During IgL chain rearrangement in mouse pre–B cells, DNA breaks inflicted by RAG proteins induce Pim2 to promote cell survival and limit proliferation; thus, DNA breaks effectively stand in for the prosurvival cytokine IL-7, whose signaling is attenuated during this stage of B cell development.
Abstract
Interleukin 7 (IL-7) promotes pre–B cell survival and proliferation by activating the Pim1 and Akt kinases. These signals must be attenuated to induce G1 cell cycle arrest and expression of the RAG endonuclease, which are both required for IgL chain gene rearrangement. As lost IL-7 signals would limit pre–B cell survival, how cells survive during IgL chain gene rearrangement remains unclear. We show that RAG-induced DNA double-strand breaks (DSBs) generated during IgL chain gene assembly paradoxically promote pre–B cell survival. This occurs through the ATM-dependent induction of Pim2 kinase expression. Similar to Pim1, Pim2 phosphorylates BAD, which antagonizes the pro-apoptotic function of BAX. However, unlike IL-7 induction of Pim1, RAG DSB-mediated induction of Pim2 does not drive proliferation. Rather, Pim2 has antiproliferative functions that prevent the transit of pre–B cells harboring RAG DSBs from G1 into S phase, where these DNA breaks could be aberrantly repaired. Thus, signals from IL-7 and RAG DSBs activate distinct Pim kinase family members that have context-dependent activities in regulating pre–B cell proliferation and survival.
B cell development relies on coordinate signals from cell surface antigen and cytokine receptors. IgH chain antigen receptor genes are assembled in pro–B cells and pair with the surrogate light chain (λ5 and VpreB) to form the pre–B cell receptor (BCR) (Herzog et al., 2009). Signals from the pre-BCR drive cellular expansion, transit from the pro–B to pre–B cell stage, and initiation of IgL chain gene assembly (Herzog et al., 2009). At the pre–B cell stage, successful assembly of an IgL chain gene and subsequent pairing with the IgH chain leads to the expression of a BCR, which signals termination of antigen receptor gene assembly and promotes transition to the immature B cell stage (Bassing et al., 2002; Herzog et al., 2009).
Antigen receptor genes are assembled through V(D)J recombination, a reaction initiated by the RAG1 and RAG2 proteins, which together form the RAG endonuclease (Fugmann et al., 2000). V(D)J recombination is restricted in developing lymphocytes to the G1 phase of the cell cycle, in part as a result of the degradation of RAG2 in S phase (Desiderio et al., 1996). RAG introduces DNA double-strand breaks (DSBs) at the border of two recombining gene segments (V, D, or J) and their flanking RAG recognition sequences, termed recombination signals (Fugmann et al., 2000). RAG DNA cleavage forms two blunt signal ends and two hairpin-sealed coding ends. These DNA ends are processed and joined by the nonhomologous end-joining (NHEJ) pathway of DNA repair to form a signal joint and a coding joint, which completes formation of the second exon of antigen receptor genes (Rooney et al., 2004). The DNA breaks generated during this process activate the ATM kinase, which promotes canonical DNA damage responses, including cell death pathways which ultimately kill cells with persistent unrepaired RAG DSBs (Gapud and Sleckman, 2011).
In addition to pre-BCR and BCR signals, IL-7 receptor signals are important to promote survival and proliferation of developing B cells (Milne and Paige, 2006). The IL-7 receptor activates the JAK–STAT (STAT5) pathway, which induces the expression of Pim1 (Goetz et al., 2004). Pim1 is a member of a family of constitutively active serine-threonine kinases, which also includes Pim2 and Pim3, and supports lymphocyte proliferation and survival (Amaravadi and Thompson, 2005; Nawijn et al., 2011). The importance of IL-7 receptor signals is indicated by the block in B cell development observed in mice deficient for IL-7, IL-7 receptor, or STAT5 (Malin et al., 2010b; Milne and Paige, 2006). Pim1-deficient mice exhibit defects in B cell development, albeit to a lesser extent than mice compromised for IL-7 signaling (Domen et al., 1993). The incomplete developmental block in Pim1−/− mice suggests that other signaling pathways can contribute to pre–B cell proliferation and survival. In this regard, in developing T cells, IL-7 activates both Pim1 and the Akt kinase to support cell survival (Li et al., 2004). Whether Akt has a similar prosurvival function in developing B cells is not known.
Although IL-7 signals are critical for developing pre–B cells, these signals must be attenuated during IgL chain gene assembly as: (1) IL-7 signals suppress RAG gene expression in pre–B cells, (2) proliferative signals from IL-7 promote the degradation of RAG2 as cells pass into S phase, (3) persistent proliferative signals promote genomic instability by transiting cells with RAG DSBs into S phase before these DNA breaks can be repaired by NHEJ, and (4) IL-7 signals negatively regulate IgL gene accessibility (Billips et al., 1995; Desiderio et al., 1996; Amin and Schlissel, 2008; Johnson et al., 2008; Malin et al., 2010a). Recent studies have suggested that IL-7 signaling in pre–B cells may be attenuated through the pre-BCR–mediated expression of CXCR4, which may promote migration of pre–B cells away from IL-7–producing stromal cells in the bone marrow (Tokoyoda et al., 2004; Johnson et al., 2008).
