Abstract
Lack of enteral feeding, with or without parenteral nutritional support, is associated with increased intestinal permeability and translocation of bacteria. Such translocation is thought to be important in the high morbidity and mortality rates of patients who receive nothing by mouth. Recently, Paneth cells, important constituents of innate intestinal immunity, were found to be crucial in host protection against invasion of both commensal and pathogenic bacteria. This study investigates the influence of food deprivation on Paneth cell function in a mouse starvation model. Quantitative PCR showed significant decreases in mRNA expression of typical Paneth cell antimicrobials, lysozyme, cryptdin, and RegIIIγ, in ileal tissue after 48 hours of food deprivation. Protein expression levels of lysozyme and RegIIIγ precursor were also significantly diminished, as shown by Western blot analysis and IHC. Late degenerative autophagolysosomes and aberrant Paneth cell granules in starved mice were evident by electron microscopy, Western blot analysis, and quantitative PCR. Furthermore, increased bacterial translocation to mesenteric lymph nodes coincided with Paneth cell abnormalities. The current study demonstrates the occurrence of Paneth cell abnormalities during enteral starvation. Such changes may contribute to loss of epithelial barrier function, causing the apparent bacterial translocation in enteral starvation.
The intestine is challenged with the task of protecting the body's internal milieu against bacterial invasion. To this end, the gut is equipped with an epithelial lining connected by tight junctions, a mucus layer, gut-associated lymphoid tissue, and Paneth cells. Paneth cells are highly specialized epithelial cells located in the crypts of the small intestine, and play an important role in gut innate immunity.1 These cells sense bacterial presence and secrete granules containing antimicrobial peptides, including lysozyme, RegIIIγ, and cryptdins (the murine counterparts of human α-defensins) both constitutively and in response to activation by bacteria or their products.2 Using a murine cell ablation model, Paneth cells were shown to be crucial in host protection against invasion of both commensal and pathogenic microbiota.3 In addition, our group has recently shown the additive importance of Paneth cells in preventing bacterial translocation in situations of physical intestinal barrier loss.4
Enteral starvation and total parenteral nutrition, as applied to critically ill patients, are reported to result in increased gut wall permeability, a compromised immune system, and bacterial translocation.5, 6, 7, 8, 9, 10 Because autophagy, a process induced on starvation,11, 12, 13 influences the generation of Paneth cell granules,14 we hypothesize that enteral starvation impairs Paneth cell function, contributing to starvation-associated gut compromise.
In this study, the effects of food deprivation on Paneth cell function were investigated using a mouse starvation model. We provide evidence that lack of enteral feeding results in Paneth cell autophagy, decreased expression of antimicrobial products (ie, lysozyme, cryptdin, and RegIIIγ), and the presence of atypical secretory granules. Our results propose compromised Paneth cells to be involved in the reduced protection against bacterial translocation in enteral starvation.
Materials and Methods
Materials
The following primary antibodies were used: rabbit anti-mouse RegIIIγ (provided by Dr. Lora V. Hooper, The Howard Hughes Medical Institute, University of Texas, Southwestern Medical Center, Dallas, TX), rabbit anti-mouse lysozyme antiserum (provided by Dr. Tomas Ganz, David Geffen School of Medicine at UCLA, Los Angeles, CA), rabbit anti-rat PAP-III,15 mouse anti–rat LC3 (Cosmo Bio Co, Tokyo, Japan), and mouse anti–β–actin (Sigma, St Louis, MO). The horseradish peroxidase (HRP)–conjugated secondary antibodies goat anti–rabbit and rat anti–mouse were obtained from Jackson (West Grove, PA), Alexa 488–conjugated goat anti–rabbit was obtained from Invitrogen (Carlsbad, CA), and biotin–conjugated secondary swine anti–rabbit and streptavidin–biotin HRP complex were obtained from DakoCytomation (Glostrup, Denmark). The bicinchoninic acid protein assay kit and Supersignal West Pico were from Thermo Fisher Scientific Inc. (Rockford, IL), Moloney murine leukemia virus reverse transcriptase was from Life Technologies (Paisley, UK), and Colombia III blood agar base plates and Chocolate PolyviteX agar plates were from BioMérieux (Marcy L'Etoile, France). All other reagents were obtained from Sigma, unless otherwise mentioned.
Animals and Tissue Collection
Male C57BL/6 mice (15 per group), obtained from Charles River (Maastricht, The Netherlands), were housed (three mice per cage) under controlled environmental conditions, with free access to food and water until the age of 12 weeks. At this point, mice were housed separately in metabolic cages to prevent coprophagia and were fasted for 48 hours. The unfasted control group was similarly housed. The ambient temperature was 21°C. After 24 hours, food–deprived animals were warmed by infrared light,16 keeping the ambient temperature at 24°C to 25°C. During the experiment, all animals had free access to water. All animals survived 48 hours of food restriction. Fasted mice lost approximately 25% of their initial body weight, whereas control mice maintained a constant weight of approximately 25 g. Mice were anesthetized with 100 mg/kg ketamine and 10 mg/kg xylazine and sacrificed by bleeding. Mesenteric lymph nodes (MLNs) and the ileum were instantly harvested. The experiments were approved by the Animal Care Committee of Maastricht University, Maastricht (permit no. 2008-163).
