When fused to a rabies virus glycoprotein peptide that shuttles small RNAs across the blood–brain barrier, an HCMV noncoding RNA that protects mitochondrial Complex I activity, delivered intravenously, shows therapeutic efficacy in a rat model of Parkinson’s disease.
Abstract
Parkinson’s disease (PD) is a neurodegenerative disorder that results in the loss of nigrostriatal dopamine neurons. The etiology of this cell loss is unknown, but it involves abnormalities in mitochondrial function. In this study, we have demonstrated that the administration of a novel noncoding p137 RNA, derived from the human cytomegaloviral β2.7 transcript, can prevent and rescue dopaminergic cell death in vitro and in animal models of PD by protecting mitochondrial Complex I activity. Furthermore, as this p137 RNA is fused to a rabies virus glycoprotein peptide that facilitates delivery of RNA across the blood–brain barrier, such protection can be achieved through a peripheral intravenous administration of this agent after the initiation of a dopaminergic lesion. This approach has major implications for the potential treatment of PD, especially given that this novel agent could have the same protective effect on all diseased neurons affected as part of this disease process, not just the dopaminergic nigrostriatal pathway.
Parkinson’s disease (PD) is a common progressive neurodegenerative disorder of the central nervous system (CNS), which has as part of its core pathology the loss of the nigrostriatal dopaminergic neurons. The causes and mechanisms of such selective neuronal loss are not well defined, but recent studies have highlighted an important role for mitochondrial dysfunction, especially Complex I (Schapira et al., 1990; Schapira, 2006; Dawson et al., 2010). In fact, some of the most widely used experimental models of PD, such as 6-hydroxydopamine (6-OHDA) and rotenone, mediate at least part of their toxicity through this pathway (Dabbeni-Sala et al., 2001; Sherer et al., 2003). On this basis, the possibility that protection of mitochondrial function could limit neuronal loss and act therapeutically has been suggested as a possible treatment for PD. We have previously identified a novel RNA expressed during human CMV (HCMV) infection that functions to prevent cell death (Reeves et al., 2007). This viral noncoding RNA, termed the β2.7 transcript, is essential to maintain high levels of energy production in infected cells (Reeves et al., 2007). The mechanism by which β2.7 protects infected cells is novel and appears to be mediated by a direct interaction between β2.7 and Complex I (Reeves et al., 2007). We therefore sought to investigate, using both in vitro and in vivo models to imitate dopaminergic cell loss in PD, whether a truncated form of β2.7 (p137) containing the putative TRL4 subdomain (Bergamini et al., 1998) still prevents dopaminergic neuronal death.
Over the past decade, much research has been done on the delivery of therapeutic gene products to restore the impaired dopaminergic system in experimental models of PD. Many of these attempts have been based on viral vectors involving either knockin (overexpression) gene therapy (Luo et al., 2002) or knockout (interference) gene silencing (Outeiro et al., 2007). However, several problems are associated with such delivery systems, such as the invasive nature of the intracerebral procedure to administer therapeutic agents and the nonspecific expression of these agents outside neural cells. Recently, Kumar et al. (2007) described a method to deliver short interfering RNA (siRNA) to the brain using a small peptide derived from the rabies virus glycoprotein (RVG). This peptide binds to the acetylcholine receptor (AChR) exclusively expressed in CNS cells (Hanham et al., 1993). Although the RVG peptide itself has no RNA binding affinity, a derivative containing nonamer arginine residues (RVG9R) binds RNA efficiently and delivers the RNA cargo across the blood–brain barrier after peripheral administration (Kumar et al., 2007). We therefore also sought to test whether the transvascular administration of the p137 RNA could be successfully delivered in this way to prevent dopaminergic cell loss in models of PD.
RESULTS AND DISCUSSION
The RVG9R–p137 system protects dopaminergic cells in both in vitro and in vivo models of PD
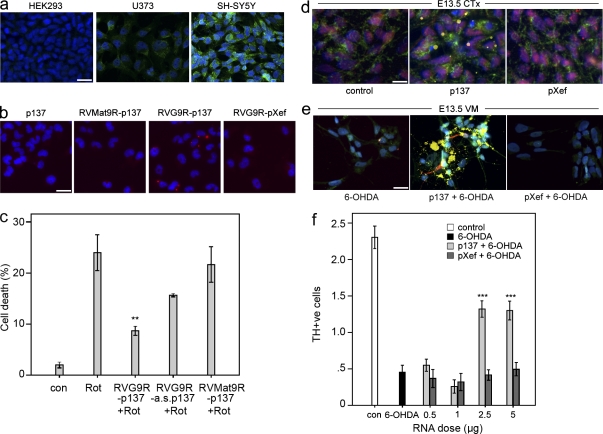
Our initial experiments clearly showed that the p137 RNA complexed with RVG9R peptide could protect SH-SY5Y cells from exposure to rotenone, a highly selective inhibitor to mitochondrial Complex I (Fig. 1 c; Betarbet et al., 2000). Conjugation with the RVG9R peptide enabled the delivery of p137 RNA into neurons of both dopaminergic and nondopaminergic systems (Fig. 1, d and e) and protected primary fetal dopaminergic cells from a 6-OHDA insult (Fig. 1 f). Such protection was not observed using a range of RNA and peptide controls of various sizes, which included RVG9R–antisense p137, RVMat9R–p137 (comprising a control peptide unable to bind to AChR), or control RVG9R–pXef , which encodes the Xenopus laevis elongation factor 1α (Fig. 1). Similarly, p137 RNA complexed with RVG9R peptide could also be delivered to the α3/α5 nicotinic AChR+ U373 cell line (Fig. 1, a and b). In contrast, incubation of p137 RNA alone, with the RVMat9R–p137 complex, or with the RVG9R–pXef complex failed to result in p137 delivery to U373 cells (Fig. 1 b).
Figure 1.
Delivery of RVG9R–p137 to cultured cells in vitro prevents neurotoxin-induced cell death. (a) Representative photomicrographs on the expression of α3/α5 nicotinic AChR (green) on HEK293, U373, and SH-SY5Y cells. Nuclei were counterstained with Hoechst (blue). (b) Representative photomicrographs showing the delivery of unlabeled p137 RNA in U373 cells treated with p137 alone, control RVMat9R peptide–conjugated p137, RVG9R–p137, or RVG9R–pXef, as detected using FISH to detect p137 RNA (red). (c) Treatment of RVG9R–p137 but not RVG9R complexed with the control antisense p137 RNA (a.s. p137), or the control RVMat9R peptide complexed with the p137 RNA significantly protected SH-SY5Y cells from rotenone (Rot)-mediated cell death analyzed by TUNEL staining. (d and e) FITC-tagged p137 RNA could be detected in both cholinergic (d) and dopaminergic (e) neurons after incubation with rat E13.5 primary cortical (CTx) and VM cells, respectively. FITC signals (yellow) were colocalized with Hoechst (blue), AChR (green), and ChAT/TH (red). (f) Numbers ofTH+ cells in untreated VM cultures (con) or in VM cultures insulted with 6-OHDA, in the presence of increasing concentrations of RVG9R–p137 or RVG9R–pXef, were quantified. The proportion of TH+ cells was calculated against the total number of Hoechst+ cells. (a–f) Data were obtained from three (a–c), two (d and e), and six (f) independent experiments. Data represent mean ± SEM; **, P < 0.005; ***, P < 0.001, compared with the neurotoxin only group. Bars: (a and b) 30 µm; (d) 25 µm; (e) 40 µm.