Despite the importance of attenuating IL-7 proliferative signals, lost IL-7 survival signals could limit the time for pre–B cells to assemble a functional IgL chain gene. IgLκ gene assembly occurs through the rearrangement of a Vκ gene segment to one of four Jκ gene segments. The structure of the locus is such that multiple Vκ to Jκ rearrangements can, and often do, occur on a single allele. A single rearrangement can take several hours to complete and, thus, pre–B cells may have to survive for an extended period of time as they iteratively assemble IgLκ genes in an attempt to express a functional IgLκ chain (Casellas et al., 2001). Moreover, cells that fail to generate an IgLκ chain gene often go on to attempt IgLλ chain gene assembly (Arakawa et al., 1996). What promotes the survival of pre–B cells that are assembling IgL chain genes is largely unknown.
In response to RAG DSBs, the activation of ATM induces a broadly functional genetic program (Bredemeyer et al., 2008). In this paper, we develop an experimental approach designed to elucidate the function of signaling pathways activated by RAG DSBs in the regulation of B cell development. We find that RAG DNA breaks generated during IgL chain gene rearrangement promote the survival of pre–B cells that have lost IL-7 signals. This cytokine-independent survival requires the induction of Pim2 expression by RAG DSBs. When compared with IL-7 induction of Pim1, Pim2 has distinct context-dependent survival and proliferative activities in cells undergoing V(D)J recombination. Our findings establish that signals from RAG DSBs are coordinated with those from the IL-7 receptor to regulate the survival and proliferation of developing pre–B cells.
RESULTS AND DISCUSSION
Pim1 and Akt provide survival signals in response to IL-7
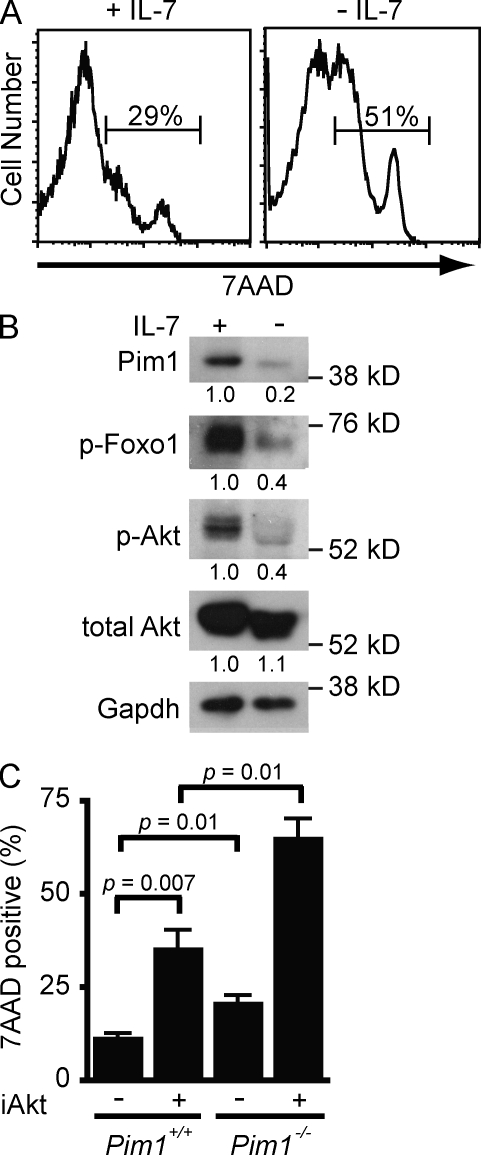
Large proliferating pre–B cells require the continuous presence of IL-7 (Fig. 1 A; Milne and Paige, 2006). IL-7 induces expression of the Pim1 kinase, which promotes survival as indicated by the enhanced uptake of 7AAD in Pim1−/− pre–B cells compared with wild-type (Pim1+/+) pre–B cells in the presence of IL-7 (Fig. 1, B and C; Domen et al., 1993). However, survival is not completely abrogated in Pim1−/− pre–B cells, suggesting that IL-7 regulates additional prosurvival pathways. In this regard, the Akt kinase is activated in pre–B cells by IL-7 as indicated by its autophosphorylation and the phosphorylation of Foxo-1, a known target of Akt in pre–B cells (Fig. 1 B; Amin and Schlissel, 2008). Inhibition of Akt kinase activity with Akt inhibitor VIII leads to an increase in pre–B cell death in the presence of IL-7 (Fig. 1 C). Additionally, treatment of Pim1−/− pre–B cells with this inhibitor leads to a synergistic increase in cell death (Fig. 1 C). Together, these findings demonstrate that Pim1 and Akt function to regulate pre–B cell survival in response to IL-7.
Figure 1.
Akt and Pim1 promote IL-7-dependent survival. (A) Cell death assessed by 7AAD uptake in wild-type pre–B cells cultured in IL-7 (+IL-7) and 48 h after IL-7 withdrawal (−IL-7). (B) Western blot analysis of Pim1, phosphorylated Foxo1 (p-Foxo1), phosphorylated Akt (p-Akt), and total Akt in whole cell lysates from wild-type pre–B cells in IL-7 (+) and 48 h after IL-7 withdrawal (−). Gapdh is shown as a protein loading control. Numbers represent band intensity relative to +IL-7 and standardized to loading controls. (C) Cell death assessed by 7AAD uptake in wild-type (Pim1+/+) and Pim1−/− pre–B cells cultured for 48 h in media containing IL-7 and 5 µM Akt inhibitor VIII (+) or DMSO (−) for 48 h. Shown is the mean and standard deviation from three independent experiments. Data in A and B are representative of at least three independent experiments.
