- ABA
abscisic acid
- ARF10
auxin response factor 10
- GA
gibberellic acid
- MTF
membrane-bound transcription factor
- NAC
NAM/ATAF1/2/CUC2
- YUC3
YUCCA3
References
- 1.Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- 2.Finkelstein R, Reeves W, Ariizumi T, Steber C. Molecular aspects of seed dormancy. Annu Rev Plant Biol. 2008;59:387–415. doi: 10.1146/annurev.arplant.59.032607.092740. [DOI] [PubMed] [Google Scholar]
- 3.Finch-Savage WE, Cadman CS, Toorop PE, Lynn JR, Hilhorst HW. Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 2008;51:60–78. doi: 10.1111/j.1365-313X.2007.03118.x. [DOI] [PubMed] [Google Scholar]
- 4.Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, et al. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 1996;10:655–661. doi: 10.1046/j.1365-313x.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- 5.Peng J, Harberd NP. The role of GA-mediated signaling in the control of seed germination. Curr Opin Plant Biol. 2002;5:376–381. doi: 10.1016/s1369-5266(02)00279-0. [DOI] [PubMed] [Google Scholar]
- 6.Weijers D, Jurgens G. Funneling auxin action: specificity in signal transduction. Curr Opin Plant Biol. 2004;7:687–693. doi: 10.1016/j.pbi.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 7.Quint M, Gray WM. Auxin signaling. Curr Opin Plant Biol. 2006;9:448–453. doi: 10.1016/j.pbi.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abel S. Auxin is surfacing. ACS Chem Biol. 2007;2:380–384. doi: 10.1021/cb7001158. [DOI] [PubMed] [Google Scholar]
- 9.Birgit K, Marc AC, Gerhard LM. Plant hormone interactions during seed dormancy release and germination. Seed Sci Res. 2005;15:281–307. [Google Scholar]
- 10.Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 11.Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, et al. CYP707A1 and CYP707A2, which encode abscisic acid 8'-hydroxylases, are indispensable for proper control of seed dormancy and germination in Arabidopsis. Plant Physiol. 2006;141:97–107. doi: 10.1104/pp.106.079475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SG, Lee AK, Yoon HK, Park CM. A membrane-bound NAC transcription factor NTL8 regulates gibberellic acid-mediated salt signaling in Arabidopsis seed germination. Plant J. 2008;55:77–88. doi: 10.1111/j.1365-313X.2008.03493.x. [DOI] [PubMed] [Google Scholar]
- 13.Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, et al. Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 2006;48:354–366. doi: 10.1111/j.1365-313X.2006.02881.x. [DOI] [PubMed] [Google Scholar]
- 14.Liu PP, Montgomery TA, Fahlgren N, Kasschau KD, Nonogaki H, Carrington JC. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007;52:133–146. doi: 10.1111/j.1365-313X.2007.03218.x. [DOI] [PubMed] [Google Scholar]
- 15.Perruc E, Kinoshita N, Lopez-Molina L. The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 2007;52:927–936. doi: 10.1111/j.1365-313X.2007.03288.x. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- 17.Kim YS, Kim SG, Park JE, Park HY, Lim MH, Chua NH, Park CM. A membrane-bound NAC transcription factor regulates cell division in Arabidopsis. Plant Cell. 2006;18:3132–3144. doi: 10.1105/tpc.106.043018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seo PJ, Kim MJ, Park JY, Kim SY, Jeon J, Lee YH, et al. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 2010;61:661–671. doi: 10.1111/j.1365-313X.2009.04091.x. [DOI] [PubMed] [Google Scholar]
- 19.Seo PJ, Kim SG, Park CM. Membrane-bound transcription factors in plants. Trends Plant Sci. 2008;13:550–556. doi: 10.1016/j.tplants.2008.06.008. [DOI] [PubMed] [Google Scholar]