Based on the in vitro results, we tested the ability of RVG9R–p137 to protect dopaminergic cell loss against 6-OHDA neurotoxicity in vivo. Adult male rats were stereotactically injected with peptide–RNA complex containing 2.5 µg RNA into the dopaminergic cell bodies at the level of the substantia nigra pars compacta (SNc) 3 d before an acute, intranigral 6-OHDA insult. We observed the expected functional (Table 1) and phenotypical (not depicted) deficits in those rats that were given RVG9R–pXef pretreatment, whereas rats pretreated with equivalent doses of RVG9R–p137 showed significant attenuation of the lesioning effects.
Table 1.
Summary of scores in the drug-induced rotation tests, forelimb cylinder test, and forelimb stepping tests in the RVG9R–p137 and RVG9R–pXef receiving groups
| Groups | Treatment (no. of rats) | Amphetamine-induced rotation | Apomorphine-induced rotation | Contralateral forelimb use | Contralateral forelimb stepping (no. of adjustment steps) |
| turns/min | turns/min | % | |||
| Pre-lesion treatment (intranigral) | pXef (10) | 6.45 ± 1.04 | 4.63 ± 1.16 | 10.37 ± 3.57 | 2.00 ± 0.31 |
| p137 (10) | 0.32 ± 0.69c | −1.57 ± 1.08b | 40.24 ± 7.95c | 11.22 ± 0.81c | |
| Pre-lesion treatment (intravenous) | pXef (10) | 10.93 ± 6.80 | 4.34 ± 0.71 | 1.47 ± 2.31 | 2.33 ± 1.03 |
| p137 (12) | 1.61 ± 4.53a | 0.35 ± 1.32c | 41.61 ± 19.96b | 10.33 ± 2.81c | |
| Post-lesion treatment (intranigral) | pXef (9) | 4.72 ± 1.65 | 0.85 ± 0.72 | 16.98 ± 16.74 | 6.22 ± 1.20 |
| p137 (10) | 0.44 ± 1.82b | −0.53 ± 0.40 | 40.56 ± 12.74a | 7.91 ± 1.38 | |
| Post-lesion treatment (intravenous) | pXef (6) | 5.75 ± 3.41 | 0.70 ± 0.31 | 19.07 ± 11.89 | 4.17 ± 1.94 |
| p137 (6) | 0.82 ± 0.77b | −0.12 ± 0.28a | 57.03 ± 8.19c | 7.50 ± 1.05a |
p137 encodes the neuroprotective domain of the human cytomegaloviral β2.7 transcript. Data shown for all treatment groups are derived from one observation, involving all lesioned animals, from four independent experiments over a time span of 2 yr, using multiple batches of RVG9R peptide and RNAs. Data represent mean ± SEM.
P < 0.05.
P < 0.01.
P < 0.001.
As the RVG peptide was originally engineered to enable its RNA cargo to cross the blood–brain barrier and target its delivery to the brain, we examined the possibility of using a transvascular approach for RVG9R–p137 administration. Rats were treated with an intravenous injection of 300 µg of RVG9R–p137, via the tail vein, 3 d before an intranigral 6-OHDA lesion. Consistent with our observations using intranigral delivery, rats pretreated transvascularly with RVG9R–p137 performed significantly better than those receiving RVG9R–pXef on a battery of behavioral tests (Table 1). This correlated well with immunohistochemical analysis, showing that the RVG9R–p137 pretreatment significantly protected the nigrostriatal dopaminergic system from the lesion (not depicted).
RVG9R–p137 mediates neuroprotection by protecting mitochondrial Complex I activity
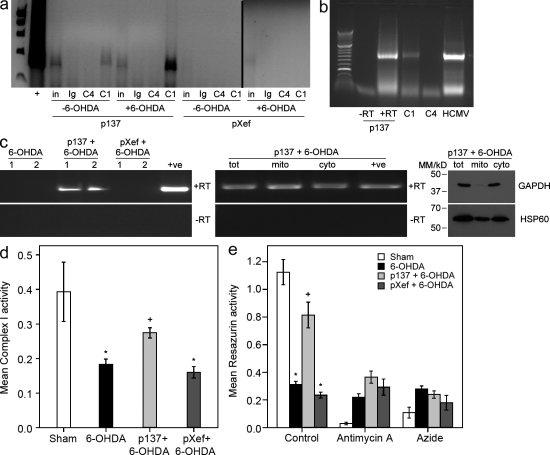
To verify that the RVG9R–p137-mediated neuroprotection was associated with an interaction between p137 and mitochondrial Complex I, we analyzed whether p137 was physically interacting with Complex I in the nigral tissue of rats receiving this treatment. We used reversible cross-linking of RNA–protein conjugates combined with Complex I immunoprecipitation. Native Complex I from the nigra of rats pretreated with RVG9R–p137 clearly resulted in the coprecipitation with p137 (Fig. 2 a). This interaction was specific for native (assembled in the mitochondria) Complex I, as immunoprecipitation of mitochondrial Complex IV showed no such interaction with p137 RNA. This was recapitulated when using primers to amplify the full-length p137 product from RNA immunoprecipitations (Fig. 2 b), arguing that full-length p137 was associated with Complex I. We observed that the specific interaction between p137 and Complex I was more pronounced in RVG9R–p137-pretreated rats subsequently exposed to 6-OHDA compared with the unlesioned animals (Fig. 2 a). There was no interaction between pXef and Complex I using pXef-specific PCRs of the same cDNAs used (Fig. 2 a).
