ZBTB1, a BTB-ZF family member, is essential for T cell development and for complete B and NK cell differentiation.
Abstract
In this study, we describe a chemically induced mouse mutation that caused a complete and cell-intrinsic T cell deficiency. Development of other lymphoid lineages was also partially impaired and was severely compromised under competitive conditions. Positional cloning, retroviral transduction, and a somatic reversion event revealed that the causative mutation lay within Zbtb1 (zinc finger and BTB domain containing 1), a gene conserved throughout vertebrate evolution. Our data establish ZBTB1 as a critical determinant of T cell development and lymphopoiesis in general, most likely by acting as a transcriptional regulator.
Lymphocytes arise from multipotent progenitors in the bone marrow (Kondo et al., 1997), deviating from the myeloid lineage and completing their differentiation under the influence of a suite of transcriptional regulators (Busslinger, 2004; Rothenberg and Taghon, 2005), including Ikaros (Georgopoulos et al., 1994), Pax5 (Nutt et al., 1999), and Notch1 (Radtke et al., 1999). The transcriptional regulators that guide this process are of special interest because many are also involved in lymphoid tumorigenesis (Ye et al., 1993; Weng et al., 2004; Mullighan et al., 2007, 2008), whereas others can be manipulated for directed reprogramming of cell fate (Cobaleda et al., 2007).
Among all known transcriptional regulators of lymphopoiesis, the BTB-ZF (Broad complex, Tramtrack, and Bric à brac–zinc finger) family of proteins is well represented. These proteins influence lymphoid development from the common lymphoid progenitor (CLP) stage (Kosan et al., 2010), to the T versus B fate decision (Maeda et al., 2007), to CD4+ versus CD8+ T cell selection (He et al., 2005), and even as late as the differentiation of Tfh (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009) and NKT (Kovalovsky et al., 2008; Savage et al., 2008) effector lineages.
In this study, we have used chemical mutagenesis in mice to reveal the physiological function of the BTB-ZF protein ZBTB1 (zinc finger and BTB domain containing 1). ZBTB1 was essential for T cell development and also for the development of B and NK cells under competitive conditions.
RESULTS AND DISCUSSION
Severe T cell lymphopenia
While surveying the descendants of chemically mutagenized mice for lymphocyte deficiencies (Siggs et al., 2011), we identified a single male mouse devoid of T cells. This phenotype, nicknamed scanT because of the low (or scant) frequency of T cells, was transmitted as a recessive trait (Fig. 1, A and B). scanT mutant mice were born at the expected Mendelian ratio, were typically fertile, and were outwardly normal in behavior.
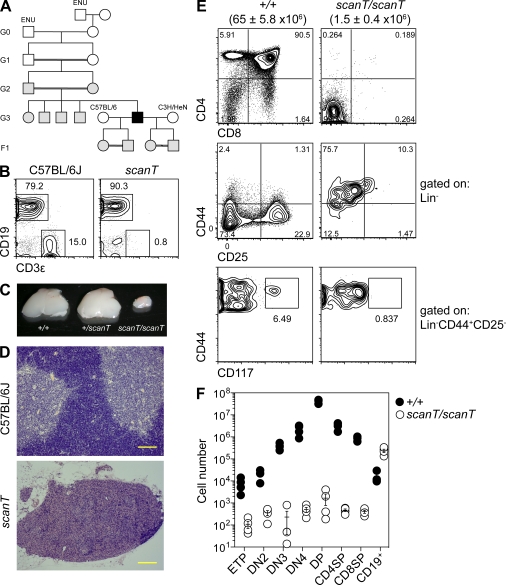
Figure 1.
T cell aplasia in scanT mice. (A) Initial generations of the scanT pedigree. Black symbols, T-deficient phenotype; gray symbols, wild-type phenotype; open symbols, not tested. (B) Percentages of B (CD19+) and T (CD3ε+) cells in the peripheral blood of 8-wk-old mice as measured by flow cytometry. (C–F) Relative size (C), histological appearance (H&E; D), and cellular composition (E and F) of the thymus of wild-type and mutant siblings at 8 wk of age. Lineage markers in E and F were CD11b, CD3ε, B220, Ter119, Ly6G, NK1.1, and CD8α. Subsets in F were gated as follows: ETP (Lin−CD44+CD25−CD117+), DN2 (Lin−CD44+CD25+), DN3 (Lin−CD44−CD25+), DN4 (Lin−CD44−CD25−), DP (CD4+CD8α+), CD4SP (CD4+CD8α−), and CD8SP (CD4−CD8α+). Data are representative of one (A and D), two to three (E and F), or more than three (B and C) independent experiments. Error bars represent standard error, and symbols in F represent individual mice. Numbers in parentheses in E represent mean thymic cellularity and standard error. Bars, 100 µm.
scanT thymi were severely hypoplastic (Fig. 1 C), containing around 2% of the number of wild-type cells, and lacked corticomedullary definition (Fig. 1 D). Thymocyte development was impaired from the early thymic precursor (ETP) stage and beyond (Fig. 1, E and F). Of the lymphocytes that were present in the thymus, almost all were B cells, representing a 20-fold increase over wild-type numbers (Fig. 1 F).