IHC
Ileum samples for immunohistochemistry (IHC) were fixed immediately in 4% formaldehyde and embedded in paraffin, after which tissue sections (3 μm thick) were cut. These sections were stained using H&E or immunostained for Paneth cell marker lysozyme. Oligosaccharides in Paneth cell granules were stained using lectin Triticum vulgaris. After deparaffination, endogenous peroxidase was quenched with 0.3% H2O2 in methanol. For oligosaccharide staining, antigen retrieval was obtained in citrate buffer (Dako REAL Target Retrieval Solution; DakoCytomation). Sections were blocked in 5% bovine serum albumin, and biotin-conjugated lectin T. vulgaris was incubated overnight at 4°C. For lysozyme staining, after blocking nonspecific antibody binding with 5% bovine serum albumin, sections were incubated with specific antibodies to mouse lysozyme at room temperature for 60 minutes. Next, an appropriate biotin-conjugated secondary antibody was applied. Immunoreaction was detected using the streptavidin-biotin HRP system and 3,3′-diaminobenzidine tetrahydrochloride (DakoCytomation) and 3-amino-9-ethylcarbazole for oligosaccharides and lysozyme, respectively. Nuclei were counterstained with hematoxylin. Stained sections were mounted in aqueous mounting medium (DakoCytomation). No significant staining was detected in sections incubated with control rabbit serum instead of the primary antibody. The histological features shown are representative of all tissue samples studied.
For quantification of the number of Paneth cells, sections of all test and control mice were stained with H&E, and cells were counted in 20 crypts in representative microscopic fields (original magnification, ×200) (n = 15 per group). Villus and crypt lengths were measured in H&E-stained sections of all control and starved mice. Per group, 15 villi and 15 crypts were measured in digitized images at ×200 magnification.
Immunofluorescence
Immunofluorescence was performed as adapted from Matsumoto et al.15 In short, ileum samples were fixed immediately in 4% formaldehyde and embedded in paraffin, after which tissue sections (3 μm thick) were cut. After deparaffination, antigen retrieval was obtained in citrate buffer (Dako REAL Target Retrieval Solution, DakoCytomation). Sections were blocked in 5% bovine serum albumin and incubated with anti-PAPIII antibody overnight. Next, an Alexa 488–conjugated secondary antibody was applied, and nuclei were visualized with DAPI. Sections were mounted using fluorescent mounting medium (DakoCytomation). Images shown are representative for all tissue samples studied.
Electron Microscopy
For electron microscopy (EM), ileal tissue was immersed in 3% glutaraldehyde fixative buffered in 0.09 mol/L KH2PO4 at pH 7.4 and supplemented with 1.4% sucrose. Next, samples were washed in 0.09 mol/L KH2PO4 buffer containing 7.5% sucrose and transferred to a 1% OsO4 fixative solution buffered with veronal acetate buffer (pH 7.4) plus 1.5% ferrocyanide. After washing in veronal acetate buffer plus 7% sucrose for 5 minutes at 4°C, dehydration was performed in graded ethanol series, followed by embedding in Epon (Burlington, VT). Semithin sections (1 μm thick) were stained with toluidine blue for Paneth cell localization. Tissue sections were examined with a Philips CM 100 electron microscope (Philips, Eindhoven, The Netherlands) at an accelerating voltage of 80 KV.
In Vivo Intestinal Permeability Assay
Thirty minutes before sacrifice, animals were anesthetized to perform the in vivo intestinal permeability assay, as adapted from Costantini et al.17 In short, a midline laparotomy was performed, and a 5-cm segment of distal ileum was isolated between silk ties. Fluorescein isothiocyanate (FITC)–dextran (4.4 kDa), 25 mg, in a solution of 125 mg/mL PBS was injected intraluminally. The intestine was returned to the abdominal cavity, and the skin was closed. Thirty minutes after injection, blood was obtained through vena cava puncture. The FITC-dextran concentration in the plasma was analyzed using a Spectramax M2 fluorescence spectrophotometer (Molecular Devices, Sunnyvale, CA). A standard curve was obtained by serial dilutions of FITC-dextran in mouse plasma.
Western Blot Analysis
Dissected ileal segments were immediately snap frozen in liquid nitrogen and stored at −80°C until further processing for Western blot analysis. Tissue samples were homogenized in lysis buffer containing 200 mmol/L NaCl, 10 mmol/L Tris buffer, 5 mmol/L EDTA, 10% glycerol, and 1% NP-40 using a mini–bead beater (Biospec, Bartlesville, OK). Samples were centrifuged at 18,000 × g for 15 minutes at 4°C. The protein concentration of supernatants was determined using a bicinchoninic acid protein assay kit. For Western blot analysis of lysozyme and RegIIIγ, 10 μg of protein was heated at 100°C for 5 minutes in a reducing SDS sample buffer, separated by SDS-PAGE on a 15% polyacrylamide gel, and transferred onto a polyvinylidene fluoride membrane (Immobilon P; Millipore, Bedford, MA). After the transfer of proteins, blocking and antibody incubation of the membrane were performed in Tris-buffered saline–0.05% Tween supplemented with 5% and 3% nonfat dry milk, respectively. Primary antibody incubation occurred overnight at 4°C with anti-lysozyme (which recognizes mouse lysozymes P and M) or anti-mouse RegIIIγ. To confirm equal protein loading and transfer, membranes were reprobed with anti-β-actin antibodies. For LC3, 10 μg of protein was heated and separated on a 4% to 20% gradient gel (Biorad, Hercules, CA) and transferred onto a polyvinylidene fluoride membrane. Blocking and antibody incubation were performed in TBS–0.1% Tween supplemented with 3% nonfat dry milk. Mouse anti-rat LC3 antibody was incubated for 1 hour at room temperature. After washing with Tris-buffered saline–0.05% Tween, membranes were incubated with an appropriate HRP-conjugated secondary antibody for 90 minutes at room temperature. Signals were captured on blue X-ray film (SuperRX; Fuji, Tokyo, Japan) using the chemiluminescent substrate Supersignal West Pico. The band intensity was semiquantitatively analyzed using Quantity One (Biorad). No signal was detected in membranes incubated with control serum instead of the primary antibody. Figures shown are representative for all tissue samples studied.