Figure 2.
p137 interacts with mitochondrial Complex I in the substantia nigra and protects Complex I activity from a 6-OHDA–induced deficit. (a) Single cell suspensions of the nigra from rats intranigrally pretreated with RVG9R–p137 (p137) or control RVG9R–pXef (pXef), and subsequently left unlesioned (−6-OHDA) or lesioned (+6-OHDA), were analyzed for p137–Complex I interaction by RNA immunoprecipitation. Immunoprecipitates using control Ig antibody, anti–Complex IV antibody (C4), or anti–Complex I antibody (C1) were analyzed by p137 RT-PCR for the presence of coimmunoprecipitated p137 RNA. Starting samples (∼5% of samples used for immunoprecipitation) were also directly amplified by RT-PCR to determine input levels of p137 (in). RNA immunoprecipitations were performed twice independently, from two different in vivo experiments, and the data shown are from one of these experiments, which has been analyzed three times and showed the same patterns of PCR bands. (b) Single cell suspensions of the nigra of rats intranigrally pretreated with RVG9R–p137 were analyzed for the interaction of full-length p137 with Complex I by RNA immunoprecipitation. Immunoprecipitates of cells using anti–Complex I or anti–Complex IV antibody were analyzed by a p137-specific RT-PCR for the presence of coimmunoprecipitated p137 RNA. For positive controls, full-length p137 RNA with (+) or without (−) RT was amplified as well as RNA from HCMV-infected fibroblasts (HCMV). Results were obtained from the same samples as shown in panel a, and this full-length p137 PCR was independently performed twice on the same samples with equivalent results. (c) p137 RNA was analyzed by RT-PCR from a new group of rats treated with 6-OHDA or rats treated with RVG9R–p137 or RVG9R–pXef, before 6-OHDA insult (left). Duplicate samples are shown with (+) and without (−) RT. Additionally, RNA from equivalent amounts of mitochondrial (mito) and cytosolic (cyto) fractions or total sample (tot), from 6-OHDA rats pretreated with RVG9R–p137, was isolated and analyzed for the presence of p137 RNA by RT-PCR with (+) and without (−) RT (middle). The relative purity of the fractions was also assessed by Western blot analysis (right) using antibodies to HSP60 (mitochondrial marker) and GAPDH (cytosolic marker). This PCR and immunoblotting analysis was performed once. As we believed it was necessary to analyze as pure a mitochondrial fraction as possible for the presence of p137 RNA, we used an extremely stringent mitochondrial isolation protocol, which resulted in some contamination of the cytosolic fraction with mitochondrial proteins. (d) Complex I activity (shown as arbitrary units) was analyzed in nigral tissues from sham-treated rats (n = 4), rats treated with 6-OHDA (n = 4), and rats receiving RVG9R–pXef (n = 3) or RVG9R–p137 (n = 5) 3 d before 6-OHDA lesion. Results were derived from one analysis, but equivalent trends were observed in two other independent assays. (e) A resazurin reduction assay (shown as relative OD590) was used to assess mitochondrial respiration. Mitochondria were isolated from the nigra of sham-treated rats, rats treated with 6-OHDA, and rats treated with RVG9R–p137 or RVG9R–pXef before 6-OHDA insult (n = 3 per group). Where designated, assays were also performed on the mitochondrial samples in the presence of the mitochondrial respiration inhibitors antimycin A and sodium azide, which are specific for inhibiting Complex III and Complex IV, respectively. Data represent mean ± SEM; *, P < 0.05 compared with sham; and +, P < 0.05 compared with 6-OHDA.
To confirm this interaction between p137 and mitochondria, we also analyzed the distribution of p137 between the mitochondrial and cytosolic fractions of the isolated nigra from rats receiving RVG9R–p137 (Fig. 2 c). This showed the presence of p137 RNA by RT-PCR specifically in RVG9R–p137-treated rats, as expected (Fig. 2 c, left). We then fractionated mitochondria from the nigra of these rats, confirmed purity (Fig. 2 c, right) by Western blot detection of GAPDH (cytosol specific) and HSP60 (mitochondria specific), and analyzed levels of p137 RNA (Fig. 2 c, middle).Taking into account contamination of the cytosol with mitochondrial markers but little contamination of the mitochondrial fraction with cytosol, this analysis showed that at least half of the p137 delivered was associated with mitochondria. This is consistent with mitochondria being a major but not exclusive site of action of p137 as the cytosolic fraction also contains noticeable levels of p137. This is not unexpected given that the viral β2.7 RNA is likely to be multifunctional and is known to interact with other cellular targets (Reeves et al., 2007); what is important is that its major action here is through a selective interaction with mitochondrial Complex I.
We then examined whether RVG9R–p137 delivery offered protection at the level of enzyme activity of Complex I in the SNc in the presence of the 6-OHDA insult (Fig. 2 d). Semipurified mitochondria were isolated from the nigra of rats treated with RVG9R–p137 or RVG9R–pXef before an acute 6-OHDA lesion. Univariate analysis of variance showed that there was a significant difference in Complex I activity between groups (P = 0.013), and post-hoc analysis revealed that there was a significant reduction in Complex I activity in rats receiving an acute 6-OHDA insult. A similar attenuation on Complex I activity was seen in the lesioned rats receiving RVG9R–pXef pretreatment. In contrast, mitochondria from rats pretreated with RVG9R–p137 demonstrated a significant protection of Complex I enzyme activity. Such protection in mitochondrial function was also confirmed using a resazurin reduction–based assay (Fig. 2 e). Respiration in purified mitochondria from rats treated with 6-OHDA was substantially inhibited compared with mitochondria from sham-treated rats, as expected. In contrast, rats pretreated with RVG9R–p137 demonstrated a protected mitochondrial function that was not observed in the RVG9R–pXef-pretreated rats.
Collectively, these results suggest that RVG9R–p137 pretreatment clearly mediates a robust protective effect to enhance dopaminergic cell survival against an acute 6-OHDA lesion. Such protection is associated with a specific interaction between p137 and Complex I, as well as the attenuation of 6-OHDA–mediated impairment of Complex I activity and mitochondrial respiratory function.
Repeated RVG9R–p137 treatment attenuates 6-OHDA toxicity in a progressive lesion model without eliciting a host immune response
Although our data showed that we could prevent cell death using RVG9R–p137 when delivered before 6-OHDA lesion, it is clinically more relevant to show protective effects after a lesion had been initiated. Therefore, we investigated whether the RVG9R–p137 treatment could mediate a neuroprotective function to attenuate cell loss after the onset of lesion by using a progressive lesion model of PD. It has been well reported that a partial and retrograde nigrostriatal dopaminergic degeneration can be achieved by 6-OHDA injection into the ventrolateral striatum (Kirik et al., 1998; Blandini et al., 2007), which causes a progressive cell death over a 2–6-wk period. Similar to the aforementioned pretreatment experiments, a significant correction of behavioral deficits (Table 1), as well as a rescue of nigral and striatal tyrosine hydroxylase (TH) expression (not depicted), was observed with repeated intranigral administrations of RVG9R–p137 starting 3 d after the intrastriatal 6-OHDA lesion. Such protection was not observed in rats receiving RVG9R–pXef under an equivalent treatment regimen. Clearly, RVG9R–p137 treatment was able to attenuate 6-OHDA toxicity in a progressive lesion model when delivered directly to the SNc after the initiation of lesion.