Mutation of a previously uncharacterized zinc finger protein
We mapped the scanT genetic lesion first by genome-wide linkage to chromosome 12 (Fig. 2 A) and then by fine mapping to a 5-Mbp interval between markers D12Mit33 and D12Mit4 (Fig. 2 B). Because none of the 35 annotated protein-encoding genes in the interval (Table S1) had previously been implicated in T cell development, we sequenced the coding exons and flanking splice junctions of three (Zbtb1, Zbtb25, and Ppp2r5e) based on their predominant expression in lymphocytes (http://biogps.gnf.org/). 93.4% of all target nucleotides (6,081/6,511) were covered on both strands of wild-type and scanT samples with a Phred quality score of >30, and a single missense transition was identified in Zbtb1 (C74R; Fig. 2 C). PolyPhen-2 (Adzhubei et al., 2010) assigned a score of 0.954 to this mutation, predicting a deleterious effect with 93% confidence. 43.2% of the total coding critical region (59,447/137,702 nt) was also covered at least three times by SOLiD 3 sequencing, with no additional mutations found.
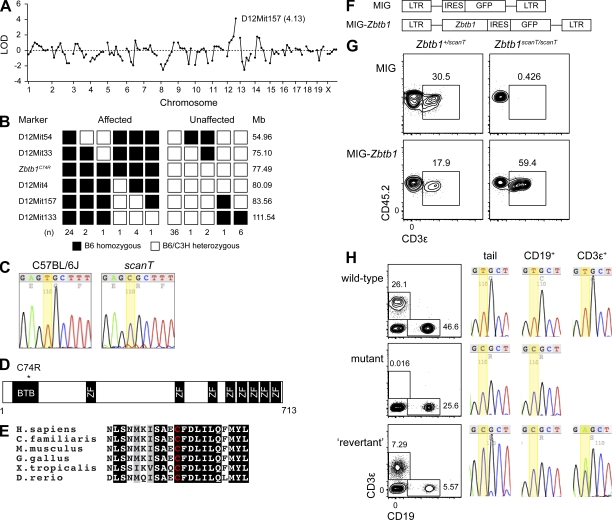
Figure 2.
Identification of a missense mutation in ZBTB1. (A and B) Chromosomal mapping (A) and fine mapping (B) of the scanT phenotype. LOD, logarithm of odds score. (C) DNA sequence chromatograms of the mutated nucleotide in Zbtb1 (thymine to cytosine), resulting in the substitution of arginine for cysteine at codon 74. (D) Predicted ZBTB1 protein domain structure. (E) Conservation of ZBTB1 sequence across multiple vertebrates, with the amino acid corresponding to ZBTB1C74 highlighted in red. ZBTB1 counterparts were not found in nonvertebrate proteomes. (F) Schematic of the MIG-Zbtb1 and control retroviral constructs. MIG, MSCV-IRES-GFP; LTR, long terminal repeat. (G) Bone marrow cells from scanT mice or heterozygous controls (CD45.2+) were transduced with a retroviral construct expressing Zbtb1 and transferred into irradiated CD45.1+ recipients (∼2 × 106 cells/mouse). Transduced CD3ε+ cells were identified in blood 4 mo after transfer by coexpression of GFP. (H) Phenotypic reversion associated with somatic mutation of Zbtb1. DNA was prepared from tail clippings and flow-sorted CD19+ and CD3ε+ lymph node cells and sequenced at the locus mutated in the scanT pedigree. Contour plots were obtained from blood. Data in G are representative of three independent experiments. (C and H) Yellow highlighting indicates the position of the nucleotide mutated in the scanT pedigree..
Zbtb1 encodes a 713-aa member of the POK (POZ [Poxviruses and zinc finger] and Krüppel) or BTB-ZF family of transcriptional regulators and had no previously described function. Mouse ZBTB1 is 94% identical to its counterpart in man. Two characteristic domains are shared by BTB-ZF family members: an N-terminal BTB/POZ domain and a series of C-terminal C2H2 Krüppel-type zinc finger motifs. ZBTB1 itself contains an N-terminal BTB domain, eight zinc finger motifs, and two nuclear localization sequences (Matic et al., 2010). The residue mutated in the scanT pedigree (C74) is predicted to lie within the A3 helix of the BTB domain (Fig. 2 D and Fig. S1; Stogios et al., 2005) and is highly conserved across the vertebrate lineage (Fig. 2 E). Two unique transcripts are predicted to arise from the Zbtb1/ZBTB1 locus, both of which harbor the scanT mutation, yet only one of which (Ensembl release 65 accession no. ENST00000394712) is predominantly expressed in T cells and other leukocytes in mouse and man (Fig. S2). The other (Ensembl release 65 accession no. ENST00000358738) is known to be transcribed in HeLa cells by the nonconventional single-polypeptide nuclear RNA polymerase IV (Kravchenko et al., 2005).
