Abstract
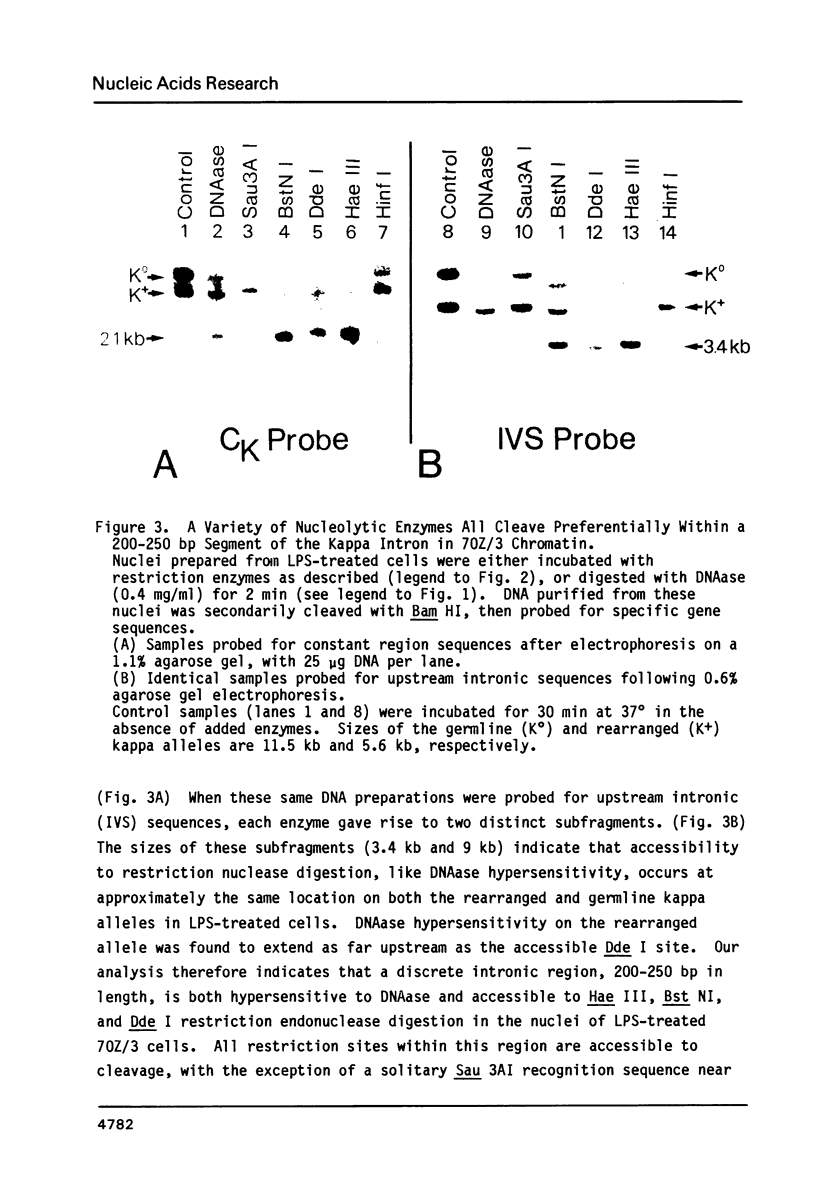
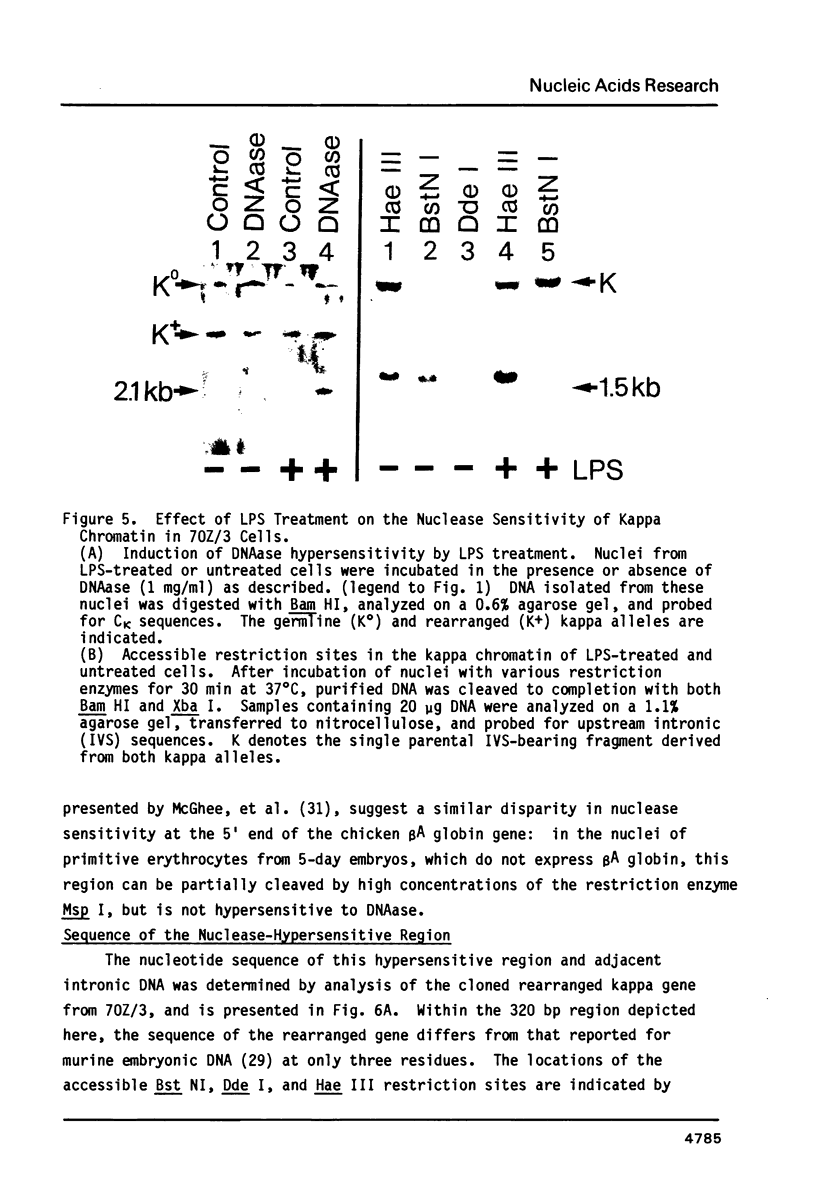
A discrete chromatin region inside the active immunoglobulin kappa gene is preferentially accessible to cleavage by nucleolytic enzymes. This region comprises 200-250 bp of DNA, and is situated within the large intron of the gene, approximately 600 bp upstream from the constant region coding sequence. The local chromatin structure of this region correlates with tissue-specific kappa gene expression: it is resistant to nucleolytic digestion in the inactive kappa genes of murine brain and liver nuclei, but becomes uniquely sensitive to cleavage by deoxyribonuclease I or by a variety of restriction endonucleases in the chromatin of kappa-producing cells. Nuclease sensitivity at this site occurs in both rearranged and unrearranged kappa alleles, and can be maintained in the absence of ongoing kappa transcription. The nucleotide sequence of the hypersensitive region has been selectively conserved in evolution, and includes both a 7 bp inverted repeat sequence and a short segment homologous to the transcriptional enhancer elements of certain eukaryotic viruses. Molecular events occurring at this locus may play a role in the regulation of kappa gene expression, perhaps by influencing the activity of promoter sequences several kilobases upstream.
