Abstract
Functionally relevant femtosecond to picosecond dynamics in enzyme active sites can be difficult to measure because of a lack of spectroscopic probes that can be located in the active site without altering the behavior of the enzyme. We have developed a new NAD+ analog 3-Picolyl Azide Adenine Dinucleotide (PAAD+), which has the potential to be a general spectroscopic probe for NAD-dependent enzymes. This analog is stable and binds in the active site of a typical NAD-dependent enzyme formate dehydrogenase (FDH) with similar characteristics to natural NAD+. It has an isolated infrared transition with high molar absorptivity that makes it suitable for observing enzyme dynamics using 2D IR spectroscopy. 2D IR experiments show that in aqueous solution, the analog undergoes complete spectral diffusion within hundreds of femtoseconds consistent with the water hydrogen bonding dynamics that would be expected. When bound to FDH in a binary complex, it shows picosecond fluctuations and a large static offset, consistent with previous studies of the binary complexes of this enzyme. These results show that PAAD+ is an excellent probe of local dynamics and that it should be a general tool for probing the dynamics of a wide range of NAD-dependent enzymes.
Keywords: two-dimensional infrared spectroscopy, enzyme dynamics, NAD analogs, formate dehydrogenase
Introduction
Enzyme motions can span from femtosecond to millisecond timescales both on the exterior of the protein as well as in the active site.1–4 Characterizing these protein motions is essential to understanding the structure-dynamics-function relationship in enzymes. There is a growing interest in understanding enzyme motions at femtosecond to picosecond time scales as it has been suggested that protein dynamics modulate the activation barrier and influence the complex energy landscape of the catalyzed reaction.5–9 The importance of fast dynamics at enzyme active sites is also invoked to explain anomalous kinetic isotope effects and their temperature dependence.10–13 These results were interpreted in the context of Marcus-like models that link environmental reorganization of the active site to the catalyzed H-transfer reactions and specifically suggest a role for femtosecond to picosecond dynamics that modulate the donoracceptor distance and, thus, the reaction rate. The potential for such motions to influence the catalyzed reaction is a subject of much debate in the community. Such dynamics were, until recently, experimentally inaccessible, but, with the advent of nonlinear vibrational techniques like 2D IR spectroscopy, it is now possible to access these motions directly.14–17
Several proteins have previously been studied by nonlinear vibrational spectroscopies including myoglobin,18, 19 hemoglobin,20 neuroglobin,21 native and unfolded cytochrome c,22 cytochrome P450,23 horseradish peroxidase,24 HIV-1 reverse transcriptase,25 carbonic anhydrase26 and formate dehydrogenase.27, 28 In all of these studies a small molecule or ion is bound to the protein and serves as a vibrational reporter of the protein dynamics. Unfortunately, the probes in these studies cannot readily be extended to a broad range of enzymes as the chromophores used in those studies are system specific and, therefore, lack generality. Another approach that is commonly used to make proteins accessible to vibrational spectroscopy is site-specific labeling either with isotopes or non-natural amino acids in which a spectroscopic label has been incorporated.29–35 Such modifications are sometimes challenging for large proteins, can result in limited quantities of material, and, in some cases, can compromise the integrity of the native protein structure. More importantly, the protocol for incorporating the spectroscopic label must be developed anew for every new protein to be studied. Although these challenges do not prohibit the widespread application of these approaches, they do make them sufficiently difficult that these labeling schemes have not been widely adopted for 2D IR applications yet. Thus there is a need for general spectroscopic reporters that can bind to the active sites of many enzymes with minimal perturbation to the native structure of the protein and that are suitable for use with 2D IR spectroscopy.
One approach to address this problem is to label a cofactor or coenzyme that can be used in a wide range of systems. Mid–IR active analogs of NAD+ represent an excellent target for developing such a probe of protein dynamics because NAD+ is a ubiquitous cofactor for many enzymes. As a cofactor, it binds in the active site and should be a good reporter of protein dynamics in the binding pocket of enzymes. Chromophores substituted at the 3-position of the nicotinamide ring of NAD+ are likely to preserve their biological function and bind to the active site in a way that is similar to native NAD+.36–39 Probes that have strong transition moments and transition frequencies that are well separated from other absorption bands are ideal for spectroscopic studies. We have recently shown that an azido-derivatized analog of NAD+, azido-NAD+, has the potential to be a general probe to investigate the active site dynamics of NAD-dependent enzymes.40 This analog exhibits binding and kinetic properties that are similar to native NAD+ for a number of enzymes, suggesting that the analog binds to the enzyme with minimal perturbations to the active site structure. For azido-NAD+ we have also studied its spectroscopic characteristics.41 The azido group of the analog is sensitive to hydrogen bond fluctuations in water as would be expected, but this probe has a weak transition moment compared to that for the azide anion, which limits its utility. In general, enzymes have poor solubility and are difficult to study with 2D IR even with strong chromophores. Because the 2D IR signal scales as εı, having a probe with higher molar absorptivity would make it much easier to study enzyme motions. An NAD+ analog retaining all the other characteristics of azido-NAD+ but with a high molar extinction coefficient would access the active site dynamics of a wide range of proteins. Such a probe would be ideal for 2D IR spectroscopy of enzyme dynamics.
Here, we report the synthesis and characterization of picolyl azide adenine dinuceotide (PAAD+), a derivative of NAD+ with a high molar absorptivity for the azido antisymmetric stretching vibration. We report the spectral dynamics of this analog in water, which are governed by hydrogen bond fluctuations and are similar to those for azido-NAD+ and other small chromophores in water. We also show that the binary complex of PAAD+ with the enzyme formate dehydrogenase (FDH) exhibits dynamics that are consistent with what we would expect for this complex based on our previous work with this enzyme. These results reveal the potential of this analog to report enzyme dynamics in FDH and a wide range of other NAD- and NADP-dependent enzymes.
Experimental Methods
Chemicals
3-picolyl chloride hydrochloride, sodium azide, sodium biocarbonate, NADase, NAD+, and all solvents were obtained from Sigma Aldrich and used as received. Formate dehydrogenase (FDH, Candida boidinii) is obtained from Roche Diagnostics, Germany.
Synthesis of 3-picolyl azide
In a round bottom flask, 3.4 g (26 mmols) of 3-picolyl chloride hydrochloride dissolves in 50 mL of water and reacts with 2.6 g (40 mmols) of sodium azide. The reaction continues for 16 hours at 50 °C and then sodium biocarbonate quenches it. An oily product is obtained after extraction with methylene chloride. The 1H-NMR shows peaks at 4.16 (2H,s), 7.3–7.38 (1H, m), 7.72–7.76 (1H, m), 8.6 (2H, s). 13C-NMR shows peaks at 123.4, 135.5, 130.9, 141.1, 141.9, 51.9. This compound is soluble in a mixture of 25% DMF in water and has an infrared absorption peak at 2107 cm−1 that has a full width at half maximum (FWHM) of 26 cm−1.