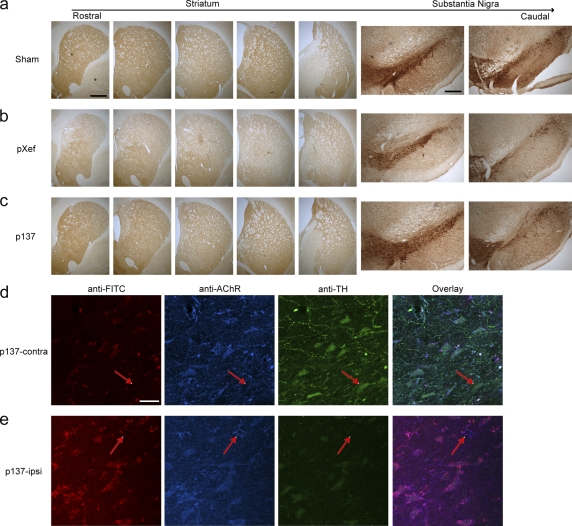
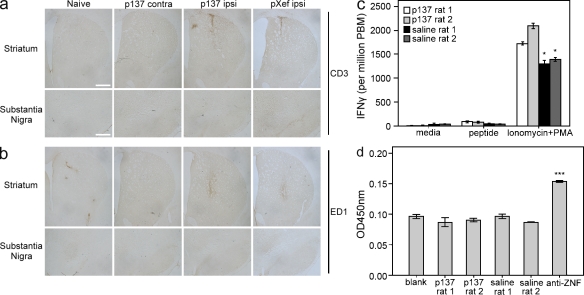
We then evaluated whether a comparable neuroprotective effect of RVG9R–p137 could be obtained by an intravenous route of administration. Rats were given four separate injections of 300 µg RVG9R–p137 via the tail vein 1–2 d after the lesion, with a further injection 8 d after the lesion, and with this last treatment, some animals were given FITC-tagged RVG9R–p137 for confocal microscopy analysis to evaluate the distribution of p137. Consistent with our observations using repeated intranigral delivery, a significant neuroprotection was detected only in animals receiving RVG9R–p137 treatment (Fig. 3, a–c; and Table 1). Densitometric analysis of striatal TH expression revealed significantly higher TH levels in rats receiving RVG9R–p137 (85.3 ± 3.9% of the contralateral side) compared with those given RVG9R–pXef (51.4 ± 4.1% of the contralateral side; P < 0.001). This correlated with the unbiased stereological counting showing that the number of dopaminergic cells in the SNc (6,182 ± 221 per contralateral side) was 54.2 ± 8.0% in the RVG9R–pXef, compared with 71.2 ± 3.8% in the RVG9R–p137-treated group (P < 0.01). Fluorescent detection of FITC, TH, and α3/α5 nicotinic AChR in the nigra revealed that many, but not all TH-expressing neurons were α3/α5 nicotinic AChR positive (Fig. 3 d, arrows) and that detection of p137 RNA was exclusively restricted to the AChR-expressing cells (Fig. 3, d and e). p137 could also be found associated with cells other than dopaminergic neurons (Fig. 3 e, arrows), supporting the possibility that RVG9R-mediated p137 delivery could also be used to target cell populations other than the dopaminergic system. No FITC signal could be found in rats given untagged RVG9R–p137 treatment (not depicted). There was a tendency for the p137 RNA to be recruited toward the site of a more severe lesion, i.e., the ipsilateral nigra, after intravenous RVG9R–p137 delivery. This preferential recruitment of p137 corresponds well with our immunoprecipitation data (Fig. 2 a). To determine whether repeated intravenous injections with a large RNA–protein complex stimulates a host immune reaction, we compared the level of CD3 and ED1 expression between sections from RVG9R–p137 (ipsilateral/contralateral sides), RVG9R–pXef (ipsilateral side), and naive animals. Immunostaining revealed no overt changes in the levels of T cell infiltration (CD3; Fig. 4 a) and microglial activation (ED1; Fig. 4 b) after repeated delivery of RVG9R–p137, other than those elicited by the lesioning procedure itself. This is consistent with a lack of immune responses observed by Kumar et al. (2007). We also ruled out the possibility that RVG9R–p137 treatment generated peptide-specific T cell or antibody responses. PBMCs from rats treated with RVG9R–p137 by repeated tail vein injections and subsequently stimulated with RVG9R peptide failed to elicit an IFN-γ response. In contrast, positive controls stimulated with ionomycin and PMA generated a strong response, as expected (Fig. 4 c). We also analyzed serum from the same animals for RVG9R-specific antibodies by ELISA (Fig. 4 d) and found that there was no difference in the antibody responses to RVG9R peptide between rats injected with RVG9R–p137 and saline. Overall, it is clear that the RVG9R–p137 complex delivered transvascularly does protect TH+ neurons in a progressive PD model and that this treatment is nonimmunogenic.
Figure 3.
Intravenous delivery of RVG9R–p137 attenuates the loss of dopaminergic expression in a progressive rat model of PD, and the expression of FITC-tagged p137 colocalized with AChR and TH-expressing cells in the nigra. (a–c) Representative pictures of the rostrocaudal levels of the striatum and substantia nigra from rats receiving sham (a), RVG9R–pXef (pXef; b), or RVG9R–p137 (p137; c), delivered via the tail vein, after the initiation of a progressive intrastriatal 6-OHDA lesion. (d and e) The pattern of p137 detection in the contralateral (p137-contra) and ipsilateral (p137-ipsi) substantia nigra after intravenous delivery of FITC-tagged RVG9R–p137 was examined by fluorescent confocal microscopy for p137 RNA (red), α3/α5 nicotinic AChR (blue), TH (green), and an overlay of the three channels. Arrows in d highlight the fact that not all TH neurons are AChR+, but p137 expression is restricted to AChR+ cells. Arrows in e highlight the fact that p137 expression can also be found in AChR+, nondopaminergic cells. Immunohistochemical analysis was performed once on all animals receiving repeated intravenous RVG9R–p137 (n = 6) and RVG9R–pXef (n = 6) treatments. The data shown are from one representative animal. Bars: (a–c, left) 1,000 µm; (a–c, right) 500 µm; (d and e) 40 µm.
Figure 4.