RAG DSBs activate DNA damage responses in pre–B cells
To elucidate the signaling pathways downstream of RAG DSBs in pre–B cells, we generated mice that carry IgH and Bcl2 transgenes and are deficient in RAG1 or the NHEJ factor Artemis (RAG1−/−:IgH:Bcl2 and Artemis−/−:IgH:Bcl2 mice, respectively; Fig. S1). Artemis is a nuclease that is required to open hairpin sealed coding ends (Rooney et al., 2004). Thus, cells deficient in Artemis cannot make coding joints and mice deficient in Artemis have a block in B cell development at the pro–B cell stage as a result of their inability to generate a functional IgH chain gene (Rooney et al., 2004). Mice deficient in RAG1 cannot generate RAG DSBs and thus also exhibit a block at the pro–B cell stage (Fugmann et al., 2000). However, the inclusion of an IgH transgene in RAG1−/−:IgH:Bcl2 and Artemis−/−:IgH:Bcl2 mice allows for pre-BCR expression and transit from the pro–B cell (B220+CD43+) to the pre–B cell stage (B220+CD43−) where further development is blocked as a result of the inability to generate a functional IgL chain gene (Fig. S1; Bassing et al., 2002; Herzog et al., 2009).
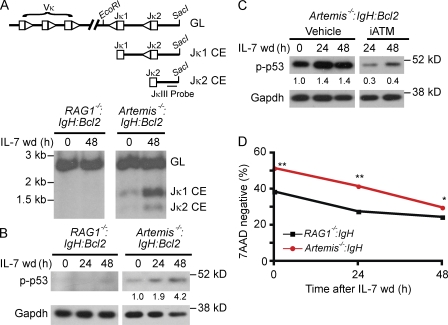
Southern blot analysis of genomic DNA from Artemis−/−:IgH:Bcl2 pre–B cells cultured in IL-7 reveals low levels of unrepaired Jκ coding ends generated by RAG cleavage because IL-7 suppresses, but does not abrogate, V(D)J recombination (Fig. 2 A; Milne et al., 2004). IL-7 withdrawal enhances RAG expression, leading to increased generation of Jκ coding ends in Artemis−/−:IgH:Bcl2 pre–B cells (Fig. 2 A). These DNA DSBs lead to the phosphorylation of p53 (Fig. 2, B and C). p53 phosphorylation was not observed in RAG1−/−:IgH:Bcl2 pre–B cells or in Artemis−/−:IgH:Bcl2 pre–B cells treated with the ATM inhibitor KU55933 (Fig. 2, B and C). Thus, RAG DSBs activate ATM and downstream DNA damage responses in pre–B cells.
Figure 2.
RAG DSBs support pre–B cell survival. (A) A schematic of the germline (GL) IgLκ locus and unrepaired Jκ1 and Jκ2 coding ends (CEs) are shown. Vκ and Jκ gene segments (rectangles) and their associated recombination sequences (triangles) are shown as are the relative positions of the EcoRI and SacI sites and the JκIII probe used for Southern blot analyses. Southern blot analysis of genomic DNA from RAG1−/−:IgH:Bcl2, and Artemis−/−:IgH:Bcl2 pre–B cells cultured in IL-7 (0 h) and after IL-7 withdrawal (wd, 48 h). Hybridizing bands generated by the GL IgLκ locus and Jκ1 and Jκ2 un-repaired CEs are indicated. (B) Western blot analysis of phosphorylated p53 (p-p53) after withdrawal of IL-7 for the indicated times. Gapdh is shown as a protein loading control. Numbers represent band intensity relative to Artemis−/−:IgH:Bcl2 at time 0 h and standardized to loading controls. No quantitation is presented for RAG1−/−:IgH:Bcl2 as no bands are present. (C) Western blot analysis of p-p53 after IL-7 withdrawal in Artemis−/−:IgH:Bcl2 treated with vehicle (DMSO) or the ATM inhibitor KU55933. Gapdh is shown as protein loading control. Numbers represent band intensity relative to time 0 h and standardized to loading controls. (D) Viability assessed by 7AAD uptake after withdrawal of IL-7 for the indicated times. Shown is the mean and standard deviation for three replicates. **, P < 0.0001; *, P = 0.001. Data in A, B, and D are representative examples of at least three independent experiments. Data in C are representative of two independent experiments.
Pre–B cells with RAG DSBs exhibit enhanced survival
Phosphorylation of the proapoptotic transcription factor p53 in response to RAG DSBs should promote cell death in Artemis-deficient compared with Rag-deficient pre–B cells. To determine the effect of RAG DSBs on pre–B cell survival, we generated RAG- and Artemis-deficient mice that did not contain the Bcl2 transgene (Rag1−/−:IgH and Artemis−/−:IgH mice, respectively). Surprisingly, Artemis−/−:IgH pre–B cells exhibited increased viability compared with RAG1−/−:IgH pre–B cells at early time points (24 h) after withdrawal of IL-7 (Fig. 2 D). Moreover, Artemis−/−:IgH pre–B cells have increased survival in IL-7 (0 h), where RAG DSBs are also generated and p53 is phosphorylated (Fig. 2). These data demonstrate that RAG DSBs activate prosurvival pathways that may antagonize the proapoptotic effects of p53 in pre–B cells.