Quantitative PCR
RNA was extracted from snap frozen ileal tissue using TRI reagent, according to the manufacturer's protocol. The RNA concentration was determined by Nanodrop (Thermo Fisher Scientific Inc.), and 1 μg of RNA was used as a template for reverse transcription using oligo dT primer and Moloney murine leukemia virus reverse transcriptase in a cDNA synthesis reaction. Quantitative real-time PCR (qPCR) was performed using the MyIQ System (Biorad). Gene expression levels of lysozyme, cryptdin, and RegIIIγ were determined using IQ5 software (Biorad) with a ΔCT relative quantification model. The geometric mean of the expression levels of two reference genes, cyclophilin A and β2-microglobulin, was calculated and used as a normalization factor. Used primers are depicted in Table 1.
Table 1.
Primers Used for qPCR Assays
| Gene product | Forward primer | Reverse primer |
|---|---|---|
| Lysozyme | 5′-GGATCAATTGCACTGCTCTG-3′ | 5′-CAGTTCCGAATATACTGGGAC-3′ |
| Cryptdin | 5′-TGAAGACACTAATCCTCCTCTCTGC-3′ | 5′-GCTCCTCAGTTTTAGTCTCTTCATCTGTA-3′ |
| RegIIIγ | 5′-GTATGATGCAGATATGGCCTG-3′ | 5′-ATATTGGCCACTGTTACCAC-3′ |
| LC3 | 5′-AAGATCCCAGTGATTATAGAGCGA-3′ | 5′-ATTGCTGTCCCGAATGTCTC-3′ |
| Cyclophilin A | 5′-TTCCTCCTTTCACAGAATTATTCCA-3′ | 5′-CCGCCAGTGCCATTATGG-3′ |
| β2-Microglobulin | 5′-CTTTCTGGTGCTTGTCTCACTGA-3′ | 5′-GTATGTTCGGCTTCCCATTCTC-3′ |
Bacterial Translocation
Translocation of luminal bacteria was assessed as previously described.18 In brief, MLNs were collected aseptically in 2 mL of preweighed thioglycolate broth tubes (Becton Dickinson Microbiology Europe, Maylan, France). After weighing, tissue specimens were homogenized and subsequently transferred onto both Columbia III blood agar base plates supplemented with 5% v/v sheep blood (duplicate plates) and Chocolate PolyviteX agar plates. After 48 hours of incubation, colonies were counted, adjusted to tissue weight, and calculated as number of colony-forming units per gram of tissue. Colonies were identified to the species level using conventional techniques to discriminate between gut and skin flora.
Statistical Analysis
Statistical analysis was performed using Prism 5.02 for Windows (GraphPad Software Inc., San Diego, CA). Gaussian distribution was tested using the Kolmogorov-Smirnov test. A two-tailed Student's t-test was used for between-group comparisons when groups were distributed normally. Otherwise, a two-tailed Mann-Whitney U-test was used. Data are represented as mean ± SEM. Differences were considered statistically significant at P < 0.05.
Results
Morphological and Functional Changes in the Starved Intestine
First, the overall effects of enteral starvation on intestinal morphological characteristics and integrity were assessed. H&E staining of ileal sections showed no apparent changes in small-intestinal structure after 48 hours of food deprivation (Figure 1A). Villus length and crypt depth did not differ between control and starved mice (Figure 1B). However, the intestinal permeability of starved mice, as assessed by measuring the systemic plasma concentration of intraluminally injected 4.4-kDa FITC-dextran, was significantly increased compared with controls (Figure 1C, P < 0.05).
Figure 1.
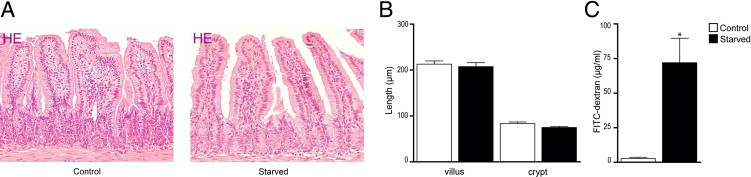
Morphological and functional changes in the starved intestine. A: H&E staining of ileum from control (left panel) and starved (right panel) mice showed no apparent changes in intestinal architecture. B: Villus length and crypt depth were similar in the ileum of starved mice compared with controls. C: Assessment of ileal permeability demonstrated a significant increase in plasma 4.4-kDa FITC-dextran of starved mice compared with controls. *P < 0.05. The histological features are representative for all tissue samples studied (n = 15 per group).