Synthesis of 3-picolyl azide adenine dinuceotide (PAAD+)
The synthetic protocol is similar to that for azido-NAD+.40 In short, NADase from pig brain effects the base exchange of picolyl azide for NAD+ at 37 °C under dark conditions. The product, PAAD+ is obtained in high purity (> 95 %) as white powder with similar solubility and stability as native NAD+ in water. An LC-MS analysis reveals one prominent peak with m/z 675.9, which is assigned to PAAD+. The compound shows an infrared absorption at 2119 cm−1 with a FWHM of 22 cm−1 in water.
Enzymatic studies
PAAD+ is an inhibitor for the enzyme FDH. The measurements of the dissociation constant, Kd, for PAAD+ with FDH are carried out on a MicroCal VP-ITC isothermal calorimeter. For these measurements, FDH is titrated with PAAD+. The concentration of FDH is 0.1 mM and the concentration of PAAD+ is approximately 1 mM. All solutions are degassed for 15 minutes prior to the experiments to prevent bubble formation. Aliquots of 14 µL of the ligand solution are added to the enzyme solution. Heats of dilution are subtracted from the raw data, and the data are analyzed using a single-site binding model42 to determine the dissociation constant.
Infrared Pump-Probe Spectroscopy
An amplified Ti:Sapphire laser system (Spectra Physic, Spitfire) generates 800 nm pulses of 80 fs duration with approximately 4 mJ of energy per pulse at a 1 kHz repetition rate. A portion of this output, about 800 µJ per pulse, pumps an optical parametric amplifier (OPA) based on a β-barium borate (BBO) crystal (θ = 27°, type II). The OPA generates two tunable near-infrared pulses, signal and idler, with a combined pulse energy of 270 µJ. The signal and idler are tuned such that the difference in the energies of the photons from each beam is equivalent to the energy of the mid-infrared light needed to excite the azido-NAD+ sample, 2140 cm−1. These are then mixed in a AgGaS2 difference frequency generation (DFG) crystal (θ = 50°, type II) that produces nearly transform limited pulses with a pulse duration of approximately 100 fs and a pulse energy of 6 µJ.
A wedged CaF2 substrate separates a small portion of the infrared light for use as a probe beam. The remaining infrared light is used as a pump beam. An infrared pulse shaper based on a germanium acousto-optic modulator built according to the design of Shim et al43, 44 chops the pump beam on alternating laser shots. A half-wave plate and polarizer in the pump beam set the polarization of the pump at the magic angle relative to the probe beam to remove the reorientation contribution to the signal. A computer-controlled translation stage varies the time delay between pump and probe pulses. The pump and probe beams are aligned parallel to one another separated by about 2 cm. A 90° off-axis parabolic mirror with an effective focal length of 100 mm focuses the beams into the sample where they have a spot size of approximately 80 µm. After a second parabolic mirror recollimates the beams, an iris isolates the probe beam. A zero-dispersion stretcher with a slit at the Fourier plane frequency narrows a portion of the 800 nm light from our femtosecond amplifier to a transform-limited bandwidth of approximately 1 cm−1. The narrowed 800 nm beam upconverts the infrared probe beam in a MgO-doped LiNbO3 crystal (θ = 46.5°, type I) into the visible. An imaging spectrometer (300 mm focal length, 1200 grooves/mm grating) disperses the upconverted probe beam and a CMOS camera with 1024 pixels measures the spectrum of each individual laser shot. Using adjacent probe spectra, we calculate the change in optical density caused by the pump beam.
2D IR spectroscopy
In heterodyned 2D IR spectroscopy, we measure the vibrational echo response as a function of one frequency variable ω3 and two time variables τ, the evolution time, and T, the waiting time. Three femtosecond infrared pulses focus into the sample and generate the vibrational echo response. The time delay between the first and second pulses is τ and that between the second and third pulses is T. For heterodyned detection, the signal overlaps with a fourth, local oscillator, pulse and passes through a monochromator to select the frequency ω3. A liquid-nitrogen-cooled mercury cadmium telluride (MCT) detector measures the interference between the signal and local oscillator. We scan the monochromator to generate the ω3 spectrum for each value of τ. As τ is stepped for a fixed value of T, the interference between signal and local oscillator produces an interferogram at each monochromator frequency. A numerical Fourier transform of each interferogram gives the ω1 spectrum at each value of ω3. A liquid sample holder made of CaF2 windows separated by a 25 µm spacer holds the sample which contains a solution with an approximately 30 mM concentration of the PAAD+ chromophore either in buffer alone or with the enzyme FDH in slight excess relative to PAAD+.
At each waiting time, we generate a 2D spectrum that functions as a correlation map showing how well correlated are the frequencies of the oscillators in our sample at time T with their initial values. If the frequencies of the oscillators have not changed much during the waiting time, then the line shape will be elongated along the diagonal. As the protein samples different conformations as a result of the enzyme dynamics, however, the line shape will rotate toward the horizontal axis. The evolution of this line shape reports the spectral diffusion dynamics of the chromophore that arise due to the structural fluctuations of the system that cause the molecules to lose the memory of their initial frequencies. The frequency-frequency correlation function (FFCF), which is given by <δω(t)δω(0)>, connects the experimental results to the underlying dynamics. This function shows the correlation between the frequencies of the ensemble of oscillators at time zero and a time T later. We employ the center line slope (CLS) method to determine the FFCF from the 2D IR data as described by Kwak et al.45, 46 In this analysis, frequency slices are taken from the 2D IR spectrum for fixed values of ω1. We then locate the frequency ω3 at which the signal goes through its maximum for each slice in ω1. This collection of points in ω3 and ω1 is called the center line, and the slope of this line, the CLS, as a function of the waiting time, T, is proportional to the FFCF. Plotting the CLS as a function of T, we fit this curve to a sum of exponentials with an offset retaining the fewest terms necessary for accurately fitting the decay. The time constants of this decay are the time scale for the decay of the FFCF as well and reflect the time scale of the structural fluctuations. To determine the absolute amplitude of the FFCF, we take the time constants of the exponentials and their relative amplitudes from the fit of the CLS decay and use those to fit the infrared absorption line shape with the absolute amplitude of the FFCF and a motionally narrowed contribution to the line shape, if necessary, as adjustable parameters.