Pim2 promotes survival downstream of RAG DSBs
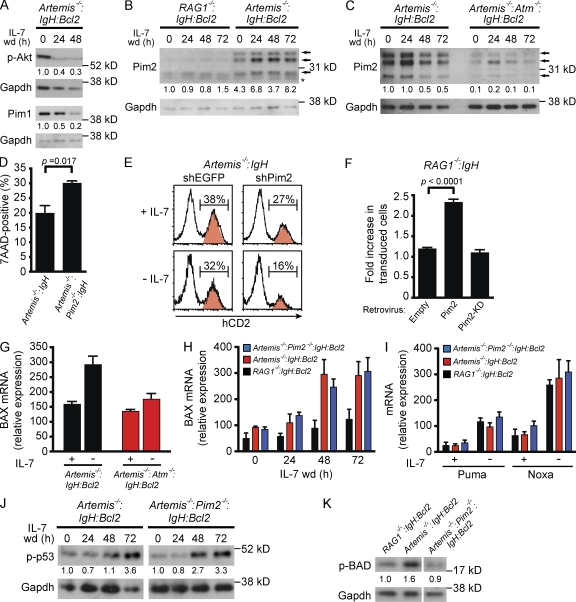
Upon withdrawal of IL-7, pre–B cells lose Pim1 and Akt survival signals leading to cell death (Fig. 1). However, early after the initiation of V(D)J recombination, pre–B cells exhibit enhanced survival suggesting that RAG DNA breaks may generate compensatory survival signals (Fig. 2 D). In contrast to IL-7 signals, RAG DSBs do not activate Akt or induce Pim1 expression (Fig. 3 A). Rather, these DNA breaks induce Pim2 kinase expression as indicated by Western blot analysis of Artemis−/−:IgH:Bcl2 and RAG1−/−:IgH:Bcl2 pre–B cells in the presence or absence of IL-7 (Fig. 3 B). RAG DSBs in pre–B cells with a compound deficiency in Artemis and ATM (Artemis−/−:ATM−/−:IgH:Bcl2) do not induce Pim2 expression (Fig. 3 C). Thus, whereas Pim1 and Akt activities are down-regulated upon IL-7 withdrawal, Pim2 expression is induced after ATM activation by RAG DSBs during IgL chain gene assembly.
Figure 3.
Pim2 induction by RAG DSBs promotes pre–B cell survival. (A) Western blot analysis of p-Akt and Pim1 in Artemis−/−:IgH:Bcl2 pre–B cells after withdrawal of IL-7 for indicated times. Gapdh is shown as a protein loading control. Numbers represent band intensity relative to time 0 h and standardized to loading controls. (B and C) Western blot analysis of Pim2 expression at indicated times after IL-7 withdrawal. Arrows indicate the three Pim2 isoforms and the asterisk indicates a nonspecific band. Gapdh is shown as a protein loading control. Numbers represent band intensity relative to RAG1−/−:IgH:Bcl2 (B) or Artemis−/−:IgH:Bcl2 (C) at time 0 h and standardized to loading controls. (D) Cell death assessed by 7AAD uptake 24 h after withdrawal of IL-7 from Artemis−/−:IgH and Artemis−/−:Pim2−/−:IgH pre–B cells. Shown is the mean and standard deviation for three replicates. (E) Artemis−/−:IgH pre–B cells were transduced with retroviral vector (red peaks) expressing hCD2 and shEGFP (control) or hCD2 and shPim2 then subsequently withdrawn from IL-7 as mixed co-cultures with nontransduced cells (white peaks). Histograms demonstrate percentage of transduced cells (hCD2+, red peak) before (+IL-7) and 72 h after (−IL-7) withdrawal of IL-7. The experiment was conducted in duplicate with P = 0.008 for shPim2 compared with shEGFP at 72 h. (F) RAG1−/−:IgH pre–B cells were transduced with empty retroviral vector (empty), retrovirus-expressing Pim2 (Pim2), or retrovirus-expressing kinase-dead version of Pim2 (Pim2-KD). IL-7 was withdrawn from mixed cultures of transduced and nontransduced cells. Retrovirally transduced cells were followed by Thy1.1 expression. Data represents the fold increase in the percentage of retrovirally transduced cells at 72 h after IL-7 withdrawal relative to the percentage in the presence of IL-7. Shown is the mean and standard deviation for duplicate cultures. (G) BAX mRNA expression assessed by RT-PCR in cells cultured in IL-7 (+) and 48 h after (−) IL-7 withdrawal. Shown is the mean and standard deviation for three replicates. (H) BAX mRNA expression assessed by RT-PCR at indicated times after IL-7 withdrawal. Shown is the mean and standard deviation for three replicates. (I) PUMA and NOXA mRNA expression assessed by RT-PCR in cells cultured in IL-7 (+) and 72 h after (−) IL-7 withdrawal. Shown is the mean and standard deviation for three replicates. (J) Western blot analysis of p-p53 after withdrawal from IL-7 for the indicated times. Gapdh is shown as a protein loading control. Numbers represent band intensity relative to time 0 h for each genotype and standardized to loading controls. (K) Western blot analysis of phosphorylated Bad (p-Bad) 96 h after IL-7 withdrawal. Gapdh is shown as a protein loading control. Numbers represent band intensity relative to RAG1−/−:IgH:Bcl2 and standardized to loading controls. All data are representative of at least three experiments.