Delivery of RVG9R–p137 via the intravenous route stimulates no apparent immune responses after repeated administration. (a) The striatum or substantia nigra of rats repeatedly treated with RVG9R–pXef or RVG9R–p137 via the tail vein after the initiation of intrastriatal 6-OHDA lesion. Immunostaining using anti-CD3 was performed on naive rats, the contralateral side of rats receiving RVG9R–p137 (p137 contra), the ipsilateral side of rats receiving RVG9R–p137 (p137 ipsi), and the ipsilateral side of rats receiving RVG9R–pXef (pXef ipsi). The minor up-regulation of CD3 signal was caused by the surgical procedure in generating the lesion. (b) Immunostaining for ED1 was performed as in panel a, and again the only apparent up-regulation in ED1 response was caused by the lesioning procedure. Immunohistochemical analysis was carried out once on all animals receiving no lesion (n = 3), repeated intravenous treatments of RVG9R–p137 (n = 6), and RVG9R–pXef (n = 6). The data shown are from one representative animal of each group. Bars: (a and b, top) 1,000 µm; (a and b, bottom) 500 µm. (c) PBMCs from duplicate rats receiving RVG9R–p137 or saline by repeated tail vein injection were stimulated with media alone, RVG9R peptide (peptide), or ionomycin + PMA (positive control). Cells expressing IFN-γ were then assayed by ELISPOT, read on an AID ELISPOT reader, and enumerated using ImageJ. (d) Sera from the same duplicate rats receiving RVG9R–p137 or saline, used in c for the IFN-γ ELISPOT analysis, were also analyzed for RVG9R-specific antibodies using RVG9R-coated ELISA plates. Wells coated with ZNF785 peptide (anti-ZNF) were used as positive controls for peptide-specific detection. ELISA plates (OD) were read at 450 nm on a Multitek ELISA plate reader. (c and d) Assays were performed from three independent experiments. Data represent mean ± SEM; *, P < 0.05; and ***, P < 0.001 compared with immune responses stimulated in animals receiving repeated RVG9R–p137 treatments.
Since the first demonstration that mitochondrial Complex I activity was impaired in the nigra of patients with PD (Schapira et al., 1990), there has been interest in the etiological role of mitochondria in PD. Therapeutic agents that enhance mitochondrial functions, such as coenzyme Q10, have been reported to have a moderate effect in slowing disease progression in patients with early PD (Shults et al., 2002). Of late, several Mendelian genetic causes for PD have been identified, including abnormalities in the α-synuclein, LRRK2 (leucine-rich repeat kinase 2), DJ-1, PINK1 (PTEN-induced putative kinase 1), and parkin genes. Many of these genes are thought to have effects on mitochondrial function, reinforcing the view that mitochondrial dysfunction may lie at the heart of this disorder, even though the majority of patients with PD have an idiopathic disease origin. As such, it would have been useful to study the effects of RVG9R–p137 in transgenic models of PD, except that none of these models show consistent dopaminergic cell loss (Dawson et al., 2010), hence our use of neurotoxin-based models. These models, although a poor imitation of the clinical condition, have nevertheless been important in the development of a whole range of therapeutic agents such as dopaminergic cell transplants and, more controversially, glial cell line–derived neurotrophic factor (Gill et al., 2003; Lang et al., 2006). The defining pathological feature of PD is traditionally considered to be the loss of the nigrostriatal pathway, even though the disease is now recognized to have a range of nonmotor features and extranigral pathology. Indeed, this has even led some to consider PD as a multistage disease with nigral pathology only emerging halfway through that disease cascade (Braak et al., 2006). This, coupled with the heterogeneous nature of PD, may limit the potential benefits of current cell replacement (Freed et al., 2001; Olanow et al., 2003) and neuroprotective (Nutt et al., 2003; Lang et al., 2006) therapies that only target neurons of the dopaminergic system. The RVG9R–p137 treatment reported in this paper may therefore offer a more useful alternative by targeting mitochondrial stress in both dopaminergic and nondopaminergic cells affected by the disease process, all of which could be reached through the systemic delivery of the agent assuming that the relevant cell populations affected express the right AChR. This has also been reinforced by a recent study, showing that the RVG peptide can target siRNA delivery to different CNS cells such as neurons, astrocytes, and oligodendrocytes via transvascular administration (Alvarez-Erviti et al., 2011).
Although we did not examine the expression of p137 outside the brain after transvascular delivery, Kumar et al. (2007) used flow cytometry to analyze the RVG9R-mediated GFP expression in the peripheral tissues and concluded that the cargo delivery was restricted to the CNS. Such a CNS-targeted method of transvascular delivery suggests that our RVG9R–p137 complex may be helpful for clinical application in the future treatment of PD and related neurodegenerative conditions. However, more work is needed to determine the pharmacokinetic profile of the RVG9R–p137 conjugate, and work is on-going to improve the half-life of siRNA in vivo, such as lipid encapsulation (Morrissey et al., 2005) and chemical modification for polyethylene glycol addition (Dassie et al., 2009).
In conclusion, we have developed a novel therapeutic agent that specifically targets Complex I and when given is neuroprotective in a range of model systems of dopaminergic cell loss mimicking PD, while not inducing any immune reactions. As such, this agent has great potential as a disease-modifying therapy for this common, incurable neurodegenerative condition.
MATERIALS AND METHODS
Peptides.
Peptide RVG9R (YTIWMPENPRPGTPCDIFTNSRGKRASNGGGGRRRRRRRRRR) comprises a short peptide derived from the RVG and nine C-terminal arginine residues that were d-arginine (Kumar et al., 2007). We also used a control peptide, RVMat9R, in which the RVG sequences were replaced with amino acids from the rabies virus matrix protein, which does not bind to the AChR (Kumar et al., 2007). At least five separate batches of peptide for use in the experiments described in this paper were synthesized by Pro-Immune Ltd. and supplied as a lyophilized powder. For use experimentally, it was suspended in water at a concentration of 20 mg/ml and stored at −70°C.
RNAs.
All RNAs were transcribed from linear DNA templates in vitro using a MEGAscript polymerase kit (Invitrogen; as described by the manufacturer). β2.7 RNA (containing nucleotides 4,577–3,782 of the β2.7 gene in HCMV strain AD169) was transcribed with T7 from p137 plasmid (Bergamini et al., 1998) after linearization with XhoI. Alternatively, control antisense p137 RNA was generated by linearization with HindIII and transcription using SP6. Additional control RNA was derived from the pTri-Xef (Invitrogen), a prelinearized plasmid template encoding the Xenopus laevis elongation factor 1α gene. FITC-labeled p137 RNA was produced using a fluorescein RNA labeling mix (Roche) in conjunction with the MEGAscript kit, substituting the kit’s supplied NTP mixtures with the fluorescein NTP mix (as described by the manufacturer). Multiple batches of control and specific RNAs were used over the course of these experiments. To generate the p137-specific probe (antisense) for fluorescence in situ hybridization (FISH), the p137 plasmid was linearized with HindIII and used as a template for in vitro transcription as described above except that Cy3-UTP (GE Healthcare) was used as described by the manufacturer to incorporate Cy3 label into the in vitro transcribed p137 RNA.
RNA–peptide complex.
RNAs, RVG9R, and RVMat9R were combined in a 10:11 (peptide/RNA) molar ratio, with serum-free MEM making up the total volume. For visualization of peptide–RNA complex on cells, a spike of FITC-labeled RNA (1:20 labeled/unlabeled for in vitro experiments and 1:4 labeled/unlabeled for in vivo imaging) was included during peptide–RNA complex formation.
Cell cultures.