To confirm that the mutation in Zbtb1 was responsible for the scanT phenotype, wild-type Zbtb1 was retrovirally transduced into scanT hematopoietic progenitors (Fig. 2 F). GFP+ T cells were recovered from recipients of Zbtb1-transduced scanT progenitors (Fig. 2 G), presumably as a consequence of restored thymic development. These data indicated that the mutation in Zbtb1 could account for T cell deficiency in scanT mice.
In a minority of cases (3/16 in a 3-mo-old cohort of Zbtb1scanT homozygotes), T cells were detected at diminished frequencies in the blood of germline mutant mice. We hypothesized that these cells arose from a somatic mutation that suppressed the scanT phenotype, considering the strong selective advantage that such a mutation would confer and given that a similar phenomenon is known to occur in human T cell–deficient SCID (Hirschhorn et al., 1996). To test for somatic suppressor mutations, DNA was isolated from sorted T and B cells. Sequencing of the mutated Zbtb1 locus revealed a heterozygous mutation within the T cell compartment of one phenotypic revertant (but not the other two; Fig. 2 H), indicating that most, if not all, T cells in this mouse were clonally derived. This mutation either occurred after the CLP stage or did not confer a selective advantage in the B cell lineage because it was not observed in DNA from CD19+ cells (Fig. 2 H). The mutation in question occurred at the same nucleotide as the scanT mutation (the first base of codon 74), yet rather than a reversion to the wild-type codon (TGC, encoding cysteine) changed the codon to serine (AGC). Given the physical similarities of serine and cysteine, we propose that Zbtb1C74S is permissive to T cell development and T cell clonal expansion, whereas Zbtb1C74R is not.
Defects in lymphoid but not myeloid development
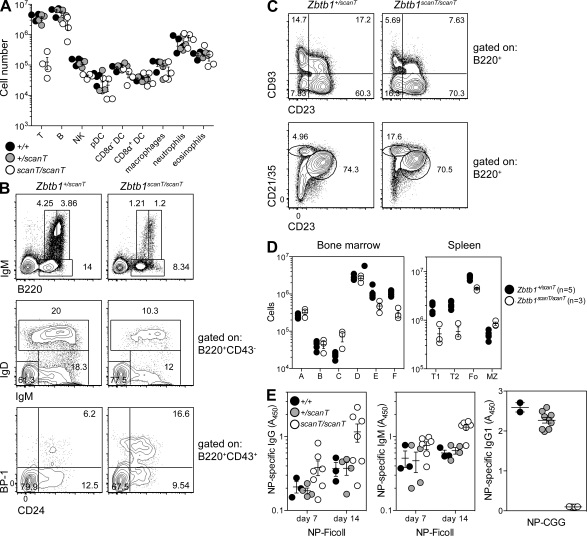
The effects of Zbtb1 mutation were not restricted to the T cell lineage. Numbers of all lymphocyte lineages (T, B, and NK) were reduced in the spleens of mutant mice, whereas numbers of myeloid cells were not (Fig. 3 A). An examination of B cell development in mutant bone marrow revealed an accumulation of B cell progenitors at the Hardy Fraction C stage, followed by a reduction of progenitors at each subsequent stage (Fig. 3, B and D). In the mutant spleen, numbers of transitional and follicular B cells were significantly reduced by an average of 72% and 44%, respectively (Fig. 3, C and D). The relatively minor reduction in follicular as compared with transitional B cell numbers may indicate mild compensatory expansion in the follicular compartment. Mutant B cells nevertheless responded to T-independent (but not T dependent) immunization (Fig. 3 E), implying a developmental rather than a functional B cell defect.
Figure 3.
scanT mice have generalized deficiencies of lymphoid but not myeloid cells. (A) Total numbers of major lymphoid and myeloid cell subsets in the spleen of 8-wk-old sex-matched littermates, gated as follows: T (CD3ε+), B (CD19+), NK (NK1.1+CD3ε−), plasmacytoid DCs (pDC; Lin−PDCA1+SiglecH+), CD8α− DCs (Lin−CD11c+CD11b+CD8α−), CD8α+ DCs (Lin−CD11c+CD11b−CD8α+), macrophages (Lin−CD11b+F4/80+SSClo), neutrophils (Lin−CD11b+F4/80−Ly6Ghi), and eosinophils (Lin−CD11b+F4/80+SSChi). Lineage markers included CD19, TCR-β, NK1.1, and the viability dye 7-AAD. (B–D) Frequencies (B and C) and numbers (D) of major B cell subsets in bone marrow and spleen. Subsets in bone marrow were gated as follows: A (CD11b−CD3e−Ter119−Ly6G−NK1.1−IgM−B220+CD19−CD24−BP-1−), B (B220+IgM−CD43+CD24+BP-1−), C (B220+IgM−CD43+CD24+BP-1+), D (B220+CD43−IgM−IgD−), E (B220+CD43−IgM+IgD−), and F (B220+CD43−IgM+IgD+). Subsets in spleen were gated as follows: T1 (B220+CD93+CD23−), T2 (B220+CD93+CD23+), Fo (follicular; B220+CD93−CD23+CD21/35int), and MZ (marginal zone; B220+CD93−CD23−CD21/35hi). (E) NP (4-hydroxy-3-nitrophenylacetyl)-specific antibodies in the serum of mice immunized with NP-Ficoll or NP-CGG at 7 or 14 d (NP-Ficoll) or 14 d (NP-CGG) after immunization, presented as absorbance at 450 nm (A450). (A–E) Data are representative of one (E) or two (A–D) independent experiments with four or more mice per group. Error bars represent standard error, and symbols represent individual mice.