Results
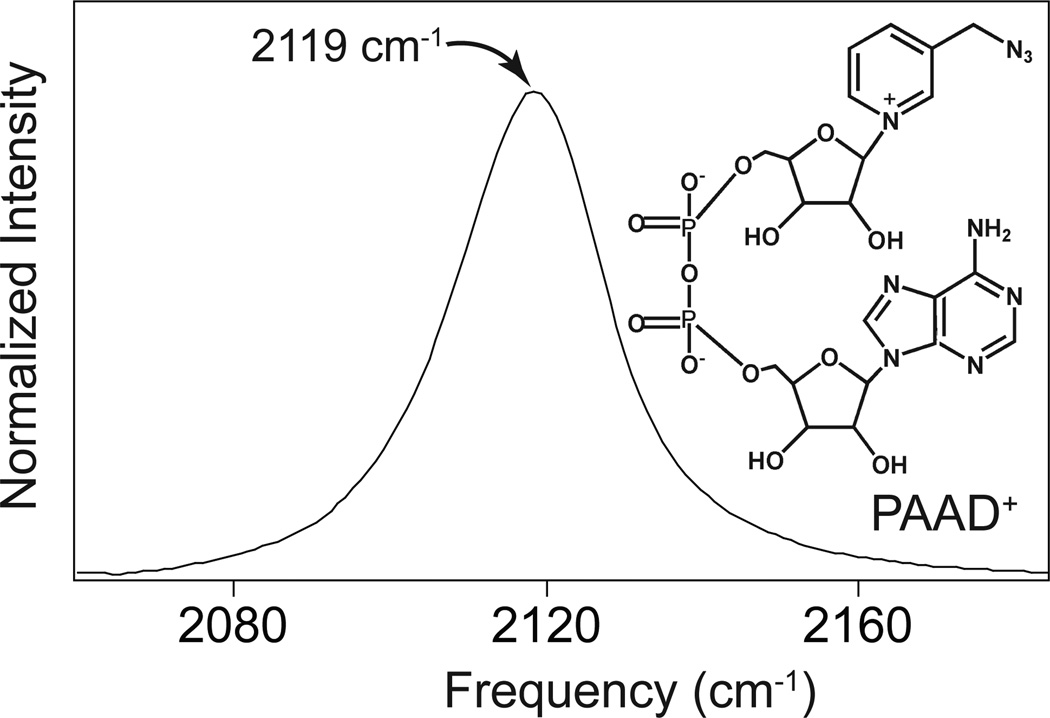
The preparation of the analog PAAD+ is analogous to the synthesis of azido-NAD+. PAAD+ is stable, soluble in water, and can be obtained in high purity. This molecule is structurally analogous to native NAD+ except that an azido group with a methylene linker replaces the amide in the nicotinamide ring. As with native NAD+, PAAD+ has a characteristic UV absorption at 260 nm with an extinction coefficient similar to that for the native cofactor. This transition results from the electronic absorption of the adenine moiety present in both molecules. Unlike native NAD+, however, this analog has a strong absorbance in the infrared near 5 µm. Figure 1 shows the linear infrared absorption spectrum and structure of PAAD+. The molar absorptivity of PAAD+ is ~2000 M−1cm−1 at 2119 cm−1, which is similar to that for the azide anion. This large molar absorptivity circumvents the major limitation of the azido-NAD+ analog we have reported previously.
Figure 1.
Infrared spectrum and structure of PAAD+.
PAAD+ is an inhibitor for FDH. Although this result is not surprising given that the redox potential of the picolyl azide ring is likely to be rather different from the nicotinamide ring, it does mean that the binding geometry of PAAD+ in the enzyme is ambiguous. Fortunately, our 2D IR results show clear indications that the picolyl azide is bound inside the protein and is not solvent exposed as described below. The binding studies of the binary complex of PAAD+ with FDH by ITC result in a Kd of 7 µM, which is comparable to the KM for NAD+ of 37 µM,47, 48 so it is likely that this analog binds in the active site in a way that is similar to that for the natural coenzyme, NAD+.
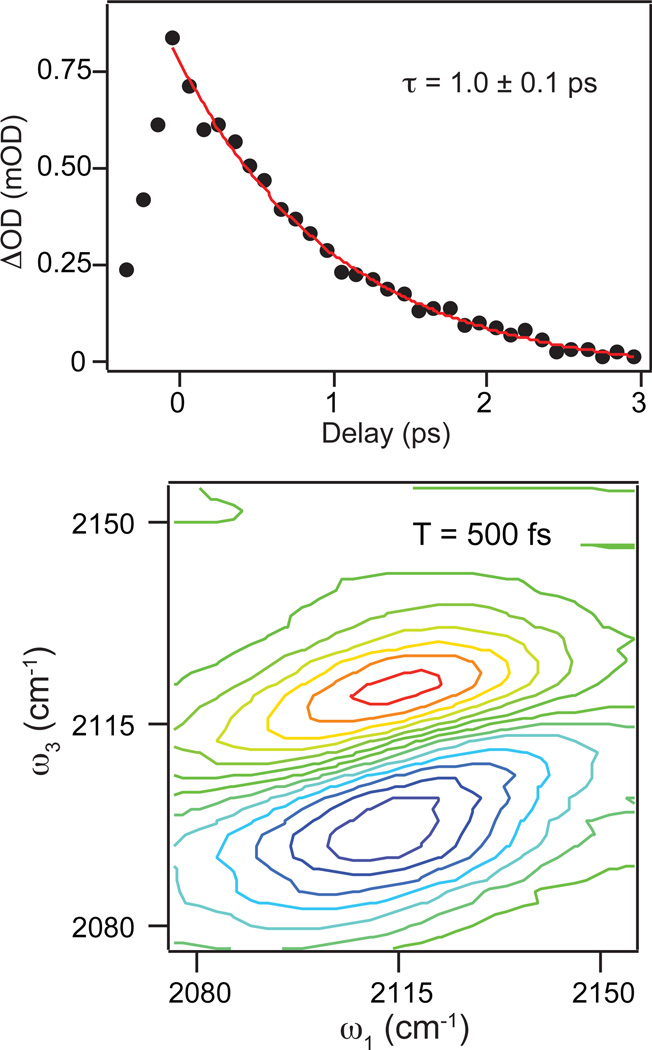
The top panel of Figure 2 shows the decay of the excited-state-absorption contribution to the dispersed pump-probe spectrum measured in the magic-angle polarization geometry. The black dots represent the measured data and the red line is a fit to the decay with a time constant of 1.0 ± 0.1 ps. The ground-state-bleach signal (data not shown) decays with a similar time constant, but exhibits an additional weak contribution to the decay likely resulting from the weakly anharmonically coupled states that are populated in the relaxation of the excited state population.
Figure 2.
(Top) Decay of the excited state absorption contribution to the frequency-dispersed pump-probe measurements for PAAD+ in water measured with relative pump and probe polarizations at the magic angle. The excited state decays to zero with a single exponential time constant of 1.0 ±0.1 ps. (Bottom) 2D IR spectrum of PAAD+ in water at T = 500 fs illustrating the full 2D IR lineshape of the raw spectrum.
