Gfi1b negatively regulates Rag expression through direct binding to the Rag locus and through inhibition of Foxo1; mice lacking both Gfi1b and Gfi1 exhibit a block in B cell development.
Abstract
Precise regulation of Rag (recombination-activating gene) expression is crucial to prevent genomic instability caused by the generation of Rag-mediated DNA breaks. Although mechanisms of Rag activation have been well characterized, the mechanism by which Rag expression is down-regulated in early B cell development has not been fully elucidated. Using a complementary DNA library screen, we identified the transcriptional repressor Gfi1b as negative regulator of the Rag locus. Expression of Gfi1b causes repression of Rag1 and Rag2 in cell lines and primary mouse cells. Conversely, Gfi1b-deficient cell lines exhibit increased Rag expression, double-strand breaks and recombination, and cell cycle defects. In primary cells, transcription of Gfi1b inversely correlates with Rag transcription, and simultaneous inactivation of Gfi1 and Gfi1b leads to an increase in Rag transcription early in B cell development. In addition, deletion of Gfi1 and Gfi1b in vivo results in a severe block in B cell development. Gfi1b orchestrates Rag repression via a dual mechanism. Direct binding of Gfi1b to a site 5′ of the B cell–specific Erag enhancer results in epigenetic changes in the Rag locus, whereas indirect inhibition is achieved through repression of the trans-activator Foxo1. Together, our experiments show that Gfi family members are essential for normal B cell development and play an important role in modulating expression of the V(D)J recombinase.
The adaptive immune response depends on the generation of a diverse repertoire of B and T cell antigen receptors. This is achieved through V(D)J recombination, whereby site-specific gene rearrangement takes place at Ig and TCR loci to generate unique sets of antigen receptor genes within developing lymphocytes (Tonegawa, 1983). V(D)J recombination depends on the generation of double-strand DNA breaks at recombination signal sequences (RSSs) that flank rearranging gene segments. These breaks are introduced by a recombinase containing the Rag1 and Rag2 proteins. Because these proteins generate double-strand DNA breaks, their expression represents a threat to genomic integrity. Therefore, Rag gene expression is tightly regulated in both a lineage and a stage-specific manner.
In developing B cells, Rag genes are expressed during heavy chain and light chain gene rearrangement but down-regulated during the proliferative burst that separates these stages. Once a functional, self-tolerant BCR has been generated, Rag expression is shut off as B cells progress to the mature stage. However, if the BCR recognizes self-antigen, then Rag expression is maintained and receptor editing ensues (Schlissel, 2003; Sukumar and Schlissel, 2011). Although the mechanism of Rag-mediated DNA cleavage has been extensively studied, comparatively little is known about the factors that regulate the dynamic pattern of Rag gene expression during the course of development.
Rag1 and Rag2 are physically linked in the genome and coordinately expressed. Several regulatory elements have been identified, including a B lineage–restricted enhancer element, Erag (Hsu et al., 2003), and T lineage–restricted silencer and anti-silencer elements (Yu et al., 1999; Yannoutsos et al., 2004). E2A (Bain et al., 1994) and FoxP1 (Hu et al., 2006) bind to Erag and contribute to its activity, whereas the forkhead family transcription factor Foxo1 is critical for activating Rag expression at all stages of B cell development (Amin and Schlissel, 2008; Dengler et al., 2008). However, the process by which Rag is down-regulated at specific stages during B cell development and how it is kept inactive in nonlymphoid-lineage hematopoietic cells have not been fully explored.
We took advantage of the fact that Abelson murine leukemia virus (AMuLV) infection of total bone marrow results in the outgrowth of transformed cells that are “frozen” at the pro– to pre–B cell transition state of B cell development. These cells cycle continuously in culture, but upon treatment with the Abl kinase inhibitor STI-571 undergo a process that mimics the developmental transition to the pre–B cell stage (Muljo and Schlissel, 2003), providing a useful model system for the study of both leukemic transformation and early B cell development. We sought to identify factors critical for Rag down-regulation by conducting an unbiased retroviral cDNA library screen in AMuLV-transformed pro–B cells. This screen revealed a novel negative regulator of Rag expression, the transcriptional repressor Growth factor–independent 1b (Gfi1b).
RESULTS
A cDNA library screen identifies Gfi1b as a negative regulator of Rag expression
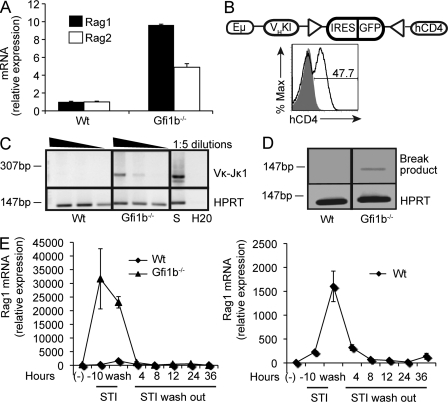
To identify factors that down-regulate Rag expression during B cell development, we used a constitutively Rag1 high AMuLV-transformed pro–B cell line, hereafter referred to as RAG1-GFPhigh, generated from a mutant mouse in which GFP was targeted to the main coding exon of Rag1 (Kuwata et al., 1999). We infected RAG1-GFPhigh reporter cells with a retroviral cDNA library generated from murine bone marrow pro– and pre–B cells (Amin and Schlissel, 2008). Infected, GFP negative reporter cells were sorted and their cDNA inserts rescued by PCR amplification and recloned into the original retroviral vector. After four rounds of enrichment, the resulting cDNAs were isolated and sequenced. A cDNA encoding the zinc finger transcriptional repressor Gfi1b was isolated on 17 occasions.