A cell-intrinsic defect
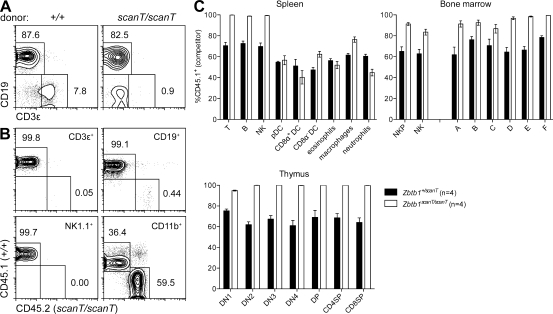
To distinguish between potential hematopoietic and nonhematopoietic origins of the Zbtb1 mutant phenotype, we created radiation chimeras. Rag1 mutants reconstituted with Zbtb1 mutant bone marrow developed B cells but not T cells (Fig. 4 A). This defect was intrinsic to Zbtb1 mutant T cell progenitors, rather than other radiosensitive hematopoietic cells, because T cells in mixed bone marrow chimeras were exclusively derived from wild-type donors (Fig. 4 B). In fact, all blood lymphocytes (but not CD11b+ myeloid cells) were wild-type donor derived, implying a general competitive failure of Zbtb1 mutant lymphoid progenitors (Fig. 4 B). Finer phenotyping of the spleen, bone marrow, and thymus of mixed bone marrow chimeric mice confirmed this to be the case. B, T, and NK cells in the spleen were exclusively wild-type derived (CD45.1+), whereas little or no competitive disadvantage was seen in the myeloid compartments (Fig. 4 C). Cells of wild-type origin dominated the B cell compartment from the Fraction A stage onward and similarly outcompeted Zbtb1 mutant NK progenitors in bone marrow and T cell progenitors in the thymus (Fig. 4 C).
Figure 4.
A cell-intrinsic T cell deficiency and competitive failure of lymphoid reconstitution. (A and B) Lethally irradiated Rag1 mutant mice (CD45.2+) were reconstituted with unmixed scanT or wild-type bone marrow (CD45.2+; A) or a mixture of scanT or wild-type marrow (CD45.2+) with wild-type bone marrow (CD45.1+; B). Chimerism was measured 8 wk after transplant. Panels in B have been gated on the indicated cell subset. (C) Lethally irradiated wild-type recipients (CD45.1+) were transplanted with an equal mixture of wild-type (CD45.1+) and heterozygous or homozygous mutant (CD45.2+) bone marrow. Wild-type donor chimerism (CD45.1+) was measured 8 wk later in the spleen, bone marrow, and thymus. Subsets were gated as in Fig. 3, with the addition of NK progenitors (NKP; 7-AAD−CD19−TCR-β−CD122+NK1.1−). (A–C) Data are representative of one (A) or three (B and C) independent experiments with at least three mice per group. Error bars represent standard error.
Uncompromised hematopoietic progenitor function
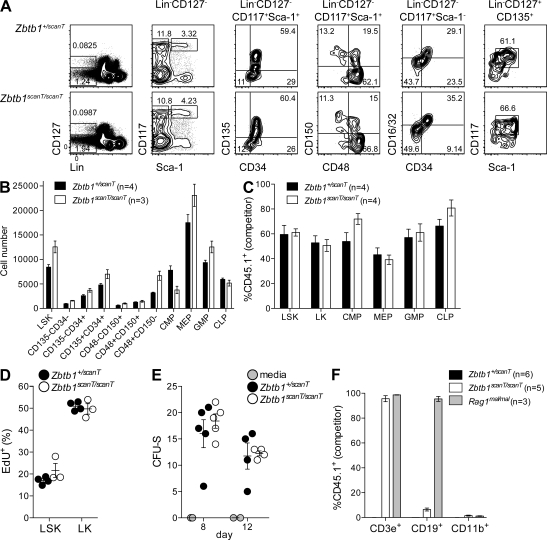
However, the scanT mutation did not appear to affect hematopoietic development before lymphoid specification. Percentages and numbers of early hematopoietic progenitors, including CLPs, were comparable in heterozygous and homozygous mutant bone marrow (Fig. 5, A and B) and were reconstituted at equivalent percentages in mixed bone marrow chimeras (Fig. 5 C). Hematopoietic stem cell (HSC) turnover, as measured by incorporation of the thymidine analogue 5-ethynyl-2′deoxyuridine (EdU), occurred at similar rates in wild-type and mutant mice (Fig. 5 D). Wild-type and Zbtb1 mutant bone marrow also generated equivalent numbers of hematopoietic colonies in the spleens of lethally irradiated recipients, both at days 8 (indicative of megakaryocyte/erythroid progenitor function) and 12 (reflective of multipotent progenitor function; Fig. 5 E; Na Nakorn et al., 2002). Myeloid engraftment did not occur in unconditioned Zbtb1 mutant recipients of wild-type bone marrow, which is consistent with the interpretation of intact HSC function (Fig. 5 F).