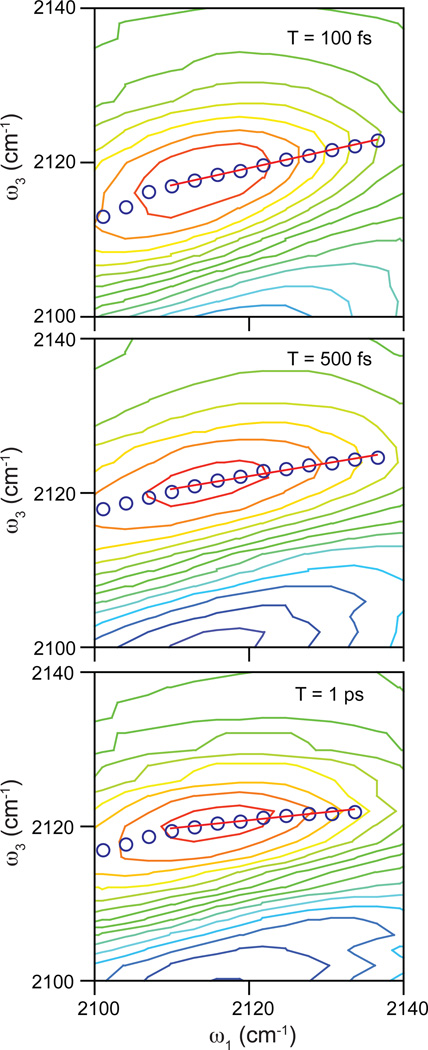
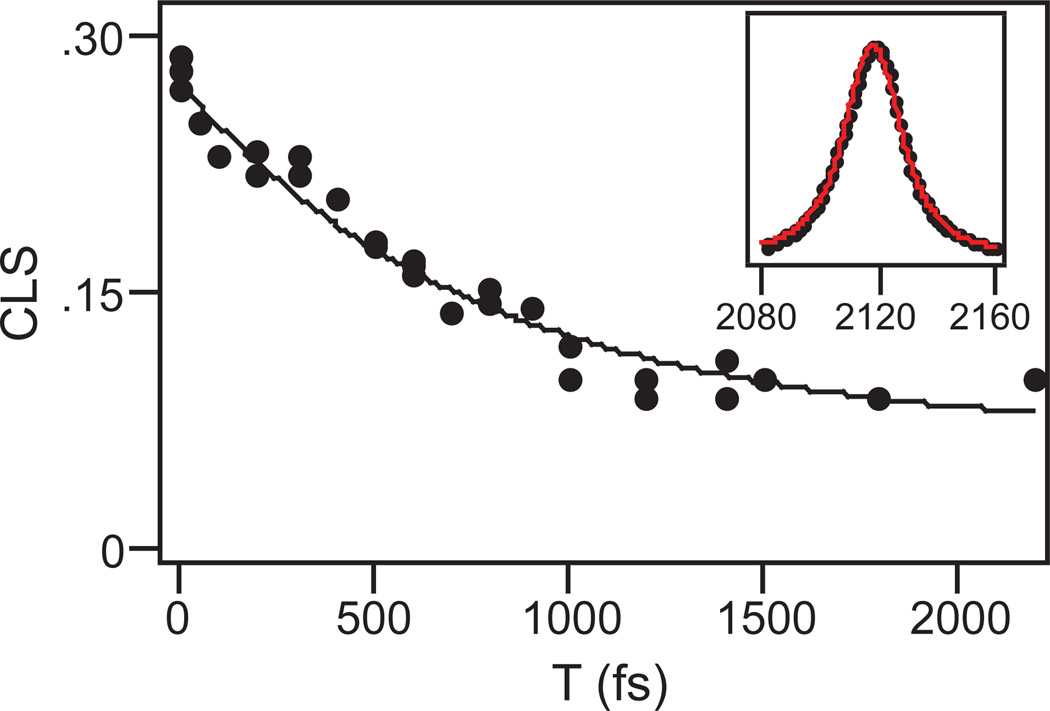
The bottom panel of Figure 2 shows a typical 2D IR spectrum of PAAD+ in water at T = 500 fs. The red contours indicate positive signals corresponding to ground state bleaching and stimulated emission signal pathways. These features are elongated along the diagonal at early waiting times and rotate toward the horizontal axis at longer waiting times. This change is a signature of spectral diffusion, in which the environmental conformations are sampled and, therefore, the frequencies of the probe vibrations change. As noted in the experimental section, we use the CLS method to determine the FFCF from the 2D IR data. Figure 3 shows this analysis for 2D IR spectra of PAAD+ in water at T = 100 fs (top), 500 fs (middle), and 1 ps (bottom). The open blue circles indicate the frequencies of the peaks in ω3 for each value of ω1 corresponding to the center line. Shown in red is the linear fit to the center line. The FFCF decay is proportional to the decay of the slope of the center line as a function of T. Figure 4 shows the decay of the CLS as a function of T for PAAD+ in water. A single exponential function with an offset fits the CLS decay well. The time constant for the exponential is 800 ± 100 fs. This model is a generalized Kubo line shape model for the FFCF, i.e. C(t) = Δ12e−t/τ + Δ22, where Δ1 and Δ2 reflect the magnitude of the frequency fluctuations and τ is the time constant for the decay of the FFCF. We get τ and the relative amplitudes of the two components from the CLS decay and the absolute amplitudes for the Δ’s are determined by fitting to the linear absorption spectrum. The parameters that result from the fitting are Δ1 = 0.91 ps−1, τ = 800 fs, and Δ2 = 0.52 ps−1. As seen in the inset of Figure 4, the experimental (black) and simulated (red) absorption spectra agree well.
Figure 3.
Representative 2D IR spectra of PAAD+ in water for waiting times of 100 fs (top), 500 fs (middle), and 1 ps (bottom) focusing on the positive feature that we analyze to extract the frequency correlations. Blue circles correspond to the center line points determined from the spectrum. The red line is the linear fit to the center line.
Figure 4.
Decay of the CLS as a function of the waiting time T for PAAD+ in water. Points are the CLS values for each value of T. The solid line is the fit to the data. The inset shows the infrared absorption spectrum of PAAD+ (black points) and the fit to the spectrum (red line).