To determine whether Pim2 promotes pre–B cell survival, we analyzed the response to RAG DSBs in pre–B cells deficient in both Artemis and Pim2 (Artemis−/−:Pim2−/−:IgH). After withdrawal of IL-7, Artemis−/−:Pim2−/−:IgH pre–B cells exhibit increased cell death compared with Artemis−/−:IgH pre–B cells (Fig. 3 D). Similarly, knockdown of Pim2 in Artemis−/−:IgH pre–B cells leads to decreased viability, as indicated by a more significant loss of Artemis−/−:IgH pre–B cells expressing a Pim2 short hairpin RNA (shRNA) compared with those expressing a control shRNA after IL-7 withdrawal (Fig. 3 E, human CD2-expressing cells). Furthermore, RAG1−/−:IgH pre–B cells transduced with a retrovirus expressing Pim2 exhibit enhanced survival after withdrawal of IL-7 compared with cells expressing the empty vector (Fig. 3 F). Expression of a kinase-deficient mutant of Pim2 (Pim2-KD) in RAG1−/−:IgH pre–B cells had no effect on survival after withdrawal of IL-7 (Fig. 3 F; Fox et al., 2003). Together, these findings demonstrate that RAG DSBs promote the expression of Pim2 and that Pim2 kinase activity augments the survival of pre–B cells undergoing IgL chain gene assembly. Consistent with this notion, Pim2−/− mice have reduced numbers of IgLλ-expressing B cells (Derudder et al., 2009).
Pim2 phosphorylates Bad in response to RAG DSBs
In pre–B cells, p53 is phosphorylated by ATM in response to RAG DSBs (Fig. 2, B and C). Upon IL-7 withdrawal, expression of the proapoptotic BAX gene, a target of p53, is up-regulated by ATM in response to RAG DSBs (Fig. 3, G and H; Vousden and Prives, 2009). The proapoptotic genes NOXA and PUMA, which function by promoting BAX activity, also exhibited increased expression upon IL-7 withdrawal (Fig. 3 I; Youle and Strasser, 2008). However, this increase was similar in Artemis−/−:IgH pre–B cells and RAG1−/−:IgH pre–B cells, demonstrating that it is not caused by RAG DSBs (Fig. 3 I). Notably, the expression of BAX, NOXA, and PUMA is not altered by Pim2 deficiency (Fig. 3, H and I). Moreover, Pim2 does not modulate p53 phosphorylation (Fig. 3 J).
How then does Pim2 inhibit pre–B cell apoptosis? Like PUMA and NOXA, the BAD protein promotes cell death by binding to antiapoptotic proteins, such as Bcl-2 and Bcl-XL, which would otherwise inhibit BAX activity (Youle and Strasser, 2008). Phosphorylation of BAD triggers release of these prosurvival factors (Youle and Strasser, 2008). In pre–B cells, BAD is phosphorylated by Pim1 and Akt in response to IL-7 (unpublished data). In the absence of IL-7, up-regulation of Pim2 by RAG DSBs maintains BAD phosphorylation (Fig. 3 K). Thus, although RAG DSBs induce BAX expression, they also promote Pim2-dependent phosphorylation of BAD, which will antagonize BAX proapoptotic activities.
Pim2 enforces the G1-S checkpoint in pre–B cells
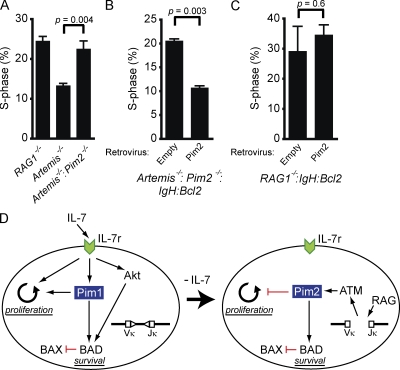
The Pim kinases have established roles in promoting proliferation downstream of cytokine receptors (Amaravadi and Thompson, 2005; Nawijn et al., 2011). Consistent with this function, Pim1 is required to promote proliferation downstream of the IL-7 receptor (unpublished data and Domen et al., 1993). In stark contrast to the pro-proliferative activities of Pim1, we find that Pim2 enforces the G1 to S checkpoint, inhibiting the proliferation of pre–B cells with RAG DSBs. In this regard, Artemis−/−:Pim2−/−:IgH:Bcl2 pre–B cell cultures have a higher percentage of S-phase cells than Artemis−/−:IgH:Bcl2 pre–B cell cultures (Fig. 4 A). Indeed, the fraction of S-phase Artemis−/−:Pim2−/−:IgH:Bcl2 pre–B cells is similar to that of RAG1−/−:IgH:Bcl2 pre–B cells, which do not have RAG DSBs and do not activate DSB-dependent checkpoint pathways (Fig. 4 A). Expression of Pim2 in Artemis−/−:Pim2−/−:IgH:Bcl2 pre–B cells leads to a decrease the percentage of S-phase cells (Fig. 4 B). However, expression of Pim2 in RAG1−/−:IgH:Bcl2 pre–B cells does not alter the percentage of S-phase RAG1−/−:IgH:Bcl2 pre–B cells (Fig. 4 C). Together, these data demonstrate that RAG DSBs in pre–B cells activate the G1-S checkpoint in a manner dependent on Pim2 and that the antiproliferative properties of Pim2 are unique to cells with DNA breaks.