U373 glioblastoma cells were grown in Earle’s MEM containing 10% fetal calf serum. SH-SY5Y neuroblastoma and HEK293 embryonic kidney cells were grown in DMEM. All media contained penicillin/streptomycin, and all cells were grown at 37°C in 5% CO2. Primary cell cultures were made of embryonic day (E) 13.5 rat ventral mesencephalic (VM) tissue. Fetal VM tissues were dissected using standard procedures and mechanically dissociated to a single cell suspension. Half of the fetal VM was plated per poly-l-lysine–coated glass coverslip in differentiation medium containing RVG9R–p137 or RVG9R–pXef at various concentrations. Further details on the culture preparation can be found in Kuan et al. (2006). The primary cells were then challenged by 6-OHDA for 24 h and fixed after 7 d in vitro growth for immunocytochemical analysis.
TUNEL assay.
TUNEL staining to detect apoptosis was performed using an In situ Cell Death Detection kit, TMR red (Roche) exactly as described by the manufacturer.
Surgical procedures.
Male Sprague-Dawley rats, weighing around 200 g at the time of initial surgery, were housed with unrestricted access to food and water. All animal experiments were performed in compliance with the UK Animals (Scientific Procedures) Act 1986. Each animal was given norocarp and terramycin before surgery, followed by injection of 18 µg/3 µl 6-OHDA to the SNc for an acute lesion (anterior–posterior, −5.3 mm; medial–lateral, −2.0 mm; dorsal–ventral, −7.2 mm) or 24 µg/4 µl to the striatum for a progressive lesion (anterior–posterior, 1.0 mm; medial–lateral, −3.0 mm; dorsal–ventral, −5.0 mm). All coordinates were based on Paxinos and Watson (1986) and are relative to bregma and the dural surface with the incisor bar set 2.3 mm below the interaural line. For the pretreatment experiments, the conjugates were directly injected into the SNc for intracranial delivery or administered via the tail vein for intravenous delivery 3 d before a single 6-OHDA lesion and on every 10th day thereafter. For the progressive lesion experiments, the conjugates were delivered intranigrally on the third day and then on every 10th day after the intrastriatal 6-OHDA lesion. Intravenous conjugate deliveries were given on the first, second, and eighth day after lesion.
Behavioral tests.
The extent of striatal dopaminergic denervation was examined by 2.5 mg/kg amphetamine– and 1.6 mg/kg apomorphine–induced rotational behavior. The number of full ipsi- and contralateral rotations was measured for 90 min after drug administration. The paw-reaching test was developed to evaluate the sensorimotor function of the animal’s forelimb. In brief, the experimenter firmly held both hindlimbs and one forelimb of the animal and moved the rat sideways across a platform over 1 m in 5 s. The unrestrained forelimb was in contact with the platform and the number of adjustment steps performed by the rats counted in the forehand direction. The animals were habituated to the handling associated with this test for three consecutive days before lesion. The cylinder test evaluated the spontaneous forelimb use during vertical exploration. In brief, the animal was allowed to move freely in a Plexiglas cylinder during a 5-min session. The observation was videotaped, with mirrors placed behind the cylinder to ensure full visibility of paw placements during this time. Data were presented as the proportion of contralateral paw touches against the total. Behavioral tests were assessed between the third and fourth week after the 6-OHDA lesion. Further details can be found in Kuan et al. (2007).
Immunohistochemistry and FISH.
Animals were sacrificed by pentobarbitone sodium and transcardially perfused 4 wk (2 wk for the rats receiving a transvascular, postlesion treatment) after lesion. The brains were removed, postfixed, and then transferred to 30% sucrose until they sank. 40-µm coronal brain sections were cut, and a 1:6 series of sections were immunostained for mouse anti-TH (1:500; Millipore), anti-AChR α3/α5 (1:1,000; Sigma-Aldrich), anti-ED1 (1:1,000; AbD Serotec), and anti-CD3 (1:500; Abcam) as described previously (Kuan et al., 2006, 2007). The cultures or sections were visualized either by immunofluorescence (Alexa Fluor 568 and Alexa Fluor 688; 1:1,000; both from Invitrogen) or diaminobenzidine. Staining omitting primary antibodies was performed in all immunohistochemical experiments to serve as a negative control.
For FISH to detect p137 RNA, cells were washed, fixed with 4% paraformaldehyde, and permeabilized with 70% cold ethanol. Cells were then rehydrated in 2× SSC (saline sodium citrate), and incubated with blocking buffer (50% formamide, 2× SSC, 10% dextran sulfate, 1× Denhardt’s solution, 2 mM ribonucleoside vanadyl complexes, 50 µg/ml of yeast tRNA, and 50 µg/ml of salmon sperm DNA) for 30 min at 37°C in a humidified incubator followed by overnight hybridization with 0.4 ng/ml Cy3-labeled antisense probe for p137 in the same buffer. After hybridization, cells were counterstained with 1 mg/ml Hoechst and mounted with Citifluor (Dako) and analyzed by fluorescence microscopy (Eclipse TE300; Nikon).
Determination of Complex I activity.
To compare changes in mitochondrial Complex I activity, mitochondria were isolated from the SNc. To do this, rats were decapitated under general anesthesia. Upon removal of the brain, a 70–90-mg nigral tissue was acutely dissected and immediately submerged in ice-cold artificial cerebrospinal flood (125 mM NaCl, 5 mM KCl, 1.25 mM NaH2PO4, 28 mM NaHCO3, 0.5 mM CaCl2, 4 mM MgCl2, 25 mM d-glucose, and 1 mM kynurenic acid). Mitochondrial preparations were obtained from brain sections using a proprietary mitochondria isolation kit (BioChain Institute) as described by the manufacturer. For analysis of Complex I activity in these mitochondria, a Complex I Enzyme Activity Microplate Assay kit (MitoSciences) was used exactly as described by the manufacturer. In all cases, samples were corrected for total mitochondrial protein assayed.
Mitochondrial respiration was also analyzed directly using a resazurin reduction assay of isolated mitochondria in a 96-well plate format, as previously described (Zhang et al., 2004). After isolation, as described in the previous paragraph, mitochondria were analyzed directly or after pretreatment with 25 µg/ml antimycin A or 650 µM sodium azide, which are specific inhibitors of mitochondrial respiration and confirmed that the mitochondrial-specific component of respiration analyses was being assayed.
RNA/protein immunoprecipitation.
Nigral tissue was prepared exactly as detailed in the previous section for preparation of mitochondria and enzymatically digested with trypsin (Worthington). The digestion was terminated with a mixture of trypsin inhibitor and DNase (both from Sigma-Aldrich). After centrifugation, the pellet was resuspended in DMEM and mechanically triturated until a single cell suspension had formed. The cells were then fixed in paraformaldehyde, quenched in glycine, and stored at −80°C until used. To determine any interaction of p137 RNA with Complex I, cells derived from the SNc of rats treated intranigrally with RVG9R–p137 or RVG9R–pXef were subject to RNA immunoprecipitation using Complex I and Complex IV immunocapture beads (MitoSciences) as described by the manufacturer. The anti–Complex I antibody used only recognizes fully formed native Complex I assembled in the mitochondria. Ig2a antibody–coated beads were also used as an additional control for the immunoprecipitation. After immunocapture of Complex I or Complex IV from the samples, the presence of interacting p137 RNA was analyzed by RT-PCR, as previously described (Reeves et al., 2007).