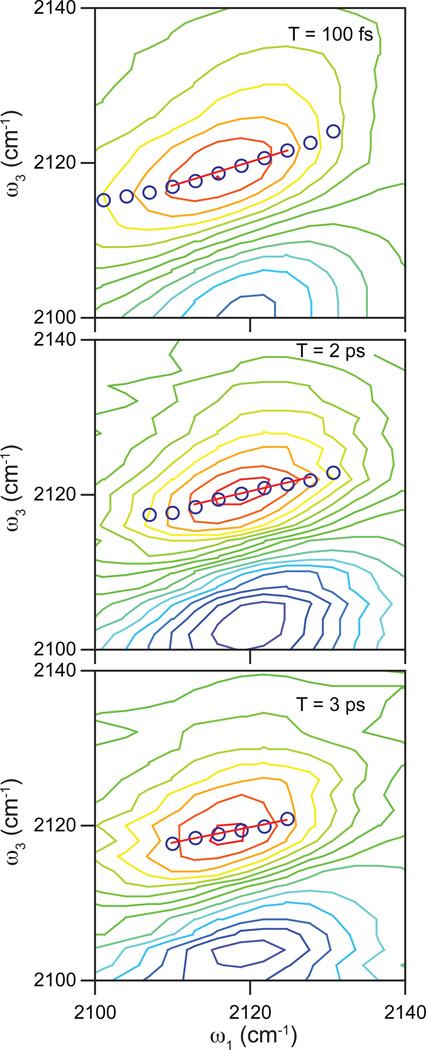
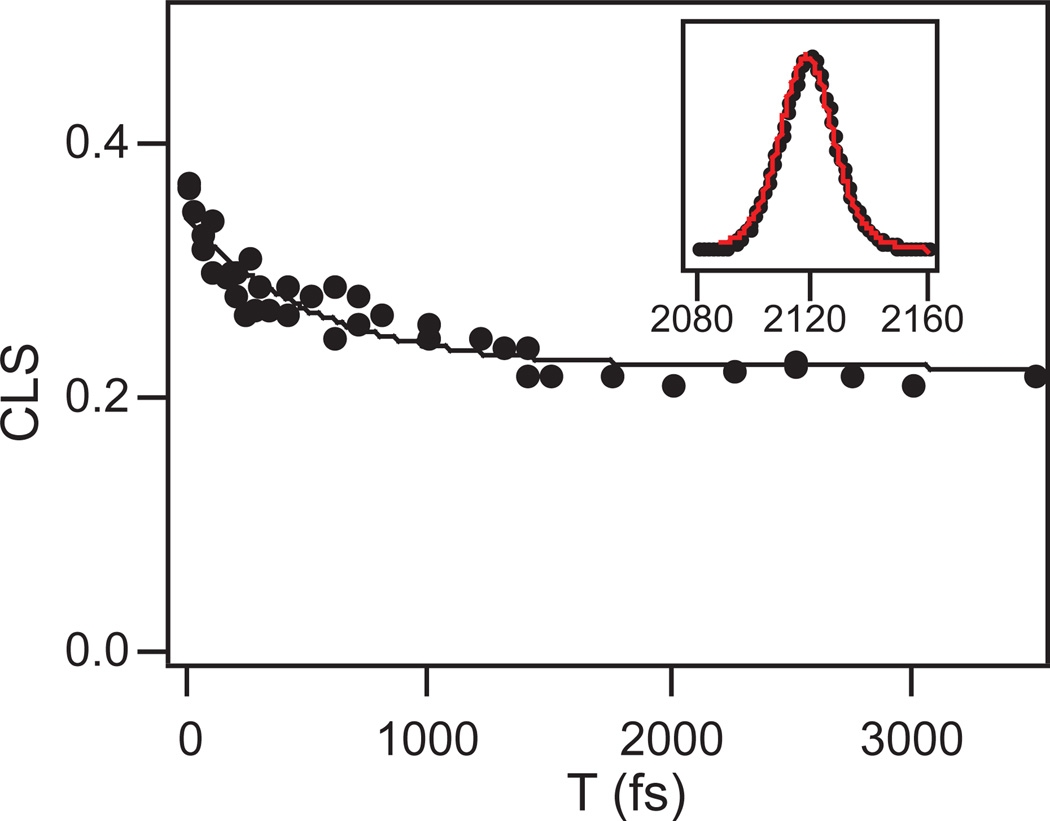
Figure 5 shows 2D IR spectra of the binary complex of FDH with PAAD+ at T = 100 fs (top), 2 ps (middle), and 3 ps (bottom). Figure 6 shows the CLS decay as a function of the waiting time, T, for the binary complex. The CLS data are presented as dots and the fit is the solid line. A single exponential with a static offset and a time constant of 500 ± 100 fs fits the decay of the CLS well. To model the FFCF in this complex, we again use a generalized Kubo line shape function of the form, C(t) = Δ12e−t/τ + Δ22. The inset in Figure 6 shows the fit to the infrared absorption spectrum using the parameters from the fit to the CLS decay and adjusting the total amplitude of the FFCF to fit the absorption line shape. The resulting FFCF parameters from the fitting are Δ1 = 0.97 ps−1, τ = 500 fs, and Δ2 = 1.34 ps−1. The simulated spectrum with these parameters is in good agreement with the linear absorption spectrum.
Figure 5.
Representative 2D IR spectra of the PAAD+-FDH binary complex for waiting times of 100 fs (top), 2 ps (middle), and 3 ps (bottom) focusing on the positive feature that we analyze to extract the frequency correlations. Blue circles correspond to the center line points determined from the spectrum. The red line is the linear fit to the center line.
Figure 6.
Decay of the CLS as a function of the waiting time T for the PAAD+-FDH binary complex. Points are the CLS values for each value of T. The solid line is the fit to the data. The inset shows the infrared absorption spectrum of PAAD+ (black points) and the fit to the spectrum (red line).
Discussion
Careful comparison of the infrared spectra of free PAAD+ in solution with that for PAAD+ bound to FDH shows no significant differences. That there is no apparent difference in these spectra does not mean, however, that they do not exhibit distinct underlying dynamics. The infrared absorption line shape is often not a good measure of the underlying dynamics that give rise to the line shape because it is determined by the integral of the correlation function, and many different FFCFs can be integrated to give the same line shape even though the associated dynamics differ. In contrast, 2D IR can probe these differences. A key feature of 2D IR is its ability to uniquely determine the FFCF. Our experimental analysis clearly shows that PAAD+ exhibits distinct spectral dynamics depending on whether it is bound to FDH or free in solution, suggesting that it binds in the active site and that it will be a suitable probe of enzyme dynamics.