Figure 4.
Pim2 attenuates IL-7-driven proliferation in cells with RAG DSBs. (A) RAG1−/−:IgH:Bcl2, Artemis−/−:IgH:Bcl2, and Artemis−/−:Pim2−/−:IgH:Bcl2 were cultured in IL-7 and the percent of cells in S phase was assessed by BrdU incorporation. Shown is the mean and standard deviation for two replicates. (B–C) Artemis−/−:Pim2−/−:IgH:Bcl2 (B) and RAG1−/−:IgH:Bcl2 (C) pre–B cells were transduced with an empty retroviral vector (empty) or retrovirus expressing Pim2 (Pim2). Cells were cultured in IL-7 and the percent of S-phase cells was assessed by BrdU incorporation 3 d after retroviral transduction. Shown is the mean and standard deviation of two replicates. Data in A, B, and C are representative of at least three experiments. (D) Model illustrating context-dependent roles of Pim1 and Pim2 in developing pre–B cells. Red lines represent inhibition and black arrows designate activation. Pim1 and Akt activity are triggered by IL-7 receptor signals. These two kinases cooperate to support pre–B cell survival through phosphorylation of BAD and subsequent inactivation of BAX. Expression of Pim1 also supports IL-7–driven proliferation. Loss of IL-7 leads to loss of Pim1 and Akt activities, expression of RAG, and initiation of IgLκ chain gene rearrangement. RAG DSBs activate ATM, which induces Pim2 expression. Pim2 phosphorylates BAD and blocks pre–B cell proliferation.
The contrasting effects of Pim1 and Pim2 on cell proliferation are context dependent, permitting pre–B cell expansion in the presence of IL-7 (Pim1) but preventing proliferation of pre–B cells that have initiated V(D)J recombination and have RAG DSBs (Pim2). In the latter context, Pim2 promotes genomic stability by preventing cells with RAG DSBs from entering S phase where these breaks could be aberrantly repaired. Importantly, we show that the induction of Pim2 by RAG DSBs inhibits pre–B cells from entering S phase even in the presence of IL-7. Thus, Pim2 would antagonize proliferative signals from any residual IL-7 present when pre–B cells have transited away from IL-7–producing stromal cells (Tokoyoda et al., 2004; Johnson et al., 2008).
We have shown that distinct signaling pathways activated by the IL-7 receptor and by RAG DSBs are functionally integrated to regulate the proliferation and survival of developing pre–B cells (Fig. 4 D). To assemble IgL chain genes, pre–B cells must attenuate IL-7 signals, including those that promote survival. Importantly, we show that RAG DSB-mediated signals support pre–B cell survival, effectively replacing lost IL-7 prosurvival signals. However, if these DSBs persist unrepaired, they would ultimately promote cell death. This temporal balance of prosurvival and proapoptotic signaling would allow pre–B cells the time needed to undergo multiple IgL chain gene rearrangements while, ultimately, eliminating any cells with persistent unrepaired RAG DSBs. These activities, coupled with the cell cycle checkpoint activity of Pim2, permit efficient IgL chain gene assembly and prevent RAG DSBs from being aberrantly resolved as chromosomal lesions, such as translocations, which could lead to cellular transformation and cancer.
MATERIALS AND METHODS
Mice.
All mice were bred and maintained under specific pathogen-free conditions at the Washington University School of Medicine and were handled in accordance with the guidelines set forth by the Division of Comparative Medicine of Washington University. Wild-type B6 mice (The Jackson Laboratory) were used as controls. All other mice are on a mixed genetic background.
Primary cell culture.
Bone marrow was harvested from 4–6-wk-old mice and cultured for 7–10 d at 2 × 106 cells/ml in media containing IL-7 at 5 ng/ml. For IL-7 withdrawal experiments, cells were resuspended in media without IL-7 and maintained at 2 × 106 cells/ml. The ATM inhibitor KU-55933 (Sigma-Aldrich) was used at 15 µM. Akt inhibitor VIII (EMD) was used at 5 µM.
cDNA expression and shRNA mediated knockdown.
cDNAs encoding Pim2 or Pim2-KD were cloned into the MSCV retrovirus containing an IRES sequence followed by Thy1.1 cDNA as a marker of transduced cells. For Pim2-KD, the lysine at residue 121 was changed to methionine by PCR-based site-directed mutagenesis of Pim2 cDNA and confirmed by sequencing (Fox et al., 2003). shRNAs were cloned into the MSCV-hCD2-mir30 vector as previously described (Amin and Schlissel, 2008). Targeted sequences are as follows: Pim2, 5′-GGGAGGTACTGCTCATTAA-3′; and p53, 5′-CCCACTACAAGTACATGTGTAA-3′. Retrovirus was produced in platE cells by transfection of the retroviral plasmid with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. Viral supernatant was collected and pooled from 24–72 h after transfection. Viral particles were concentrated by addition of PEG-8000 (Sigma-Aldrich; final concentration 8%), incubation at 4°C overnight, and centrifugation at 2,500 RPM for 20 min. Precipitated virus was resuspended at 300× concentration in sterile PBS. Cells (10 × 106/ml) were transduced with viral particles (final 6-10×) in standard media containing 5 µg/ml polybrene and centrifuged for 2 h at 1,100 RPM at room temperature. Cells were incubated overnight in virus-containing media and then placed in fresh media at 2 × 106 cells/ml. Cells expressing the retroviral construct were identified by flow cytometric assessment of Thy1.1 or hCD2 expression using a FACSCalibur (BD). Transduced cells were sorted using biotin-hCD2 or biotin-Thy1.1 (both from BD) and anti-biotin magnetic beads (Miltenyi Biotec) on MS columns (Miltenyi Biotec) according to the manufacturer’s protocol.