Decreased mRNA and Protein Expression of Paneth Cell Antimicrobials in Response to Starvation
To investigate the influence of starvation on Paneth cells, the expression of Paneth cell antimicrobials in the mouse ileum was assessed at both transcriptional and translational levels. First, IHC data showed intense staining for lysozyme in Paneth cell granules of control mouse ileum, whereas reduced Paneth cell lysozyme staining was observed in the ileum of starved mice (Figure 2A). These results were confirmed by Western blot analysis (Figure 3, A and B), in which constitutive expression of lysozyme was observed in ileum of unfasted mice. In contrast, food deprivation led to a significantly decreased lysozyme expression (P < 0.01). Corresponding results were obtained by qPCR, which also showed significant reduction in lysozyme mRNA expression in starvation (Figure 3C, P < 0.01). In mice, two forms of lysozyme are expressed, one being specific for the intestine and, in particular, the Paneth cell (lysozyme P) and the other being expressed in neutrophils and macrophages (lysozyme M).19, 20 The antibody used was generated against lysozyme P but cross-reacts with lysozyme M.20 However, the results obtained by Western blot and qPCR analyses reflect changes in Paneth cell–produced lysozyme because no difference in PMN or macrophage presence was detected, as illustrated by the IHC data on lysozyme. To gain more insight into this changed lysozyme expression in Paneth cells, the number of Paneth cells in crypts of starved mice was compared with the number of Paneth cells present in crypts of control mice. Quantification of Paneth cells revealed equal numbers in the ileum of both starved and control groups (Figure 2B), excluding the possibility that the observed decrease in lysozyme expression in Paneth cells was caused by reduced cell numbers. Next, we investigated the expression of other important antimicrobial proteins expressed by Paneth cells. Western blot data for RegIIIγ, which is represented by two bands on Western blot analysis (a high-molecular-weight precursor form of approximately 16.5 kDa and a functionally active low-molecular-weight form of 15 kDa),21 showed that expression of the inactive precursor form was significantly reduced on fasting (P < 0.05), whereas expression of the active form remained unaffected (Figure 3, A and B). qPCR results showed lowered mRNA transcription of the RegIIIγ gene (P < 0.05), confirming decreased RegIIIγ protein levels. To relate RegIIIγ results to Paneth cells, as it is also expressed by other enterocytes, we performed immunofluorescent staining (Figure 3D). The ileum from control mice showed expression of RegIIIγ in both enterocytes, at the transition of crypt to villus, and Paneth cells. In starved mice, the expression of RegIIIγ was reduced in all cells expressing this protein. Finally, we assayed the expression of cryptdin, which is exclusively produced by Paneth cells. qPCR analysis of cryptdin showed significantly decreased mRNA expression (P < 0.05) after 48 hours of starvation (Figure 3C). Together, these findings signify an inability of Paneth cells to maintain antimicrobial levels during starvation.
Figure 2.
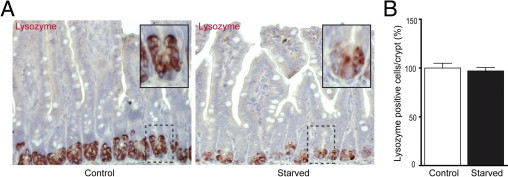
Starvation results in decreased lysozyme expression in Paneth cells. A: IHC for lysozyme in ileum from control (left panel) versus starved (right panel) mice showed reduced expression of lysozyme in Paneth cells from fasted animals. Insets: Magnifications of single crypts from both control and starved mice. B: The number of Paneth cells per crypt in the ileum of starved and control mice showed no difference. The histological features are representative for all tissue samples studied (n = 15 per group).
Figure 3.
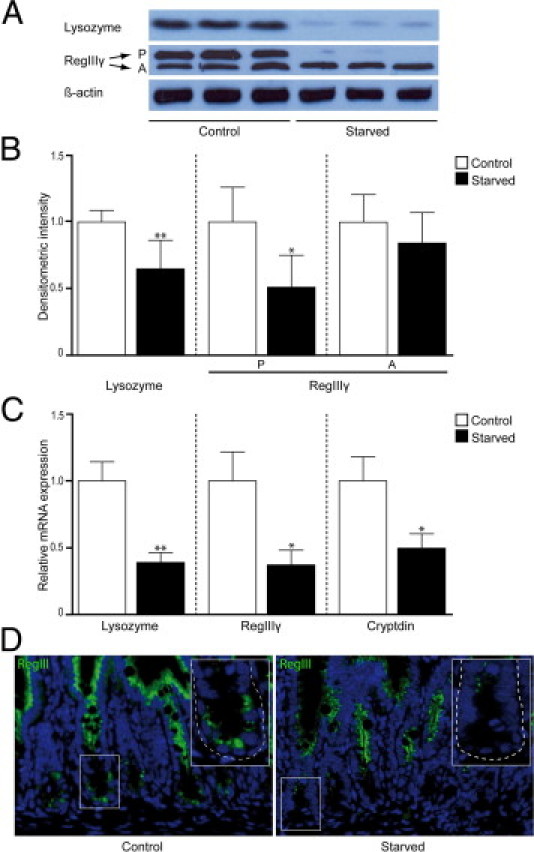
Protein and mRNA levels of Paneth cell antimicrobial proteins are decreased after 48 hours of starvation. A: Western blot analysis showed decreased band density for lysozyme (15 kDa) and the precursor (P) form of RegIIIγ (16.5 kDa) in the ileum of starved mice compared with control mice, whereas no change was observed in the active form (A) of RegIIIγ (representative bands of three mice per group are shown). β-Actin was used to correct for equal protein loading (n = 12 per group). B: Quantification of Western blots demonstrated a significant decrease in both lysozyme (*P < 0.01) expression and expression of the precursor form of RegIIIγ (**P < 0.05). C: qPCR analysis showed a 2.5-fold decrease in lysozyme gene expression (*P < 0.01), a 2.7-fold decrease in gene expression of RegIIIγ (**P < 0.05), and a twofold decrease in cryptdin (**P < 0.05) gene expression after starvation (n = 15 per group). D: Immunofluorescence for RegIIIγ in ileum from control (left panel) versus starved (right panel) mice showed an overall reduced expression of RegIIIγ in fasted animals, as well as in Paneth cells specifically. Insets: Magnifications of single crypts from both control and starved mice. The histological features are representative for all tissue samples studied (n = 15 per group).