Based on our measurements of PAAD+ in water it is logical to conclude that the frequency fluctuations of the azido stretching vibration are determined by the water hydrogen bond dynamics. The FFCF of PAAD+ in water is similar to that for other small chromophores.41, 49–53 The time scale of spectral diffusion, 800 fs, is consistent with these earlier measurements and with the conclusions of those studies that this time scale reflects the hydrogen bond fluctuations around the chromophore, in this case the azido moiety of PAAD+. In solution, NAD+ is a mixture of folded and unfolded forms with the aromatic rings stacked parallel to one another in a π-stacking interaction. One might expect this equilibrium to contribute to the frequency fluctuations of the azido vibration. The equilibrium exchange between the folded and the unfolded structure should be slow compared to the time window for our measurement suggesting that such a contribution would appear at long times. In addition, unlike azido-NAD+, PAAD+ has a methylene linker between the azide and the pyridine ring that should increase the orientational flexibility of the azido group and could also contribute to the decay of the FFCF at long waiting times. In our experiments on PAAD+ in water we do observe a small static contribution indicative of dynamics that occur beyond our experimental time scale that could reflect the slow equilibrium exchange between folded and unfolded conformers or rotations about the dihedral angles associated with the methylene group. Nevertheless, this contribution is about a fourth of the amplitude of the 800 fs component. Thus we conclude that local hydrogen bond fluctuations of water molecules in the first solvation shell around the azido group have the greatest influence on the decay dynamics of the FFCF for PAAD+ in water. In addition to the 2D IR measurements, the pump-probe experiments for PAAD+ in water show that the excited vibration relaxes in 1 ps, which is typical for azido stretching vibrations.41, 53–55 Although this time scale is short, it is adequate for sampling the dynamics on the hundreds of femtoseconds to few picoseconds time scale. Based on our 2D IR measurements, it appears that the lifetime for PAAD+ bound to FDH may be somewhat longer thus enabling the measurement of 2D IR spectra at somewhat longer waiting times than are accessible in water. We have not measured the pump-probe decay for PAAD+ bound to FDH primarily because the sample concentration would be much lower than that used for the experiments in water making that measurement substantially more difficult. Nevertheless, we can conclude that, although the population lifetime for the azido stretching vibration of PAAD+ is short, it is adequate for sampling the femtosecond to picosecond dynamics of enzymes.
PAAD+ bound to FDH shows distinct spectral diffusion dynamics from those seen in water, and the resulting FFCF agrees well with that seen in previous studies of enzyme dynamics. PAAD+ in the binary complex with FDH decays to a large static offset. The decay of the FFCF in the binary complex exhibits a 500 fs time constant, and the static contribution to the FFCF represents 65% of the overall decay. This result is consistent with the FFCFs of other small molecules bound to proteins and is expected since proteins typically exhibit dynamics at a wide range of time scales from femtoseconds to milliseconds. The motions of the protein that occur on time scales longer than the measurement time appear as a static contribution to the FFCF and frequently contribute 20–50% of the overall decay. For the FDH-PAAD+ binary complex, we collect 2D IR spectra for waiting times up to 3.5 ps indicating that the population lifetime of the bound chromophore is somewhat longer than in the free state as noted above. These results suggest that PAAD+ should be a good general probe of enzyme active-site dynamics of NAD-dependent enzymes. Based on the similarity of the dissociation constants for PAAD+ as compared to the native coenzyme, NAD+, we anticipate that PAAD+ binds to FDH in a way that is similar to NAD+ with the picolyl azide ring located in the active site where the nicotinamide ring would normally be.56 Of course PAAD+ is missing the key amide group that is present in NAD+. Although the methylene-linked azido group of PAAD+ retains the H-bond accepting ability of this amide, the H-bond donating capability is lost in this substitution. This difference between NAD+ and PAAD+ has the potential to alter the binding of PAAD+ and its interactions with the enzyme active site, which could limit the applicability of this cofactor as a probe of enzyme dynamics in some enzymes.
The FFCF of the PAAD+-FDH complex is similar to that for the FDH-azide binary complex that we have studied previously.27 Both of these complexes have a significant static component that comprises more than 50% of the initial value of the FFCF. As with the FDH-azide binary complex, the binary complex of PAAD+ with FDH likely represents a structure that is well removed from the transition-state-analog conformation represented by the ternary complex, FDH-NAD+-azide. In the ternary complex, we observed the surprising result that the FFCF decays to zero on the picosecond time scale. We explained this unusual result on the grounds that the ternary complex is a mimic of the transition state and therefore resides in a deep potential minimum in which one would anticipate a narrow conformational distribution and fast fluctuations about the average configuration. The presence of the static component in the FFCF decay of the FDH-azide binary complex was presented as evidence in support of this hypothesis since the binary complex does not resemble the transition state structure. The current result for the binary complex of FDH with PAAD+ lends further support to this conclusion. Here too the complex is removed from the transition state structure, and, again, we observe a significant static contribution to the decay. A key test of the potential of PAAD+ as a probe of enzyme dynamics is whether the ternary complex with PAAD+ and azide exhibits complete decay of the FFCF on the picosecond time scale. Such a result would demonstrate that PAAD+ does not significantly perturb the active site structure and dynamics of the transition-state-analog complex. These experiments are presently underway.
Conclusion
We report the synthesis, characterization and kinetics of an azido analog of NAD+, PAAD+. It binds to FDH with an affinity comparable to native NAD+. PAAD+ exhibits a strong infrared absorption at 2119 cm−1 associated with the azido antisymmetric stretching vibration, a region suitable for enzymatic studies. It has a high molar extinction coefficient comparable to that of the azide ion, which makes it an ideal spectroscopic probe of enzyme dynamics. Our results show that the azido antisymmetric stretch of PAAD+ is sensitive to the dynamics in its local environment. In aqueous solution, the spectral diffusion is nearly complete within hundreds of femtoseconds whereas when bound to FDH, PAAD+ shows a substantial static contribution to the FFCF. This observation is consistent with other enzymes where slow motions influence the FFCF.
The ability of PAAD+ to bind, form kinetically relevant complexes with the enzyme and to report the active-site dynamics show that PAAD+ can be a general spectroscopic tool to probe the femtosecond to picosecond dynamics of numerous NAD+ dependent and other systems, where NAD+ plays a role. Studies of this probe bound to different enzymes both wild type and mutants are currently underway.
Acknowledgments
This work was supported by NIH R01 GM79368 and NSF CHE-0644410 (CMC), and NIH R01 GM65368, BSF (2007256), and NSF CHE-0715448 (AK).