Flow cytometric analyses.
All flow cytometric analyses were performed on a FACSCalibur. Antibodies used included FITC-conjugated anti-CD45R/B220 and allophycocyanin (APC)-conjugated anti-IgM, PE-conjugated anti-CD43, APC-conjugated anti-hCD2, PE-conjugated anti-hCD2, and PE-conjugated anti-Thy1.1. All antibodies were obtained from BD. Cell viability was assessed by incubation of cells for 10 min at room temperature with 0.5 µg/ml of 7AAD (BD). Proliferation was assessed using the BrdU-FITC kit (BD) per the manufacturer’s instructions.
Southern blot.
Southern blot analyses of coding ends generated during rearrangement at the IgLκ locus were performed on genomic DNA digested with SacI and EcoRI using the JκIII probe as described previously (Bredemeyer et al., 2008).
Western blot and EMSA analyses.
Western blots were done on whole cell lysates as previously described (Bredemeyer et al., 2008). Anti–phospho(Thr308)-Akt, anti–total Akt, anti–phospho(Ser256)-Foxo1, anti–phospho(Ser15)-p53, and anti–phospho(Ser112)-BAD antibodies were obtained from Cell Signaling Technology. Anti-Pim1, anti-Pim2, and anti-Gapdh antibodies were obtained from Santa Cruz Biotechnology, Inc. Secondary reagents were horseradish peroxidase–conjugated goat anti–mouse IgG (Invitrogen) or donkey anti–rabbit IgG (GE Healthcare). Quantitation was done using ImageJ software (National Institutes of Health) and ratios of bands are all referenced to loading controls.
RT-PCR.
RNA was isolated using RNeasy (QIAGEN) and reversed transcribed using a polyT primer and SuperScriptII (Invitrogen) according to the manufacturers’ protocol. RT-PCR was performed using Brilliant II SYBR Green (Agilent Technologies) and acquired on an Mx3000P (Agilent Technologies). Each reaction was run in triplicate. Primer sequences were: Noxa (forward: 5′-ACCACCTTAAATCCAGCTGTCCCA-3′; reverse: 5′-CCCTTCAGCCCTTGATTGCTTGTT-3′), Puma (forward: 5′-AGACAAGAAGAGCAGCATCGACAC-3′; reverse: 5′-TAGGCACCTAGTTGGGCTCCATTT-3′), and Bax (forward: 5′-TGTTTGCTGATGGCAACTTC-3′; reverse: 5′-GATCAGCTCGGGCACTTTAG-3′).
Statistical analysis.
All p-values were generated via Student’s t test using Prism (GraphPad Software).
Online supplemental material.
Fig. S1 shows shows flow cytometric analysis of bone marrow from mice used to generate IL-7 cultures. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20112078/DC1.
Acknowledgments
We thank Drs. Andre Nussenzweig, Craig Bassing, and Eugene Oltz for critical review of the manuscript.
This work was supported by the National Institutes of Health grants CA136470 (B.P. Sleckman), AI074953 (B.P. Sleckman), AI47829 (B.P. Sleckman), and HL48702 (M.S. Schlissel). J.J. Bednarski was supported by a Children’s Discovery Institute Postdoctoral Fellowship and a National Institutes of Health training grant (5T32HD007499).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BCR
- B cell receptor
- DSB
- double-strand break
- NHEJ
- nonhomologous end joining
- shRNA
- short hairpin RNA
References
- Amaravadi R., Thompson C.B. 2005. The survival kinases Akt and Pim as potential pharmacological targets. J. Clin. Invest. 115:2618–2624 10.1172/JCI26273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin R.H., Schlissel M.S. 2008. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 9:613–622 10.1038/ni.1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H., Shimizu T., Takeda S. 1996. Re-evaluation of the probabilities for productive arrangements on the kappa and lambda loci. Int. Immunol. 8:91–99 10.1093/intimm/8.1.91 [DOI] [PubMed] [Google Scholar]
- Bassing C.H., Swat W., Alt F.W. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell. 109:S45–S55 10.1016/S0092-8674(02)00675-X [DOI] [PubMed] [Google Scholar]
- Billips L.G., Nuñez C.A., Bertrand F.E., III, Stankovic A.K., Gartland G.L., Burrows P.D., Cooper M.D. 1995. Immunoglobulin recombinase gene activity is modulated reciprocally by interleukin 7 and CD19 in B cell progenitors. J. Exp. Med. 182:973–982 10.1084/jem.182.4.973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer A.L., Helmink B.A., Innes C.L., Calderon B., McGinnis L.M., Mahowald G.K., Gapud E.J., Walker L.M., Collins J.B., Weaver B.K., et al. 2008. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 456:819–823 10.1038/nature07392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casellas R., Shih T.A., Kleinewietfeld M., Rakonjac J., Nemazee D., Rajewsky K., Nussenzweig M.C. 2001. Contribution of receptor editing to the antibody repertoire. Science. 291:1541–1544 10.1126/science.1056600 [DOI] [PubMed] [Google Scholar]