Autophagy and Granule Abnormalities in Paneth Cells of Starved Mice
Because starvation is associated with the occurrence of autophagy, a process that is involved in the formation of Paneth cell secretory granules,22 we next studied Paneth cells by EM, the gold standard to assess autophagy. EM revealed the presence of dark c-shaped structures in the cytoplasm, corresponding to late degradative autophagolysosomes (Figure 4A).23 Similar structures were occasionally seen in control mice but at a much lower abundance. Furthermore, enhanced mRNA levels of LC3 (P < 0.01), important in the formation of the autophagosomes, and a trend toward an increase in the ratio between phosphatidylethanolamine-conjugated LC3 (LC3-II) and LC3-I (P = 0.10) confirmed this result (Figure 4, B and C).
Figure 4.
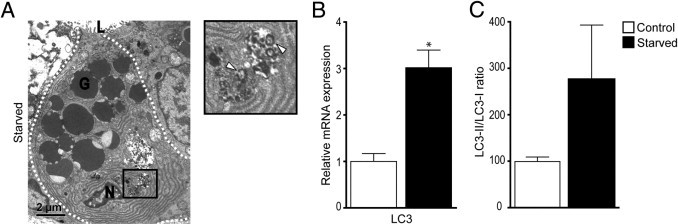
Autophagy in Paneth cells of starved mice. A: Electron microscopy showed the presence of late degradative autophagosomes (white arrowheads). G, granule; L, lumen; N, nucleus. B: qPCR showed a significant increase in LC3 gene expression in the small intestine of starved mice. *P < 0.01. C: Quantification of band density of phosphatidylethanolamine-conjugated LC3 (LC3-II) and LC3-I, as assayed by using Western blot analysis, revealed a trend for an increase in the LC3-II/LC-I ratio. P = 0.10.
In addition, ultrastructural analysis of Paneth cells by EM also revealed abnormalities in the morphological characteristics of Paneth cell granules. The peripheral electron-lucent halo surrounding the electron-dense core of granules in Paneth cells of starved mice differed from granule conformation of control mice. In starved mice, instead of the usually round delineated continuous halo (Figure 5A), the halo was disturbed and often showed interrupted droplet-like structures at the outer periphery of the granules (Figure 5A). The peripheral halo of Paneth cell granules mainly contains O-linked oligosaccharides with N-acetyl-galactosamine and N-acetyl-d-glucosamine residues and N-linked oligosaccharides with N-acetyl-d-glucosamine residues, which are easily detected by T. vulgaris lectin.24 Staining with this lectin showed less intense staining and aberrant granule morphological characteristics, suggesting alterations in the oligosaccharide composition to be present in starved mice compared with control mice (Figure 5B).
Figure 5.
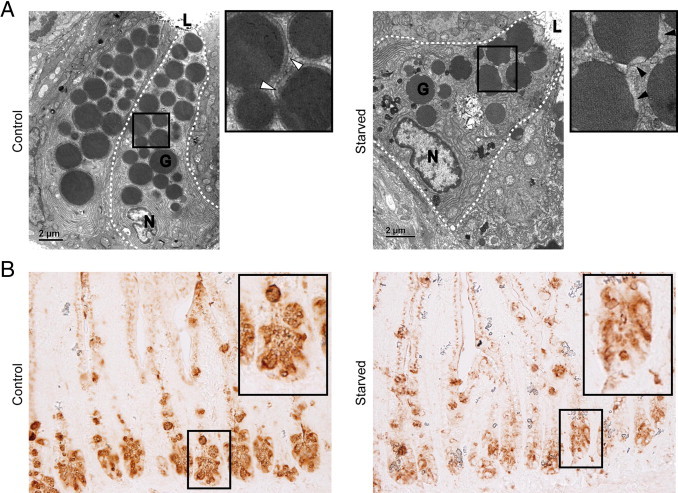
Granule morphological features and composition is aberrant in Paneth cells of starved mice. A: Electron microscopy revealed changes in the electron-lucent halo of granules in Paneth cells of starved mice (right panel, magnification, black arrowheads) compared with control mice (left panel, magnification, white arrowheads). The dashed lines demarcate Paneth cells. G, granule; L, lumen; N, nucleus (n = 3 per group). B: Staining with lectin T. vulgaris showed less intense staining and aberrant granule morphological features in Paneth cells of starved mice (n = 15 per group). Images are representative for all tissue samples studied.
Increase in Bacterial Translocation to MLNs on Starvation
Finally, considering the importance of Paneth cells in the defense of the small intestine against bacterial invasion,3 we examined the consequences of starvation on the occurrence of bacterial translocation. As shown in Figure 6, a twofold increase in colony-forming units per gram of MLN tissue (P < 0.01) was observed in starved mice. These results indicate that, on Paneth cell dysfunction, caused by starvation, bacteria are able to invade host tissue.
Figure 6.
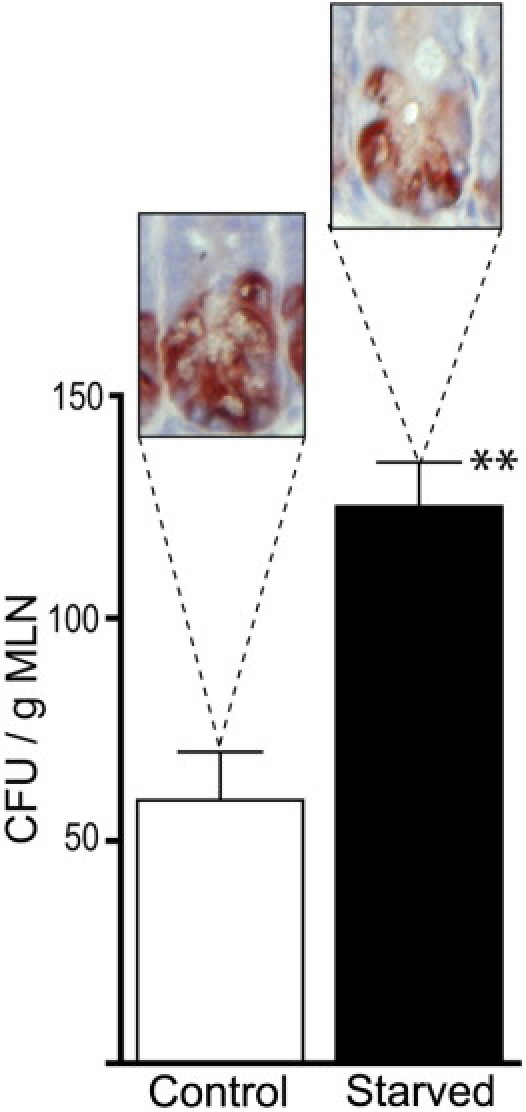
Enhanced bacterial translocation to the MLNs in starved mice. Bacterial translocation to MLNs is significantly elevated in mice with decreased lysozyme staining after 48 hours of starvation compared with controls [59.1 ± 10.8 versus 125.1 ± 9.9 colony-forming units per gram (CFU/g) of MLNs]. **P < 0.01 (n = 10 per group).