References
- 1.Agarwal PK. Role of Protein Dynamics in Reaction Rate Enhancement by Enzymes. J. Am. Chem. Soc. 2005;127(43):15248. doi: 10.1021/ja055251s. [DOI] [PubMed] [Google Scholar]
- 2.Benkovic SJ, Hammes-Schiffer S. Biochemistry - Enzyme motions inside and out. Science. 2006;312(5771):208–209. doi: 10.1126/science.1127654. [DOI] [PubMed] [Google Scholar]
- 3.Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450(7171):913. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 4.Nagel ZD, Klinman JP. A 21st century revisionist's view at a turning point in enzymology. Nat. Chem. Biol. 2009;5(8):543–550. doi: 10.1038/nchembio.204. [DOI] [PubMed] [Google Scholar]
- 5.Antoniou D, Schwartz SD. Internal enzyme motions as a source of catalytic activity: Rate-promoting vibrations and hydrogen tunneling. J. Phys. Chem. B. 2001;105(23):5553–5558. [Google Scholar]
- 6.Schwartz SD. Vibrationally enhanced tunneling and kinetic isotope effects of enzymatic reactions. Isotope Effects in Chemistry and Biology. 2006:475–498. [Google Scholar]
- 7.Benkovic SJ, Hammes GG, Hammes-Schiffer S. Free-energy landscape of enzyme catalysis. Biochemistry. 2008;47(11):3317–3321. doi: 10.1021/bi800049z. [DOI] [PubMed] [Google Scholar]
- 8.Hammes-Schiffer S. Impact of enzyme motion on activity. Biochemistry. 2002;41(45):13335. doi: 10.1021/bi0267137. [DOI] [PubMed] [Google Scholar]
- 9.Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu. Rev. Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 10.Klinman JP. An integrated model for enzyme catalysis emerges from studies of hydrogen tunneling. Chem. Phys. Lett. 2009;471(4–6):179–193. doi: 10.1016/j.cplett.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagel ZD, Klinman JP. Tunneling and Dynamics in Enzymatic Hydride Transfer. Chem. Rev. (Washington, DC, U.S.) 2006;106(8):3095. doi: 10.1021/cr050301x. [DOI] [PubMed] [Google Scholar]
- 12.Sen A, Kohen A. Enzymatic tunneling and kinetic isotope effects: chemistry at the crossroads. J. Phys. Org. Chem. 2010;23(7):613–619. [Google Scholar]
- 13.Roth JP, Klinman JP. Kinetic isotope effects. Encycl. Biol. Chem. 2004;2:522–527. [Google Scholar]
- 14.Fayer MD. Dynamics of Liquids, Molecules, and Proteins Measured with Ultrafast 2D IR Vibrational Echo Chemical Exchange Spectroscopy. Ann. Rev. Phys. Chem. 2009;60:21–38. doi: 10.1146/annurev-physchem-073108-112712. [DOI] [PubMed] [Google Scholar]
- 15.Kim YS, Hochstrasser RM. Applications of 2D IR Spectroscopy to Peptides, Proteins, and Hydrogen-Bond Dynamics. J. Phys. Chem. B. 2009;113(24):8231–8251. doi: 10.1021/jp8113978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunt NT. 2D-IR spectroscopy: ultrafast insights into biomolecule structure and function. Chem. Soc. Rev. 2009;38(7):1837–1848. doi: 10.1039/b819181f. [DOI] [PubMed] [Google Scholar]
- 17.Hochstrasser RM. Two-dimensional spectroscopy at infrared and optical frequencies. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14190–14196. doi: 10.1073/pnas.0704079104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bagchi S, Nebgen BT, Loring RF, Fayer MD. Dynamics of a Myoglobin Mutant Enzyme: 2D IR Vibrational Echo Experiments and Simulations. J. Am. Chem. Soc. 2010;132(51):18367–18376. doi: 10.1021/ja108491t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Merchant KA, Thompson DE, Xu Q-H, Williams RB, Loring RF, Fayer MD. Myoglobin-CO Conformational Substate Dynamics: 2D Vibrational Echoes and MD Simulations. Biophys. J. 2002;82(6):3277. doi: 10.1016/S0006-3495(02)75669-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finkelstein IJ, Zheng JR, Ishikawa H, Kim S, Kwak K, Fayer MD. Probing dynamics of complex molecular systems with ultrafast 2D IR vibrational echo spectroscopy. Phys. Chem. Chem. Phys. 2007;9(13):1533–1549. doi: 10.1039/b618158a. [DOI] [PubMed] [Google Scholar]
- 21.Ishikawa H, Finkelstein IJ, Kim S, Kwak K, Chung JK, Wakasugi K, Massari AM, Fayer MD. Neuroglobin dynamics observed with ultrafast 2D-IR vibrational echo spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2007;104(41):16116–16121. doi: 10.1073/pnas.0707718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim S, Chung JK, Kwak K, Bowman SEJ, Bren KL, Bagchi B, Fayer MD. Native and Unfolded Cytochrome c—Comparison of Dynamics using 2D-IR Vibrational Echo Spectroscopy. The Journal of Physical Chemistry B. 2008;112(32):10054. doi: 10.1021/jp802246h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thielges MC, Chung JK, Fayer MD. Protein Dynamics in Cytochrome P450 Molecular Recognition and Substrate Specificity Using 2D IR Vibrational Echo Spectroscopy. J. Am. Chem. Soc. 2011;133(11):3995–4004. doi: 10.1021/ja109168h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finkelstein IJ, Ishikawa H, Kim S, Massari AM, Fayer MD. Substrate binding and protein conformational dynamics measured by 2D-IR vibrational echo spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 2007;104(8):2637–2642. doi: 10.1073/pnas.0610027104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang C, Bauman JD, Das K, Remorino A, Arnold E, Hochstrasser RM. Two-dimensional infrared spectra reveal relaxation of the nonnucleoside inhibitor TMC278 complexed with HIV-1 reverse transcriptase. Proc. Natl. Acad. Sci. U.S.A. 2008;105(5):1472–1477. doi: 10.1073/pnas.0709320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill SE, Bandaria J, Fox M, Vanderaugh E, Kohen A, Cheatum CM. Exploring the Molecular Origins of Protein Dynamics in the Active Site of Human Carbonic Anhydrase II. J. Phys. Chem. B. 2009;113(33):11505–11510. doi: 10.1021/jp901321m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bandaria JN, Dutta S, Nydegger MW, Rock W, Kohen A, Cheatum CM. Characterizing the dynamics of functionally relevant complexes of formate dehydrogenase. Proc. Natl. Acad. Sci. U.S.A. 2010;107(42):17974–17979. doi: 10.1073/pnas.0912190107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bandaria JN, Dutta S, Hill SE, Kohen A, Cheatum CM. Fast enzyme dynamics at the active site of formate dehydrogenase. J. Am. Chem. Soc. 2008;130(1) doi: 10.1021/ja077599o. 22-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stafford AJ, Ensign DL, Webb LJ. Vibrational Stark Effect Spectroscopy at the Interface of Ras and Rap1A Bound to the Ras Binding Domain of RalGDS Reveals an Electrostatic Mechanism for Protein-Protein Interaction. J. Phys. Chem. B. 2010;114(46):15331–15344. doi: 10.1021/jp106974e. [DOI] [PubMed] [Google Scholar]