- Derudder E., Cadera E.J., Vahl J.C., Wang J., Fox C.J., Zha S., van Loo G., Pasparakis M., Schlissel M.S., Schmidt-Supprian M., Rajewsky K. 2009. Development of immunoglobulin lambda-chain-positive B cells, but not editing of immunoglobulin kappa-chain, depends on NF-kappaB signals. Nat. Immunol. 10:647–654 10.1038/ni.1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiderio S., Lin W.C., Li Z. 1996. The cell cycle and V(D)J recombination. Curr. Top. Microbiol. Immunol. 217:45–59 [DOI] [PubMed] [Google Scholar]
- Domen J., van der Lugt N.M., Acton D., Laird P.W., Linders K., Berns A. 1993. Pim-1 levels determine the size of early B lymphoid compartments in bone marrow. J. Exp. Med. 178:1665–1673 10.1084/jem.178.5.1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox C.J., Hammerman P.S., Cinalli R.M., Master S.R., Chodosh L.A., Thompson C.B. 2003. The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 17:1841–1854 10.1101/gad.1105003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann S.D., Lee A.I., Shockett P.E., Villey I.J., Schatz D.G. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495–527 10.1146/annurev.immunol.18.1.495 [DOI] [PubMed] [Google Scholar]
- Gapud E.J., Sleckman B.P. 2011. Unique and redundant functions of ATM and DNA-PKcs during V(D)J recombination. Cell Cycle. 10:1928–1935 10.4161/cc.10.12.16011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz C.A., Harmon I.R., O’Neil J.J., Burchill M.A., Farrar M.A. 2004. STAT5 activation underlies IL7 receptor-dependent B cell development. J. Immunol. 172:4770–4778 [DOI] [PubMed] [Google Scholar]
- Herzog S., Reth M., Jumaa H. 2009. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat. Rev. Immunol. 9:195–205 10.1038/nri2491 [DOI] [PubMed] [Google Scholar]
- Johnson K., Hashimshony T., Sawai C.M., Pongubala J.M., Skok J.A., Aifantis I., Singh H. 2008. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 28:335–345 10.1016/j.immuni.2007.12.019 [DOI] [PubMed] [Google Scholar]
- Li W.Q., Jiang Q., Khaled A.R., Keller J.R., Durum S.K. 2004. Interleukin-7 inactivates the pro-apoptotic protein Bad promoting T cell survival. J. Biol. Chem. 279:29160–29166 10.1074/jbc.M401656200 [DOI] [PubMed] [Google Scholar]
- Malin S., McManus S., Busslinger M. 2010a. STAT5 in B cell development and leukemia. Curr. Opin. Immunol. 22:168–176 10.1016/j.coi.2010.02.004 [DOI] [PubMed] [Google Scholar]
- Malin S., McManus S., Cobaleda C., Novatchkova M., Delogu A., Bouillet P., Strasser A., Busslinger M. 2010b. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat. Immunol. 11:171–179 10.1038/ni.1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne C.D., Paige C.J. 2006. IL-7: a key regulator of B lymphopoiesis. Semin. Immunol. 18:20–30 10.1016/j.smim.2005.10.003 [DOI] [PubMed] [Google Scholar]
- Milne C.D., Fleming H.E., Paige C.J. 2004. IL-7 does not prevent pro-B/pre-B cell maturation to the immature/sIgM(+) stage. Eur. J. Immunol. 34:2647–2655 10.1002/eji.200425400 [DOI] [PubMed] [Google Scholar]
- Nawijn M.C., Alendar A., Berns A. 2011. For better or for worse: the role of Pim oncogenes in tumorigenesis. Nat. Rev. Cancer. 11:23–34 10.1038/nrc2986 [DOI] [PubMed] [Google Scholar]
- Rooney S., Chaudhuri J., Alt F.W. 2004. The role of the non-homologous end-joining pathway in lymphocyte development. Immunol. Rev. 200:115–131 10.1111/j.0105-2896.2004.00165.x [DOI] [PubMed] [Google Scholar]
- Tokoyoda K., Egawa T., Sugiyama T., Choi B.I., Nagasawa T. 2004. Cellular niches controlling B lymphocyte behavior within bone marrow during development. Immunity. 20:707–718 10.1016/j.immuni.2004.05.001 [DOI] [PubMed] [Google Scholar]
- Vousden K.H., Prives C. 2009. Blinded by the Light: The Growing Complexity of p53. Cell. 137:413–431 10.1016/j.cell.2009.04.037 [DOI] [PubMed] [Google Scholar]
- Youle R.J., Strasser A. 2008. The BCL-2 protein family: opposing activities that mediate cell death. Nat. Rev. Mol. Cell Biol. 9:47–59 10.1038/nrm2308 [DOI] [PubMed] [Google Scholar]