Figure 5.
Normal development and function of hematopoietic progenitors. (A and B) Frequencies (A) and numbers (B) or hematopoietic progenitors in heterozygous and homozygous Zbtb1 mutant bone marrow. LT-HSC (long-term HSC; CD135−CD34− or CD150+CD48−), ST-HSC (short-term HSC; CD135−CD34+ or CD150+CD48+), MPP (multipotent progenitors; CD135+CD34+ or CD150−CD48+), CMP (common myeloid progenitors; CD16/32−CD34+), GMP (granulocyte/monocyte progenitors; CD16/32+CD34+), MEP (megakaryocyte/erythroid progenitors; CD16/32−CD34−), and CLP (CD117intSca-1int) were gated as indicated. (C) Chimerism of a subset of hematopoietic progenitors outlined in A, 8 wk after reconstitution of lethally irradiated CD45.1+ recipients with an equal mixture of CD45.1+ and Zbtb1+/scanT (CD45.2+) or CD45.1+ and Zbtb1scanT/scanT (CD45.2+) bone marrow cells. (D) Incorporation of the thymidine analogue EdU 4 h after injection. (E) Spleen colony forming units (CFU-S) counted 8 or 12 d after transfer of 105 bone marrow cells (or media alone) into lethally irradiated recipients. (F) Engraftment of 2 × 106 CD45.1+ bone marrow cells in unconditioned recipients 8 wk after injection. Lineage markers in A–D were CD11b, CD3ε, B220, Ter119, Ly6G, NK1.1, and CD8α. (A–F) Data are representative of one (D and F), two (A–C), and three (E) independent experiments with at least three mice per group. Error bars represent standard error, and symbols in D and E represent individual mice.
In certain aspects, Zbtb1 mutants reflect the phenotype of mice lacking Notch1. Both Zbtb1 and Notch1 mutants are profoundly T cell deficient and show an accumulation of B cells in the thymic rudiment (Radtke et al., 1999). Yet although Zbtb1 mutants have a competitive disadvantage in the B and NK lineages, Notch1 mutants do not (Radtke et al., 1999). Germline mutation of Zbtb1 appears permissive to embryonic development, whereas mutation of Notch1 is not (Swiatek et al., 1994), indicating a more specialized role for ZBTB1 in determining lymphoid fate. This is consistent with the conservation of Zbtb1 only among vertebrate genomes and might implicate the human ZBTB1 locus in genetically obscure cases of T cell–deficient SCID (Fischer, 2007).
Although there are no previous studies of a physiological role for ZBTB1, one describes some of its biochemical characteristics (Matic et al., 2010). ZBTB1, like many of its BTB-ZF counterparts, acts as a potent transcriptional repressor, as determined by the repressive activity of a Gal4-ZBTB1 fusion protein. This repressive activity, as well as the colocalization of ZBTB1 with the SMRT transcriptional repressor, is also regulated by SUMOylation (Matic et al., 2010).
Our data raise several questions about the function of ZBTB1 in T cell and lymphoid specification. In particular, why is ZBTB1 critical for T cell development but only essential for B and NK development under competition? Can the reduction in ETP numbers be explained by their failure to migrate to the thymus, by a failure to proliferate within it, or a combination of both? Programmed death of T cell progenitors is unlikely to account for this because T cells also fail to develop in the absence of the proapoptotic protein BIM (which can rescue erythropoiesis in mice deficient for the BTB-ZF relative LRF; Fig. S3; Maeda et al., 2009).
In summary, our work defines ZBTB1 as a regulator of lymphoid development, joining the ranks of several other lymphoid-promoting BTB-ZF proteins. As is the case with other BTB-ZF family members, ZBTB1 presumably acts as a transcriptional suppressor, and the determination of precisely which genes it regulates will be of central importance to our understanding of lymphopoiesis.
MATERIALS AND METHODS
Mice and positional cloning.