To assess the relative levels of localization of p137 RNA in mitochondria and the cytosol, mitochondria were isolated using the proprietary mitochondria isolation kit (BioChain Institute), and RNA isolated from each cellular subfraction by TRIZOL extraction was analyzed by p137-specific RT-PCR (Reeves et al., 2007). Relative purity of the fractions was assessed using Western blot analysis with antibodies to HSP60 (specific for mitochondria) and GAPDH (specific for cytosol).
Assessment of peptide-specific T cell and antibody responses in treated rats.
Blood was collected from rats by heart puncture (8 ml blood in 2 ml of 100 U/ml heparin sodium), and PBMCs were isolated by density barrier centrifugation using a 2.5:2 vol/vol ratio of OptiPrep (Axis-Shield) and tricine buffer (10 mM tricine and 0.145 M NaCl, pH 7.0). Heparinized rat blood was diluted 1:1 with tricine buffer and 6 ml of diluted blood layered onto 3 ml of a density barrier and centrifuged at 800 g for 20 min. The PBMCs were removed from the interface with a Pasteur pipette and diluted in PBS (Oxoid) and washed twice before resuspension and counting in RPMI 1640 10% FCS.
Rat IFN-γ ELISPOT assays were performed according to the manufacturers protocols (BD). In brief, PBMCs from two RVG- and two saline-inoculated animals were used at 1.875 × 105 and 3.75 × 105 cells per well in triplicate. Wells were stimulated with media alone (as a negative control), media containing 20 µg/ml RVG peptide, or media containing 500 ng/ml ionomycin and 5 ng/ml PMA (positive control). ELISPOT plates were incubated overnight at 37°C in 5% CO2 and then developed according to the manufacturers protocols. Plates were read on a Reader System, AID ELISPOT reader and enumerated using ImageJ (National Institutes of Health).
For antibody analysis, 96-well ELISA (Thermo Fisher Scientific) plates were coated with 0.1 µg per well of either RVG9R peptide or ZNF785 peptide (Abcam; as a positive control for peptide-specific ELISA detection) in bicarbonate buffer overnight at 4°C. Plates were washed three times in PBS 0.05% Tween-20 (PBST), and 200 µl of 10 mg/ml bovine serum albumin in PBST was added to each well for 2 h at 37°C as a blocking agent. Rat sera from saline- or RVG peptide–treated rats or rat sera specific for ZNF785 peptide was diluted 1:1,600 in PBST 3% BSA (antibody dilution buffer), and 100 µl was added to peptide-coated wells and incubated for 1 h at 37°C. Plates were washed five times in PBST. Anti–rat horseradish peroxidase was diluted 1:4,000 in antibody dilution buffer, and 100 µl was added to each well, incubated for 30 min at 37°C, and then washed five times. 50 µl TMB substrate (eBioscience) was added per well, and 50 µl of 2N sulfuric acid was added to stop color development. ELISA plates were read at 450 nm on a Multitek ELISA plate reader.
Microscopy, densitometric, stereological, and statistical analysis.
Imaging was performed using a DM6000 microscope or a TCS SP2 confocal microscope (Leica). The mean OD was determined by subtracting the OD of the background from the region of interest defined by TH staining. Results were expressed as the ratio between the OD of the ipsi- and contralateral side. Total cell numbers in the SNc were estimated by unbiased stereology sampling using the Stereo Investigator 9.10.3 (MBF Bioscience). Densitometric, stereological, and behavioral data between all groups of animals was analyzed using univariate analysis of variance, and a post-hoc Bonferroni test was used as appropriate with significance set at P < 0.05. All the analysis was performed using ImageJ 1.42q and SPSS Release 16.0.0.
Acknowledgments
We thank Dr. G. Bergamini for the TRL4 expression plasmid. We are also grateful to Mrs. Linda Teague for her excellent technical assistance.
We thank the Michael J. Fox Foundation and the National Institute for Health Research for the award of a Biomedical Research Centre grant to the University of Cambridge/Addenbrooke’s Hospital. S. Karniely was a recipient of a European Molecular Biology Organization long-term fellowship (ALTF 887-2009).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- 6-OHDA
- 6-hydroxydopamine
- AChR
- acetylcholine receptor
- CNS
- central nervous system
- FISH
- fluorescence in situ hybridization
- HCMV
- human CMV
- PD
- Parkinson’s disease
- RVG
- rabies virus glycoprotein
- siRNA
- short interfering RNA
- SNc
- substantia nigra pars compacta
- TH
- tyrosine hydroxylase
- VM
- ventral mesencephalic
References
- Alvarez-Erviti L., Seow Y., Yin H., Betts C., Lakhal S., Wood M.J. 2011. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 29:341–345 10.1038/nbt.1807 [DOI] [PubMed] [Google Scholar]
- Bergamini G., Reschke M., Battista M.C., Boccuni M.C., Campanini F., Ripalti A., Landini M.P. 1998. The major open reading frame of the beta2.7 transcript of human cytomegalovirus: in vitro expression of a protein posttranscriptionally regulated by the 5′ region. J. Virol. 72:8425–8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betarbet R., Sherer T.B., MacKenzie G., Garcia-Osuna M., Panov A.V., Greenamyre J.T. 2000. Chronic systemic pesticide exposure reproduces features of Parkinson’s disease. Nat. Neurosci. 3:1301–1306 10.1038/81834 [DOI] [PubMed] [Google Scholar]
- Blandini F., Levandis G., Bazzini E., Nappi G., Armentero M.T. 2007. Time-course of nigrostriatal damage, basal ganglia metabolic changes and behavioural alterations following intrastriatal injection of 6-hydroxydopamine in the rat: new clues from an old model. Eur. J. Neurosci. 25:397–405 10.1111/j.1460-9568.2006.05285.x [DOI] [PubMed] [Google Scholar]
- Braak H., Bohl J.R., Müller C.M., Rüb U., de Vos R.A., Del Tredici K. 2006. Stanley Fahn Lecture 2005: The staging procedure for the inclusion body pathology associated with sporadic Parkinson’s disease reconsidered. Mov. Disord. 21:2042–2051 10.1002/mds.21065 [DOI] [PubMed] [Google Scholar]