Discussion
Paneth cells play an important physiological role in maintaining the barrier of the small intestine. First, they are indispensable in crypt survival, providing essential niche signals to stem cells.25 Second, they are crucial in small-intestinal immunity by sensing and responding to enteric bacteria, protecting stem cells and limiting penetration of the intestinal wall by both commensal and pathogenic bacteria.3 To this end, Paneth cells produce and secrete cytoplasmic granules containing a battery of antimicrobial proteins.26
Because enteral starvation and the use of total parenteral nutrition result in bacterial translocation in critically ill patients,10 we hypothesized that the absence of oral food intake compromises Paneth cell functionality and its role in gut protection. Our results, which demonstrate morphological abnormalities in Paneth cells and a defect in Paneth cell antimicrobial protein production, support this hypothesis.
Herein, we show that Paneth cells of starved mice display aberrant granules, with changes in their composition. Recently, mice deficient in ATG16L1 or ATG5 protein, important in the formation and proper subcellular localization of the autophagosome,27 showed substantial structural abnormalities in Paneth cells and their granules. More specifically, the Paneth cell granule number was decreased, and Paneth cells showed distorted packaging and exocytosis of their granules. These results suggest a specific physiological role for autophagy in regulating the development and secretion of Paneth cell granules.22 Apart from its physiological function, autophagy can also be induced on starvation as an adaptive response that recycles macromolecules to provide nutrients and energy essential for survival.11, 12, 13 Interestingly, Paneth cells of starved mice showed increased induction of autophagy. We hypothesize that the deviant granules in Paneth cells of starved mice may be caused by induction of autophagy on starvation in an attempt to maintain vital cellular functions at the expense of the physiological function of autophagy (namely, the regulation of Paneth cell granule development). Starvation could, however, also affect other players that are important in the maintenance of mucosal homeostasis and Paneth cell physiological features. For example, commensal bacteria and their products are shown to induce a protective response to maintain epithelial homeostasis.28 A change in microbial composition, as occurs in starvation,29 could, therefore, reflect on the crucial balance maintained in the intestine and on Paneth cells that are influenced by microbiota and their products.2 Finally, starvation-induced changes could be caused by the loss of energy itself or the absence of nutritional/bulk stimulation.
Next to changes in granule morphological characteristics, our study revealed that protein expression of lysozyme and the precursor form of RegIIIγ were significantly lowered in the mouse ileum after 48 hours of food deprivation. However, the expression of the functionally active form of RegIIIγ did not differ between control and starved mice. Because the precursor form showed reduced expression levels, prolonged starvation may also exhaust the supply of active RegIIIγ in this model. Moreover, mRNA expression of all three antimicrobials studied was lowered in the ileum of the food-deprived group. In addition, no difference in Paneth cell number was observed in starved mice, demonstrating that the decreased expression of antimicrobial proteins was not caused by reduced cell numbers. Although autophagy generates new constituents to maintain cellular homeostasis, the deficit may not completely be abrogated, resulting, in this case, in hampered production of proteins less important in cell survival. Altogether, these data indicate that Paneth cells become dysfunctional during fasting, with a reduced capacity to transcribe antimicrobial genes and, correspondingly, hampered production of antimicrobial proteins. Next to Paneth cell malfunction, through decreased expression of antimicrobial proteins, we observed enhanced permeability, as assayed by FITC-dextran and translocation of microbiota to MLNs. The former could be both a result of the enhanced translocation of bacteria and impaired Paneth cell α-defensin release, resulting in IL-1β–induced enhanced permeability.30, 31, 32 We have previously shown the additive effect of deviant Paneth cell function on the occurrence of bacterial translocation in situations of physical intestinal barrier loss.4 These findings are in agreement with those of a previous study on Paneth cell function in vivo, in which Vaishnava et al3 elegantly showed the crucial role of Paneth cells in limiting translocation of commensal and pathogenic bacteria across the intestinal barrier in a Paneth cell ablation model. In addition, Nieuwenhuis et al33 associated Paneth cell granule abnormalities with alterations in the composition of the intestinal microbiota and bacteria that moved into close proximity with epithelial cells.
Although Paneth cells are the main producers of antimicrobial proteins in the intestine, other cells and mechanisms contribute to the maintenance of the intestinal barrier. Goblet cells also produce antibacterial products and, more important, mucus. Even if no changes in number of goblet cells occurred in starvation,16, 34 possible compromised goblet cell protein production could still contribute to starvation-associated changes. In addition, in line with our results, Goldberg et al showed a decrease in alkaline phosphatase and a reduction in lipopolysaccharide-dephosphorylating activity in the gut of mice starved for 48 hours,35 showing that other factors aimed at protecting the intestine from bacterial invasion are compromised.