Zbtb1scanT was generated on a pure C57BL/6J (The Jackson Laboratory) background by N-ethyl-N-nitrosourea (ENU) mutagenesis as previously described (Georgel et al., 2008). The index scanT mutant (C57BL/6J) was outcrossed to C3H/HeN females (Taconic), and F1 progeny were intercrossed. Blood was collected from the retroorbital plexus of 26 F2 progeny, and mice were grouped into mutant and wild-type cohorts (7 and 19 mice, respectively) based on relative percentages of CD19+ and CD3ε+ cells as measured by flow cytometry. Individual mice were typed at 128 microsatellite markers spaced across the genome (complete list available in the Supplemental material), with expected and observed genotype frequencies used to calculate LOD (logarithm of odds) scores at each marker. For fine mapping, an extra 71 F3 backcross progeny were genotyped at additional loci within the critical region. Candidate gene amplicons from wild-type and scanT genomic DNA were sequenced using a capillary sequencer (3730xl; Applied Biosystems). Genomic DNA from a single scanT homozygote was used to create a fragment library for single slide whole-genome sequencing on the SOLiD 3 platform (Applied Biosystems). Rag1maladaptive mice (C57BL/6J, MGI:3851764) were generated in-house by ENU mutagenesis, and B6.SJL-Ptprca Pep3b/BoyJ (CD45.1, backcrossed >22 generations to C57BL/6), Bcl2l11tm1.1Ast (backcrossed 13 generations to C57BL/6; Bouillet et al., 1999), and C57BL/6J males used for mutagenesis were obtained from the Jackson Laboratory. All other C57BL/6J mice were obtained from The Scripps Research Institute breeding colony. All animal procedures were performed in accordance with guidelines of the Institutional Animal Care and Use Committee of The Scripps Research Institute, and all experiments were conducted with homozygous mutants and sex-matched littermate controls.
Flow cytometry.
Blood from the retroorbital plexus of isoflurane-anesthetized mice was collected in cluster tubes (Costar) containing 20 µl of 6% (wt/vol) EDTA in water. 50 µl of blood was subjected to two rounds of red blood cell lysis with ammonium chloride before staining. Lymphocyte suspensions from bone marrow (femurs and tibias from one hind leg), spleen, and thymus were counted (Z2 Coulter Counter; Beckman Coulter) and were stained with a combination of the following mouse-specific antibodies: FITC-conjugated IgM (goat polyclonal; 1020-02; SouthernBiotech); FITC-conjugated CD24 (30-F1), CD48 (HM48-1), F4/80 (BM8), CD127 (A7R34); PE-conjugated CD122 (5H4), CD135 (A2F10), CD150 (9D1), IgD (11-26), PDCA1 (927), TCR-β (H57-597; PerCP-Cy5.5); CD117 (2B8; eFluor710), APC-conjugated CD8α (53-6.7), CD11b (M1/70), CD93 (AA4.1), IgM (II/41), Ly6G (RB6-8C5), NK1.1 (PK136), TER119 (TER-119; eBioscience); FITC-conjugated CD4 (GK1.5), CD23 (B3B4), CD25 (7D4), CD34 (RAM34), PE-conjugated BP-1 (BP-1), CD11b (M1/70), CD19 (1D3), CD21/35 (7G6), CD44 (IM7), PerCP-Cy5.5–conjugated B220 (RA3-6B2), CD8α (53–6.7), CD19 (1D3), NK1.1 (PK136), APC-conjugated CD11c (HL3) and CD43 (S7), CD44 (IM7; Horizon V500; BD); APC-conjugated SiglecH (551), CD3ε (145-2C11), APC-Cy7–conjugated Sca-1 (D7) and CD45.2 (104), PE-Cy7–conjugated CD16/32 (93) and CD19 (6D5), CD45.1 (A20; Pacific blue), and CD16/32 (93; purified; BioLegend). Samples were acquired on a FACSCalibur or LSRFortessa (BD), and data were analyzed with FlowJo software (Tree Star). 7-AAD was purchased from eBioscience, and EdU labeling and staining were performed according to the manufacturer’s instructions (Invitrogen) 4 h after a single intraperitoneal injection of 100 mg/kg EdU. For sorting experiments, lymph node or suspensions were labeled with antibodies, and target populations were sorted by a FACSAria II (BD) into serum for genomic DNA extraction.
Immunizations.
Immunizations were performed as described previously (Siggs et al., 2011).
Hematopoietic chimeras.
Recipient mice were γ-irradiated with a split dose of 11 Gy (2 × 5.5 Gy, 137Cs source). The next day, mice were injected with 2 × 106 bone marrow cells via the retroorbital plexus and were maintained on trimethoprim/sulfamethoxazole antibiotic water until sacrificed for analysis 8 wk later. For colony forming unit assays, mice received 105 bone marrow cells and were sacrificed 8 or 12 d later. Spleens were fixed overnight in Bouin’s solution (Sigma-Aldrich), and colonies were counted the next day. For unconditioned chimeras, unconditioned recipients received a single retroorbital injection of 2 × 106 bone marrow cells.
Retroviral transduction.