Full text
PDF















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981 Dec;27(2 Pt 1):299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Bentley D. L., Farrell P. J., Rabbitts T. H. Unrearranged immunoglobulin variable region genes have a functional promoter. Nucleic Acids Res. 1982 Mar 25;10(6):1841–1856. doi: 10.1093/nar/10.6.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S. Y., Folsom V., Wooley J. DNase I-hypersensitive sites in the chromatin of immunoglobulin kappa light chain genes. Proc Natl Acad Sci U S A. 1983 May;80(9):2427–2431. doi: 10.1073/pnas.80.9.2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremisi C. The appearance of DNase I hypersensitive sites at the 5' end of the late SV40 genes is correlated with the transcriptional switch. Nucleic Acids Res. 1981 Nov 25;9(22):5949–5964. doi: 10.1093/nar/9.22.5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Lomonossoff G. P., Laskey R. A. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981 Jun 25;9(12):2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkner F. G., Zachau H. G. Expression of mouse immunoglobulin genes in monkey cells. Nature. 1982 Jul 15;298(5871):286–288. doi: 10.1038/298286a0. [DOI] [PubMed] [Google Scholar]
- Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci U S A. 1981 Feb;78(2):943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P. A., Max E. E., Seidman J. G., Maizel J. V., Jr, Leder P. Cloned human and mouse kappa immunoglobulin constant and J region genes conserve homology in functional segments. Cell. 1980 Nov;22(1 Pt 1):197–207. doi: 10.1016/0092-8674(80)90168-3. [DOI] [PubMed] [Google Scholar]
- Katinka M., Vasseur M., Montreau N., Yaniv M., Blangy D. Polyoma DNA sequences involved in control of viral gene expression in murine embryonal carcinoma cells. Nature. 1981 Apr 23;290(5808):720–722. doi: 10.1038/290720a0. [DOI] [PubMed] [Google Scholar]
- Keene M. A., Corces V., Lowenhaupt K., Elgin S. C. DNase I hypersensitive sites in Drosophila chromatin occur at the 5' ends of regions of transcription. Proc Natl Acad Sci U S A. 1981 Jan;78(1):143–146. doi: 10.1073/pnas.78.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D. E., Coleclough C., Perry R. P. Functional significance and evolutionary development of the 5'-terminal regions of immunoglobulin variable-region genes. Cell. 1982 Jun;29(2):681–689. doi: 10.1016/0092-8674(82)90184-2. [DOI] [PubMed] [Google Scholar]
- Larsen A., Weintraub H. An altered DNA conformation detected by S1 nuclease occurs at specific regions in active chick globin chromatin. Cell. 1982 Jun;29(2):609–622. doi: 10.1016/0092-8674(82)90177-5. [DOI] [PubMed] [Google Scholar]
- Maki R., Kearney J., Paige C., Tonegawa S. Immunoglobulin gene rearrangement in immature B cells. Science. 1980 Sep 19;209(4463):1366–1369. doi: 10.1126/science.6774416. [DOI] [PubMed] [Google Scholar]
- Mather E. L., Perry R. P. Transcriptional regulation of immunoglobulin V genes. Nucleic Acids Res. 1981 Dec 21;9(24):6855–6867. doi: 10.1093/nar/9.24.6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Max E. E., Maizel J. V., Jr, Leder P. The nucleotide sequence of a 5.5-kilobase DNA segment containing the mouse kappa immunoglobulin J and C region genes. J Biol Chem. 1981 May 25;256(10):5116–5120. [PubMed] [Google Scholar]
- Max E. E., Seidman J. G., Leder P. Sequences of five potential recombination sites encoded close to an immunoglobulin kappa constant region gene. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3450–3454. doi: 10.1073/pnas.76.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Wood W. I., Dolan M., Engel J. D., Felsenfeld G. A 200 base pair region at the 5' end of the chicken adult beta-globin gene is accessible to nuclease digestion. Cell. 1981 Nov;27(1 Pt 2):45–55. doi: 10.1016/0092-8674(81)90359-7. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochi A., Hawley R. G., Shulman M. J., Hozumi N. Transfer of a cloned immunoglobulin light-chain gene to mutant hybridoma cells restores specific antibody production. Nature. 1983 Mar 24;302(5906):340–342. doi: 10.1038/302340a0. [DOI] [PubMed] [Google Scholar]
- Oi V. T., Morrison S. L., Herzenberg L. A., Berg P. Immunoglobulin gene expression in transformed lymphoid cells. Proc Natl Acad Sci U S A. 1983 Feb;80(3):825–829. doi: 10.1073/pnas.80.3.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige C. J., Kincade P. W., Ralph P. Murine B cell leukemia line with inducible surface immunoglobulin expression. J Immunol. 1978 Aug;121(2):641–647. [PubMed] [Google Scholar]
- Parslow T. G., Granner D. K. Chromatin changes accompany immunoglobulin kappa gene activation: a potential control region within the gene. Nature. 1982 Sep 30;299(5882):449–451. doi: 10.1038/299449a0. [DOI] [PubMed] [Google Scholar]