In conclusion, the current study demonstrates that, during enteral starvation, Paneth cell function is compromised, reflected by diminished antimicrobial protein production and aberrant Paneth cell morphological features, presumably because of the induction of processes aimed at cell survival. This improved insight into the interaction between starvation and the intestinal immune barrier supports the restricted use of a total parenteral nutrition regimen in critically ill patients.
Acknowledgments
We thank Dr. Tomas Ganz for providing the anti-lysozyme antiserum, Dr. Lora V. Hooper for providing the rabbit anti-mouse RegIIIγ antibody, Dr. Kenneth Cadwell and Dr. Thaddeus Stappenbeck for helping to choose antibodies for the study, and Hans Duimel for his excellent technical support.
Footnotes
Supported by the Profileringsfonds of the Academic Hospital Maastricht.
Current address of E.H., Department of Surgery, University Medical Center Groningen, Groningen, The Netherlands.
References
- 1.Bevins C.L., Salzman N.H. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011;9:356–368. doi: 10.1038/nrmicro2546. [DOI] [PubMed] [Google Scholar]
- 2.Ayabe T., Satchell D.P., Wilson C.L., Parks W.C., Selsted M.E., Ouellette A.J. Secretion of microbicidal alpha-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 3.Vaishnava S., Behrendt C.L., Ismail A.S., Eckmann L., Hooper L.V. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc Natl Acad Sci U S A. 2008;105:20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grootjans J., Hodin C.M., de Haan J.J., Derikx J.P., Rouschop K.M., Verheyen F.K., van Dam R.M., Dejong C.H., Buurman W.A., Lenaerts K. Level of activation of the unfolded protein response correlates with Paneth cell apoptosis in human small intestine exposed to ischemia/reperfusion. Gastroenterology. 2011;140:529–539. doi: 10.1053/j.gastro.2010.10.040. [DOI] [PubMed] [Google Scholar]
- 5.MacFie J. Enteral versus parenteral nutrition: the significance of bacterial translocation and gut-barrier function. Nutrition. 2000;16:606–611. doi: 10.1016/s0899-9007(00)00249-5. [DOI] [PubMed] [Google Scholar]
- 6.Ekelund M., Kristensson E., Ekelund M., Ekblad E. Total parenteral nutrition causes circumferential intestinal atrophy, remodeling of the intestinal wall, and redistribution of eosinophils in the rat gastrointestinal tract. Dig Dis Sci. 2007;52:1833–1839. doi: 10.1007/s10620-006-9678-z. [DOI] [PubMed] [Google Scholar]
- 7.Wildhaber B.E., Yang H., Spencer A.U., Drongowski R.A., Teitelbaum D.H. Lack of enteral nutrition: effects on the intestinal immune system. J Surg Res. 2005;123:8–16. doi: 10.1016/j.jss.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 8.Li J., Kudsk K.A., Gocinski B., Dent D., Glezer J., Langkamp-Henken B. Effects of parenteral and enteral nutrition on gut-associated lymphoid tissue. J Trauma. 1995;39:44–52. doi: 10.1097/00005373-199507000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Besselink M.G., van Santvoort H.C., Boermeester M.A., Nieuwenhuijs V.B., van Goor H., Dejong C.H., Schaapherder A.F., Gooszen H.G. Timing and impact of infections in acute pancreatitis. Br J Surg. 2009;96:267–273. doi: 10.1002/bjs.6447. [DOI] [PubMed] [Google Scholar]
- 10.MacFie J., Reddy B.S., Gatt M., Jain P.K., Sowdi R., Mitchell C.J. Bacterial translocation studied in 927 patients over 13 years. Br J Surg. 2006;93:87–93. doi: 10.1002/bjs.5184. [DOI] [PubMed] [Google Scholar]
- 11.Yang Z., Klionsky D.J. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ezaki J., Matsumoto N., Takeda-Ezaki M., Komatsu M., Takahashi K., Hiraoka Y., Taka H., Fujimura T., Takehana K., Yoshida M., Iwata J., Tanida I., Furuya N., Zheng D.M., Tada N., Tanaka K., Kominami E., Ueno T. Liver autophagy contributes to the maintenance of blood glucose and amino acid levels. Autophagy. 2011;7:727–736. doi: 10.4161/auto.7.7.15371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uddin M.N., Ito S., Nishio N., Suganya T., Isobe K. Gadd34 induces autophagy through the suppression of the mTOR pathway during starvation. Biochem Biophys Res Commun. 2011;407:692–698. doi: 10.1016/j.bbrc.2011.03.077. [DOI] [PubMed] [Google Scholar]
- 14.Cadwell K., Liu J.Y., Brown S.L., Miyoshi H., Loh J., Lennerz J.K., Kishi C., Kc W., Carrero J.A., Hunt S., Stone C.D., Brunt E.M., Xavier R.J., Sleckman B.P., Li E., Mizushima N., Stappenbeck T.S., Virgin H.W., 4th A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsumoto S., Konishi H., Maeda R., Kiryu-Seo S., Kiyama H. Analysis on expression of the regenerating gene (Reg) family members Reg-IIIβ and Reg-IIIγ in the mouse during development. J Comp Neurol. 2011 doi: 10.1002/cne.22705. [DOI] [PubMed] [Google Scholar]