The open reading frame of Zbtb1 was cloned into the BglII–EcoRI site of pMIG (MSCV2.2-IRES-GFP; Addgene #9044, deposited by W. Hahn [Dana-Farber Cancer Institute, Boston, MA]) to generate the pMIG-Zbtb1 vector. Vectors were transfected into the Plat-E packaging cell line using Fugene 6 (Roche), and retrovirus-containing supernatant was collected 24 and 48 h later. Bone marrow donor mice were primed 5 d before harvest with 150 mg/kg 5-fluorouracil (Sigma-Aldrich), and bone marrow cells were prestimulated overnight with 10 ng/ml of mouse recombinant IL-3, 10 ng/ml IL-6, 50 ng/ml SCF, 50 ng/ml TPO, and 5 ng/ml FLT3L (all from PeproTech). The next day, 3 ml/well of retroviral supernatant was centrifuged (2,000 rpm for 2 h at 32°C) onto RetroNectin-coated 6-well plates (20 µg/ml; Takara Bio Inc.), and cells were cultured on coated plates in the presence of 4 µg/ml Polybrene (Sigma-Aldrich) for 48 h. Recipient CD45.1+ mice were γ-irradiated with a split dose of 11 Gy (137Cs source) and the next day injected with ∼2 × 106 transduced bone marrow cells via the retroorbital plexus.
Histology.
Thymi were fixed in 10% neutral buffered formalin and embedded in paraffin wax, and 5-µm sections were stained with hematoxylin and eosin (H&E).
PolyPhen-2.
The deleterious effect of the scanT mutation was predicted using the HumVar-trained PolyPhen-2 server (version 2.1.0; Adzhubei et al., 2010).
Online supplemental material.
Fig. S1 compares the sequence of ZBTB1 with other BTB-ZF proteins. Fig. S2 shows the tissue-specific expression of Zbtb1 transcripts. Fig. S3 shows flow cytometric analysis of lymphocyte populations in Zbtb1;Bcl2l11 double mutant mice. Table S1 is a list of protein-encoding genes in the scanT critical region. The whole genome mapping panel for C57BL/6J versus C3H/HeN is also included in the supplemental material. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20112084/DC1.
Acknowledgments
We thank M. Gutierrez for animal care; C. Domingo, L. Hanley, and E. Pirie for ENU pedigree generation; S. Kalina and T. Robinson for genotyping; C. Ross and P. Lin for sequencing; C. Arnold for assistance with immunizations; and D. Yu (Monash University, Clayton, Victoria, Australia) for advice on retroviral transduction.
This work was supported by the Bill & Melinda Gates Foundation, the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (grant HHSN272200700038C to B. Beutler), and The General Sir John Monash Foundation (O.M. Siggs).
The authors have no conflicting financial interests.
Author contributions: O.M. Siggs designed research, performed all experiments, analyzed data, and wrote the manuscript. X. Li propagated the scanT strain by intracytoplasmic sperm injection, Y. Xia provided bioinformatics support, and B. Beutler managed the mutagenesis operation and corrected the manuscript.
Footnotes
Abbreviations used:
- BTB-ZF
- Broad complex, Tramtrack, and Bric à brac–zinc finger
- CLP
- common lymphoid progenitor
- EdU
- 5-ethynyl-2′deoxyuridine
- ENU
- N-ethyl-N-nitrosourea
- ETP
- early thymic precursor
- HSC
- hematopoietic stem cell
References
- Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. 2010. A method and server for predicting damaging missense mutations. Nat. Methods. 7:248–249 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouillet P., Metcalf D., Huang D.C.S., Tarlinton D.M., Kay T.W., Köntgen F., Adams J.M., Strasser A. 1999. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science. 286:1735–1738 10.1126/science.286.5445.1735 [DOI] [PubMed] [Google Scholar]
- Busslinger M. 2004. Transcriptional control of early B cell development. Annu. Rev. Immunol. 22:55–79 10.1146/annurev.immunol.22.012703.104807 [DOI] [PubMed] [Google Scholar]
- Cobaleda C., Jochum W., Busslinger M. 2007. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 449:473–477 10.1038/nature06159 [DOI] [PubMed] [Google Scholar]
- Fischer A. 2007. Human primary immunodeficiency diseases. Immunity. 27:835–845 10.1016/j.immuni.2007.11.012 [DOI] [PubMed] [Google Scholar]
- Georgel P., Du X., Hoebe K., Beutler B. 2008. ENU mutagenesis in mice. Methods Mol. Biol. 415:1–16 10.1007/978-1-59745-570-1_1 [DOI] [PubMed] [Google Scholar]
- Georgopoulos K., Bigby M., Wang J.H., Molnar A., Wu P., Winandy S., Sharpe A. 1994. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 79:143–156 10.1016/0092-8674(94)90407-3 [DOI] [PubMed] [Google Scholar]
- He X., He X., Dave V.P., Zhang Y., Hua X., Nicolas E., Xu W., Roe B.A., Kappes D.J. 2005. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 433:826–833 10.1038/nature03338 [DOI] [PubMed] [Google Scholar]
- Hirschhorn R., Yang D.R., Puck J.M., Huie M.L., Jiang C.K., Kurlandsky L.E. 1996. Spontaneous in vivo reversion to normal of an inherited mutation in a patient with adenosine deaminase deficiency. Nat. Genet. 13:290–295 10.1038/ng0796-290 [DOI] [PubMed] [Google Scholar]