- Perry R. P., Kelley D. E. Immunoglobulin messenger RNAs in murine cell lines that have characteristics of immature B lymphocytes. Cell. 1979 Dec;18(4):1333–1339. doi: 10.1016/0092-8674(79)90243-5. [DOI] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Rice D., Baltimore D. Regulated expression of an immunoglobulin kappa gene introduced into a mouse lymphoid cell line. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7862–7865. doi: 10.1073/pnas.79.24.7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SWARTZ M. N., TRAUTNER T. A., KORNBERG A. Enzymatic synthesis of deoxyribonucleic acid. XI. Further studies on nearest neighbor base sequences in deoxyribonucleic acids. J Biol Chem. 1962 Jun;237:1961–1967. [PubMed] [Google Scholar]
- Saragosti S., Cereghini S., Yaniv M. Fine structure of the regulatory region of simian virus 40 minichromosomes revealed by DNAase I digestion. J Mol Biol. 1982 Sep 15;160(2):133–146. doi: 10.1016/0022-2836(82)90171-1. [DOI] [PubMed] [Google Scholar]
- Sheffery M., Rifkind R. A., Marks P. A. Murine erythroleukemia cell differentiation: DNase I hypersensitivity and DNA methylation near the globin genes. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1180–1184. doi: 10.1073/pnas.79.4.1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shermoen A. W., Beckendorf S. K. A complex of interacting DNAase I-hypersensitive sites near the Drosophila glue protein gene, Sgs4. Cell. 1982 Jun;29(2):601–607. doi: 10.1016/0092-8674(82)90176-3. [DOI] [PubMed] [Google Scholar]
- Stalder J., Larsen A., Engel J. D., Dolan M., Groudine M., Weintraub H. Tissue-specific DNA cleavages in the globin chromatin domain introduced by DNAase I. Cell. 1980 Jun;20(2):451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- Storb U., Wilson R., Selsing E., Walfield A. Rearranged and germline immunoglobulin kappa genes: different states of DNase I sensitivity of constant kappa genes in immunocompetent and nonimmune cells. Biochemistry. 1981 Feb 17;20(4):990–996. doi: 10.1021/bi00507a053. [DOI] [PubMed] [Google Scholar]
- Sweet R. W., Chao M. V., Axel R. The structure of the thymidine kinase gene promoter: nuclease hypersensitivity correlates with expression. Cell. 1982 Dec;31(2 Pt 1):347–353. doi: 10.1016/0092-8674(82)90128-3. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. J., Sundin O., Bohn M. A stretch of "late" SV40 viral DNA about 400 bp long which includes the origin of replication is specifically exposed in SV40 minichromosomes. Cell. 1979 Feb;16(2):453–466. doi: 10.1016/0092-8674(79)90021-7. [DOI] [PubMed] [Google Scholar]
- Wasylyk B., Wasylyk C., Augereau P., Chambon P. The SV40 72 bp repeat preferentially potentiates transcription starting from proximal natural or substitute promoter elements. Cell. 1983 Feb;32(2):503–514. doi: 10.1016/0092-8674(83)90470-1. [DOI] [PubMed] [Google Scholar]
- Weiher H., König M., Gruss P. Multiple point mutations affecting the simian virus 40 enhancer. Science. 1983 Feb 11;219(4585):626–631. doi: 10.1126/science.6297005. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Larsen A., Groudine M. Alpha-Globin-gene switching during the development of chicken embryos: expression and chromosome structure. Cell. 1981 May;24(2):333–344. doi: 10.1016/0092-8674(81)90323-8. [DOI] [PubMed] [Google Scholar]
- Weisbrod S. Active chromatin. Nature. 1982 May 27;297(5864):289–295. doi: 10.1038/297289a0. [DOI] [PubMed] [Google Scholar]
- Weischet W. O., Glotov B. O., Schnell H., Zachau H. G. Differences in the nuclease sensitivity between the two alleles of the immunoglobulin kappa light chain genes in mouse liver and myeloma nuclei. Nucleic Acids Res. 1982 Jun 25;10(12):3627–3645. doi: 10.1093/nar/10.12.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Bingham P. M., Livak K. J., Holmgren R., Elgin S. C. The chromatin structure of specific genes: I. Evidence for higher order domains of defined DNA sequence. Cell. 1979 Apr;16(4):797–806. doi: 10.1016/0092-8674(79)90095-3. [DOI] [PubMed] [Google Scholar]
- Wu C., Gilbert W. Tissue-specific exposure of chromatin structure at the 5' terminus of the rat preproinsulin II gene. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1577–1580. doi: 10.1073/pnas.78.3.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. The 5' ends of Drosophila heat shock genes in chromatin are hypersensitive to DNase I. Nature. 1980 Aug 28;286(5776):854–860. doi: 10.1038/286854a0. [DOI] [PubMed] [Google Scholar]
- Yagi M., Koshland M. E. Expression of the J chain gene during B cell differentiation is inversely correlated with DNA methylation. Proc Natl Acad Sci U S A. 1981 Aug;78(8):4907–4911. doi: 10.1073/pnas.78.8.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]