- 16.Sokolovic M., Wehkamp D., Sokolovic A., Vermeulen J., Gilhuijs-Pederson L.A., van Haaften R.I., Nikolsky Y., Evelo C.T., van Kampen A.H., Hakvoort T.B., Lamers W.H. Fasting induces a biphasic adaptive metabolic response in murine small intestine. BMC Genomics. 2007;8:361. doi: 10.1186/1471-2164-8-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costantini T.W., Peterson C.Y., Kroll L., Loomis W.H., Eliceiri B.P., Baird A., Bansal V., Coimbra R. Role of p38 MAPK in burn-induced intestinal barrier breakdown. J Surg Res. 2009;156:64–69. doi: 10.1016/j.jss.2009.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Haan J.J., Lubbers T., Hadfoune M., Luyer M.D., Dejong C.H., Buurman W.A., Greve J.W. Postshock intervention with high-lipid enteral nutrition reduces inflammation and tissue damage. Ann Surg. 2008;248:842–848. doi: 10.1097/SLA.0b013e318188752c. [DOI] [PubMed] [Google Scholar]
- 19.Cross M., Mangelsdorf I., Wedel A., Renkawitz R. Mouse lysozyme M gene: isolation, characterization, and expression studies. Proc Natl Acad Sci U S A. 1988;85:6232–6236. doi: 10.1073/pnas.85.17.6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ganz T., Gabayan V., Liao H.I., Liu L., Oren A., Graf T., Cole A.M. Increased inflammation in lysozyme M-deficient mice in response to Micrococcus luteus and its peptidoglycan. Blood. 2003;101:2388–2392. doi: 10.1182/blood-2002-07-2319. [DOI] [PubMed] [Google Scholar]
- 21.Mukherjee S., Partch C.L., Lehotzky R.E., Whitham C.V., Chu H., Bevins C.L., Gardner K.H., Hooper L.V. Regulation of C-type lectin antimicrobial activity by a flexible N-terminal prosegment. J Biol Chem. 2008;284:4881–4888. doi: 10.1074/jbc.M808077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cadwell K., Patel K.K., Komatsu M., Virgin H.W., 4th, Stappenbeck T.S. A common role for Atg16L1, Atg5 and Atg7 in small intestinal Paneth cells and Crohn disease. Autophagy. 2009;5:250–252. doi: 10.4161/auto.5.2.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwata A., Riley B.E., Johnston J.A., Kopito R.R. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J Biol Chem. 2005;280:40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 24.Leis O., Madrid J.F., Ballesta J., Hernandez F. N- and O-linked oligosaccharides in the secretory granules of rat Paneth cells: an ultrastructural cytochemical study. J Histochem Cytochem. 1997;45:285–293. doi: 10.1177/002215549704500213. [DOI] [PubMed] [Google Scholar]
- 25.Sato T., van Es J.H., Snippert H.J., Stange D.E., Vries R.G., van den Born M., Barker N., Shroyer N.F., van D.E., Wetering M., Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 2011;469:415–418. doi: 10.1038/nature09637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mukherjee S., Vaishnava S., Hooper L.V. Multi-layered regulation of intestinal antimicrobial defense. Cell Mol Life Sci. 2008;284:3019–3027. doi: 10.1007/s00018-008-8182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fujita N., Itoh T., Omori H., Fukuda M., Noda T., Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Crawford P.A., Crowley J.R., Sambandam N., Muegge B.D., Costello E.K., Hamady M., Knight R., Gordon J.I. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci U S A. 2009;106:11276–11281. doi: 10.1073/pnas.0902366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi J., Aono S., Lu W., Ouellette A.J., Hu X., Ji Y., Wang L., Lenz S., van Ginkel F.W., Liles M., Dykstra C., Morrison E.E., Elson C.O. A novel role for defensins in intestinal homeostasis: regulation of IL-1beta secretion. J Immunol. 2007;179:1245–1253. doi: 10.4049/jimmunol.179.2.1245. [DOI] [PubMed] [Google Scholar]
- 31.Al-Sadi R.M., Ma T.Y. IL-1beta causes an increase in intestinal epithelial tight junction permeability. J Immunol. 2007;178:4641–4649. doi: 10.4049/jimmunol.178.7.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Sadi R., Ye D., Dokladny K., Ma T.Y. Mechanism of IL-1beta-induced increase in intestinal epithelial tight junction permeability. J Immunol. 2008;180:5653–5661. doi: 10.4049/jimmunol.180.8.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nieuwenhuis E.E., Matsumoto T., Lindenbergh D., Willemsen R., Kaser A., Simons-Oosterhuis Y., Brugman S., Yamaguchi K., Ishikawa H., Aiba Y., Koga Y., Samsom J.N., Oshima K., Kikuchi M., Escher J.C., Hattori M., Onderdonk A.B., Blumberg R.S. Cd1d-dependent regulation of bacterial colonization in the intestine of mice. J Clin Invest. 2009;119:1241–1250. doi: 10.1172/JCI36509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fernandez-Estivariz C., Gu L.H., Gu L., Jonas C.R., Wallace T.M., Pascal R.R., Devaney K.L., Farrell C.L., Jones D.P., Podolsky D.K., Ziegler T.R. Trefoil peptide expression and goblet cell number in rat intestine: effects of KGF and fasting-refeeding. Am J Physiol Regul Integr Comp Physiol. 2003;284:R564–R573. doi: 10.1152/ajpregu.00428.2002. [DOI] [PubMed] [Google Scholar]
- 35.Goldberg R.F., Austen W.G., Jr., Zhang X., Munene G., Mostafa G., Biswas S., McCormack M., Eberlin K.R., Nguyen J.T., Tatlidede H.S., Warren H.S., Narisawa S., Millan J.L., Hodin R.A. Intestinal alkaline phosphatase is a gut mucosal defense factor maintained by enteral nutrition. Proc Natl Acad Sci U S A. 2008;105:3551–3556. doi: 10.1073/pnas.0712140105. [DOI] [PMC free article] [PubMed] [Google Scholar]