- 30.Webb LJ, Boxer SG. Electrostatic fields near the active site of human aldose reductase: 1. New inhibitors and vibrational stark effect measurements. Biochemistry. 2008;47(6):1588–1598. doi: 10.1021/bi701708u. [DOI] [PubMed] [Google Scholar]
- 31.Fafarman AT, Boxer SG. Nitrile Bonds as Infrared Probes of Electrostatics in Ribonuclease S. J. Phys. Chem. B. 2010;114(42):13536–13544. doi: 10.1021/jp106406p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zimmermann J, Gundogdu K, Cremeens ME, Bandaria JN, Hwang GT, Thielges MC, Cheatum CM, Romesberg FE. Efforts toward Developing Probes of Protein Dynamics: Vibrational Dephasing and Relaxation of Carbon-Deuterium Stretching Modes in Deuterated Leucine. J. Phys. Chem. B. 2009;113(23):7991–7994. doi: 10.1021/jp900516c. [DOI] [PubMed] [Google Scholar]
- 33.Naraharisetty SRG, Kurochkin DV, Rubtsov IV. C-D Modes as structural reporters via dual-frequency 2DIR spectroscopy. Chem. Phys. Lett. 2007;437(4–6):262. [Google Scholar]
- 34.Getahun Z, Huang C-Y, Wang T, De Leon B, DeGrado WF, Gai F. Using nitrile-derivatized amino acids as infrared probes of local environment. J. Am. Chem. Soc. 2003;125(2):405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 35.Waegele MM, Tucker MJ, Gai F. 5-Cyanotryptophan as an infrared probe of local hydration status of proteins. Chem. Phys. Lett. 2009;478(4–6):249–253. doi: 10.1016/j.cplett.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bragg PD. Mechanism of hydride transfer during the reduction of 3-acetylpyridine adenine dinucleotide by NADH catalyzed by the pyridine nucleotide transhydrogenase of Escherichia coli. FEBS Lett. 1996;397(1):93. doi: 10.1016/s0014-5793(96)01147-7. [DOI] [PubMed] [Google Scholar]
- 37.Kirtley ME, Eby D. Interaction of nicotinamide adenine dinucleotide and its analogs with glyceraldehyde 3-phosphate dehydrogenase. Biochemistry. 1971;10(14):2677. doi: 10.1021/bi00790a004. [DOI] [PubMed] [Google Scholar]
- 38.Mal'tsev NI, Yanina MM. Synthesis and properties of a new nad analog: Aminoethylnicotina mide adenine dinucleotide. Pharmaceutical Chemistry Journal. 1979;13(8):803. [Google Scholar]
- 39.Samama JP, Marchalrosenheimer N, Biellmann JF, Rossmann MG. An Investigation Of The Active-Site Of Lactate-Dehydrogenase With Nad+ Analogs. Eur. J. Biochem. 1981;120(3):563–569. doi: 10.1111/j.1432-1033.1981.tb05737.x. [DOI] [PubMed] [Google Scholar]
- 40.Dutta S, Cook RJ, Houtman JCD, Kohen A, Cheatum CM. Characterization of azido-NAD(+) to assess its potential as a two-dimensional infrared probe of enzyme dynamics. Anal. Biochem. 2010;407(2):241–246. doi: 10.1016/j.ab.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dutta S, Rock W, Cook RJ, Kohen A, Cheatum CM. Two-dimensional infrared spectroscopy of azido-nicotinamide adenine dinucleotide in water. J. Chem. Phys. 2011;135(5) doi: 10.1063/1.3623418. 055106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turnbull WB, Daranas AH. On the value of c: Can low affinity systems be studied by isothermal titration calorimetry? J. Am. Chem. Soc. 2003;125(48):14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- 43.Shim SH, Strasfeld DB, Ling YL, Zanni MT. Automated 2D IR spectroscopy using a mid-IR pulse shaper and application of this technology to the human islet amyloid polypeptide. Proc. Natl. Acad. Sci. U.S.A. 2007;104(36):14197–14202. doi: 10.1073/pnas.0700804104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shim SH, Strasfeld DB, Zanni MT. Generation and characterization of phase and amplitude shaped femtosecond mid-IR pulses. Optics Express. 2006;14(26):13120–13130. doi: 10.1364/oe.14.013120. [DOI] [PubMed] [Google Scholar]
- 45.Kwak K, Park S, Finkelstein IJ, Fayer MD. Frequency-frequency correlation functions and apodization in two-dimensional infrared vibrational echo spectroscopy: A new approach. J. Chem. Phys. 2007;127 doi: 10.1063/1.2772269. [DOI] [PubMed] [Google Scholar]
- 46.Kwak K, Rosenfeld DE, Fayer MD. Taking apart the two-dimensional infrared vibrational echo spectra: More information and elimination of distortions. J. Chem. Phys. 2008;128(20):204505. doi: 10.1063/1.2927906. [DOI] [PubMed] [Google Scholar]
- 47.Popov VO, Lamzin VS. Biochemistry Journal. Pt 3. 1994;301:625–643. doi: 10.1042/bj3010625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tishkov VI, Popov VO. Catalytic mechanism and application of formate dehydrogenase. Biochem.-Moscow. 2004;69(11) doi: 10.1007/s10541-005-0071-x. 1252-+ [DOI] [PubMed] [Google Scholar]
- 49.Tayama J, Ishihara A, Banno M, Ohta K, Saito S, Tominaga K. Temperature dependence of vibrational frequency fluctuation of N(3)(−) in D(2)O. J. Chem. Phys. 2010;133(1) doi: 10.1063/1.3428672. [DOI] [PubMed] [Google Scholar]
- 50.Ohta K, Maekawa H, Tominaga K. Vibrational population relaxation and dephasing dynamics of Fe(CN)(6)(4-) in D2O with third-order nonlinear infrared spectroscopy. J. Phys. Chem. A. 2004;108(8):1333–1341. [Google Scholar]
- 51.Kuo CH, Vorobyev DY, Chen JX, Hochstrasser RM. Correlation of the vibrations of the aqueous azide ion with the O-H modes of bound water molecules. J. Phys. Chem. B. 2007;111(50):14028–14033. doi: 10.1021/jp076503+. [DOI] [PubMed] [Google Scholar]
- 52.Kuo CH, Hochstrasser RM. Two dimensional infrared spectroscopy and relaxation of aqueous cyanide. Chem. Phys. 2007;341(1–3):21–28. [Google Scholar]
- 53.Hamm P, Lim M, Hochstrasser RM. Non-Markovian dynamics of the vibrations of ions in water from femtosecond infrared three-pulse photon echoes. Phys. Rev. Lett. 1998;81(24):5326–5329. [Google Scholar]
- 54.Tucker MJ, Gai XS, Fenlon EE, Brewer SH, Hochstrasser RM. 2D IR photon echo of azido-probes for biomolecular dynamics. Phys. Chem. Chem. Phys. 2011;13(6):2237–2241. doi: 10.1039/c0cp01625j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Owrutsky JC, Kim YR, Li M, Sarisky MJ, Hochstrasser RM. Determination of the Vibrational-Energy Relaxation-Time of the Azide Ion in Protic Solvents by 2-Color Transient Infrared-Spectroscopy. Chem. Phys. Lett. 1991;184(5–6):368–374. [Google Scholar]
- 56.Lamzin VS, Dauter Z, Popov VO, Harutyunyan EH, Wilson KS. High Resolution Structures of Holo and Apo Formate Dehydrogenase. Journal of Molecular Biology. 1994;236(3):759–785. doi: 10.1006/jmbi.1994.1188. [DOI] [PubMed] [Google Scholar]