- Johnston R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., Crotty S. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M., Weissman I.L., Akashi K. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672 10.1016/S0092-8674(00)80453-5 [DOI] [PubMed] [Google Scholar]
- Kosan C., Saba I., Godmann M., Herold S., Herkert B., Eilers M., Möröy T. 2010. Transcription factor miz-1 is required to regulate interleukin-7 receptor signaling at early commitment stages of B cell differentiation. Immunity. 33:917–928 10.1016/j.immuni.2010.11.028 [DOI] [PubMed] [Google Scholar]
- Kovalovsky D., Uche O.U., Eladad S., Hobbs R.M., Yi W., Alonzo E., Chua K., Eidson M., Kim H.-J., Im J.S., et al. 2008. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9:1055–1064 10.1038/ni.1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravchenko J.E., Rogozin I.B., Koonin E.V., Chumakov P.M. 2005. Transcription of mammalian messenger RNAs by a nuclear RNA polymerase of mitochondrial origin. Nature. 436:735–739 10.1038/nature03848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Merghoub T., Hobbs R.M., Dong L., Maeda M., Zakrzewski J., van den Brink M.R.M., Zelent A., Shigematsu H., Akashi K., et al. 2007. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 316:860–866 10.1126/science.1140881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Ito K., Merghoub T., Poliseno L., Hobbs R.M., Wang G., Dong L., Maeda M., Dore L.C., Zelent A., et al. 2009. LRF is an essential downstream target of GATA1 in erythroid development and regulates BIM-dependent apoptosis. Dev. Cell. 17:527–540 10.1016/j.devcel.2009.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matic I., Schimmel J., Hendriks I.A., van Santen M.A., van de Rijke F., van Dam H., Gnad F., Mann M., Vertegaal A.C.O. 2010. Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol. Cell. 39:641–652 10.1016/j.molcel.2010.07.026 [DOI] [PubMed] [Google Scholar]
- Mullighan C.G., Goorha S., Radtke I., Miller C.B., Coustan-Smith E., Dalton J.D., Girtman K., Mathew S., Ma J., Pounds S.B., et al. 2007. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 446:758–764 10.1038/nature05690 [DOI] [PubMed] [Google Scholar]
- Mullighan C.G., Miller C.B., Radtke I., Phillips L.A., Dalton J., Ma J., White D., Hughes T.P., Le Beau M.M., Pui C.-H., et al. 2008. BCR-ABL1 lymphoblastic leukaemia is characterized by the deletion of Ikaros. Nature. 453:110–114 10.1038/nature06866 [DOI] [PubMed] [Google Scholar]
- Na Nakorn T., Traver D., Weissman I.L., Akashi K. 2002. Myeloerythroid-restricted progenitors are sufficient to confer radioprotection and provide the majority of day 8 CFU-S. J. Clin. Invest. 109:1579–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.-H., Dong C. 2009. Bcl6 mediates the development of T follicular helper cells. Science. 325:1001–1005 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt S.L., Heavey B., Rolink A.G., Busslinger M. 1999. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 401:556–562 10.1038/44076 [DOI] [PubMed] [Google Scholar]
- Radtke F., Wilson A., Stark G., Bauer M., van Meerwijk J., MacDonald H.R., Aguet M. 1999. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 10:547–558 10.1016/S1074-7613(00)80054-0 [DOI] [PubMed] [Google Scholar]
- Rothenberg E.V., Taghon T. 2005. Molecular genetics of T cell development. Annu. Rev. Immunol. 23:601–649 10.1146/annurev.immunol.23.021704.115737 [DOI] [PubMed] [Google Scholar]
- Savage A.K., Constantinides M.G., Han J., Picard D., Martin E., Li B., Lantz O., Bendelac A. 2008. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 29:391–403 10.1016/j.immuni.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggs O.M., Arnold C.N., Huber C., Pirie E., Xia Y., Lin P., Nemazee D., Beutler B. 2011. The P4-type ATPase ATP11C is essential for B lymphopoiesis in adult bone marrow. Nat. Immunol. 12:434–440 10.1038/ni.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stogios P.J., Downs G.S., Jauhal J.J.S., Nandra S.K., Privé G.G. 2005. Sequence and structural analysis of BTB domain proteins. Genome Biol. 6:R82 10.1186/gb-2005-6-10-r82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiatek P.J., Lindsell C.E., del Amo F.F., Weinmaster G., Gridley T. 1994. Notch1 is essential for postimplantation development in mice. Genes Dev. 8:707–719 10.1101/gad.8.6.707 [DOI] [PubMed] [Google Scholar]
- Weng A.P., Ferrando A.A., Lee W., Morris J.P., IV, Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306:269–271 10.1126/science.1102160 [DOI] [PubMed] [Google Scholar]
- Ye B.H., Lista F., Lo Coco F., Knowles D.M., Offit K., Chaganti R.S., Dalla-Favera R. 1993. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 262:747–750 10.1126/science.8235596 [DOI] [PubMed] [Google Scholar]
- Yu D., Rao S., Tsai L.M., Lee S.K., He Y., Sutcliffe E.L., Srivastava M., Linterman M., Zheng L., Simpson N., et al. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 31:457–468 10.1016/j.immuni.2009.07.002 [DOI] [PubMed] [Google Scholar]