- Dabbeni-Sala F., Di Santo S., Franceschini D., Skaper S.D., Giusti P. 2001. Melatonin protects against 6-OHDA-induced neurotoxicity in rats: a role for mitochondrial complex I activity. FASEB J. 15:164–170 10.1096/fj.00-0129com [DOI] [PubMed] [Google Scholar]
- Dassie J.P., Liu X.Y., Thomas G.S., Whitaker R.M., Thiel K.W., Stockdale K.R., Meyerholz D.K., McCaffrey A.P., McNamara J.O., II, Giangrande P.H. 2009. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 27:839–849 10.1038/nbt.1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson T.M., Ko H.S., Dawson V.L. 2010. Genetic animal models of Parkinson’s disease. Neuron. 66:646–661 10.1016/j.neuron.2010.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed C.R., Greene P.E., Breeze R.E., Tsai W.Y., DuMouchel W., Kao R., Dillon S., Winfield H., Culver S., Trojanowski J.Q., et al. 2001. Transplantation of embryonic dopamine neurons for severe Parkinson’s disease. N. Engl. J. Med. 344:710–719 10.1056/NEJM200103083441002 [DOI] [PubMed] [Google Scholar]
- Gill S.S., Patel N.K., Hotton G.R., O’Sullivan K., McCarter R., Bunnage M., Brooks D.J., Svendsen C.N., Heywood P. 2003. Direct brain infusion of glial cell line-derived neurotrophic factor in Parkinson disease. Nat. Med. 9:589–595 10.1038/nm850 [DOI] [PubMed] [Google Scholar]
- Hanham C.A., Zhao F., Tignor G.H. 1993. Evidence from the anti-idiotypic network that the acetylcholine receptor is a rabies virus receptor. J. Virol. 67:530–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik D., Rosenblad C., Björklund A. 1998. Characterization of behavioral and neurodegenerative changes following partial lesions of the nigrostriatal dopamine system induced by intrastriatal 6-hydroxydopamine in the rat. Exp. Neurol. 152:259–277 10.1006/exnr.1998.6848 [DOI] [PubMed] [Google Scholar]
- Kuan W.L., Hurelbrink C.B., Barker R.A. 2006. Increased capacity for axonal outgrowth using xenogenic tissue in vitro and in a rodent model of Parkinson’s disease. Xenotransplantation. 13:233–247 10.1111/j.1399-3089.2006.00291.x [DOI] [PubMed] [Google Scholar]
- Kuan W.L., Lin R., Tyers P., Barker R.A. 2007. The importance of A9 dopaminergic neurons in mediating the functional benefits of fetal ventral mesencephalon transplants and levodopa-induced dyskinesias. Neurobiol. Dis. 25:594–608 10.1016/j.nbd.2006.11.001 [DOI] [PubMed] [Google Scholar]
- Kumar P., Wu H., McBride J.L., Jung K.E., Kim M.H., Davidson B.L., Lee S.K., Shankar P., Manjunath N. 2007. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 448:39–43 10.1038/nature05901 [DOI] [PubMed] [Google Scholar]
- Lang A.E., Gill S., Patel N.K., Lozano A., Nutt J.G., Penn R., Brooks D.J., Hotton G., Moro E., Heywood P., et al. 2006. Randomized controlled trial of intraputamenal glial cell line-derived neurotrophic factor infusion in Parkinson disease. Ann. Neurol. 59:459–466 10.1002/ana.20737 [DOI] [PubMed] [Google Scholar]
- Luo J., Kaplitt M.G., Fitzsimons H.L., Zuzga D.S., Liu Y., Oshinsky M.L., During M.J. 2002. Subthalamic GAD gene therapy in a Parkinson’s disease rat model. Science. 298:425–429 10.1126/science.1074549 [DOI] [PubMed] [Google Scholar]
- Morrissey D.V., Lockridge J.A., Shaw L., Blanchard K., Jensen K., Breen W., Hartsough K., Machemer L., Radka S., Jadhav V., et al. 2005. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 23:1002–1007 10.1038/nbt1122 [DOI] [PubMed] [Google Scholar]
- Nutt J.G., Burchiel K.J., Comella C.L., Jankovic J., Lang A.E., Laws E.R., Jr, Lozano A.M., Penn R.D., Simpson R.K., Jr, Stacy M., Wooten G.F.; ICV GDNF Study Group. Implanted intracerebroventricular. Glial cell line-derived neurotrophic factor 2003. Randomized, double-blind trial of glial cell line-derived neurotrophic factor (GDNF) in PD. Neurology. 60:69–73 [DOI] [PubMed] [Google Scholar]
- Olanow C.W., Goetz C.G., Kordower J.H., Stoessl A.J., Sossi V., Brin M.F., Shannon K.M., Nauert G.M., Perl D.P., Godbold J., Freeman T.B. 2003. A double-blind controlled trial of bilateral fetal nigral transplantation in Parkinson’s disease. Ann. Neurol. 54:403–414 10.1002/ana.10720 [DOI] [PubMed] [Google Scholar]
- Outeiro T.F., Kontopoulos E., Altmann S.M., Kufareva I., Strathearn K.E., Amore A.M., Volk C.B., Maxwell M.M., Rochet J.C., McLean P.J., et al. 2007. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science. 317:516–519 10.1126/science.1143780 [DOI] [PubMed] [Google Scholar]
- Paxinos G., Watson C. 1986. The Rat Brain: In Stereotaxic Coordinates. Second edition Academic Press, Orlando/Sydney: 237 pp [Google Scholar]
- Reeves M.B., Davies A.A., McSharry B.P., Wilkinson G.W., Sinclair J.H. 2007. Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science. 316:1345–1348 10.1126/science.1142984 [DOI] [PubMed] [Google Scholar]
- Schapira A.H. 2006. Mitochondrial disease. Lancet. 368:70–82 10.1016/S0140-6736(06)68970-8 [DOI] [PubMed] [Google Scholar]
- Schapira A.H., Cooper J.M., Dexter D., Clark J.B., Jenner P., Marsden C.D. 1990. Mitochondrial complex I deficiency in Parkinson’s disease. J. Neurochem. 54:823–827 10.1111/j.1471-4159.1990.tb02325.x [DOI] [PubMed] [Google Scholar]
- Sherer T.B., Betarbet R., Testa C.M., Seo B.B., Richardson J.R., Kim J.H., Miller G.W., Yagi T., Matsuno-Yagi A., Greenamyre J.T. 2003. Mechanism of toxicity in rotenone models of Parkinson’s disease. J. Neurosci. 23:10756–10764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shults C.W., Oakes D., Kieburtz K., Beal M.F., Haas R., Plumb S., Juncos J.L., Nutt J., Shoulson I., Carter J., et al. ; Parkinson Study Group 2002. Effects of coenzyme Q10 in early Parkinson disease: Evidence of slowing of the functional decline. Arch. Neurol. 59:1541–1550 10.1001/archneur.59.10.1541 [DOI] [PubMed] [Google Scholar]
- Zhang H.X., Du G.H., Zhang J.T. 2004. Assay of mitochondrial functions by resazurin in vitro. Acta Pharmacol. Sin. 25:385–389 [PubMed] [Google Scholar]