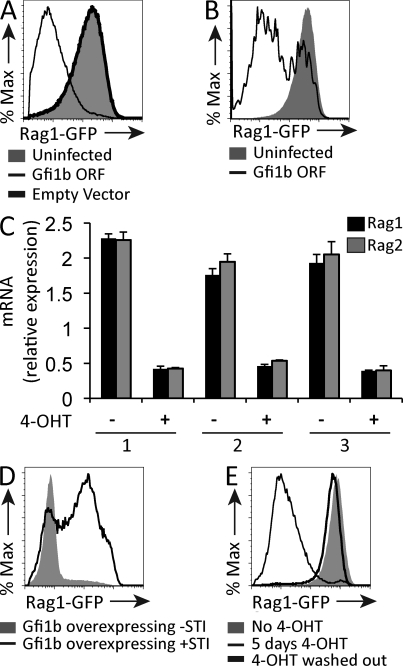
Overexpression of a Gfi1b cDNA led to a striking down-regulation of RAG1-GFP expression in RAG1-GFPhigh cells (Fig. 1 A). Gfi1b belongs to a family of transcriptional repressors that includes its close relative Gfi1 (Fiolka et al., 2006). Overexpression of Gfi1 also led to down-regulation RAG1-GFP expression in RAG1-GFPhigh cells (Fig. 1 B). We went on to engineer an overexpression vector containing the Gfi1b cDNA fused to cDNA encoding a modified human estrogen receptor (ER) hormone binding domain (Gfi1b-ER) so that Gfi1b nuclear translocation could be induced by addition of tamoxifen (4-OHT) to the culture medium (Metzger et al., 1995). Tamoxifen treatment of RAG1-GFPhigh cells expressing the Gfi1b-ER construct caused a threefold reduction of both Rag1 and Rag2 transcripts within 12 h (Fig. 1 C).
Figure 1.
Overexpression of Gfi1B causes down-regulation of RAG1-GFP transcript levels. (A) Flow cytometric analysis of RAG1-GFP expression in RAG1-GFPhigh cells isolated from a reporter mouse where GFP replaces exon1 of Rag1 and infected with a retroviral construct overexpressing the Gfi1B ORF or empty vector control. Data are representative of five independent experiments. (B) Flow cytometric analysis of RAG1-GFP expression in RAG1-GFPhigh cells overexpressing Gfi1. RAG1-GFP levels were measured 3 d after infection. Data are representative of three independent experiments. (C) Quantitative RT-PCR of Rag1 and Rag2 mRNA transcripts in RAG1-GFPhigh cells infected with a Gfi1B-ER construct and treated (+) or not (−) with tamoxifen (4-OHT) for 12 h. Numbers 1–3 indicate biological replicates. Levels are expressed relative to HPRT transcripts. Error bar represents standard deviation of triplicate PCR assays. (D) Flow cytometric analysis of RAG1-GFPhigh cells overexpressing Gfi1b cultured in the presence or absence of 2.5 µM of the Abl kinase inhibitor STI-571 (Gleevec) for 12 h. Data are representative of two independent experiments. (E) Flow cytometric analysis of RAG1-GFPhigh cells infected with a retroviral Gfi1b-ER construct. Cells were treated or not with tamoxifen (4-OHT) for 5 d and, where indicated, were washed and cultured for 9 d thereafter. Data are representative of two independent experiments.
Because Rag transcription is transiently, not permanently, down-regulated at the early pre–B cell stage, we asked whether down-regulation of Rag by Gfi1b is reversible. STI-571 causes up-regulation of Rag transcription in Abelson cells (Muljo and Schlissel, 2003). Treatment of Gfi1b-overexpressing RAG1-GFPhigh cells with STI-571 partially reversed down-regulation of Rag expression (Fig. 1 D). In addition, washing out tamoxifen from the culture medium of Gfi1b-ER–expressing cells completely restored RAG1-GFP expression (Fig. 1 E), indicating that Gfi1b’s effect on Rag transcription is reversible.
Expression of Gfi1b represses Rag transcription in primary B-lineage cells and inhibits differentiation
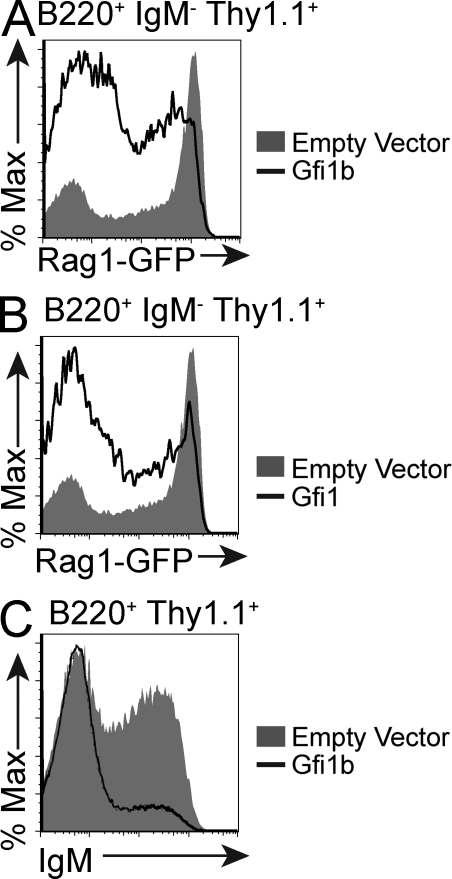
We next asked whether forced Gfi1b overexpression could modulate Rag transcription in primary developing B cells. We infected cultured total bone marrow cells isolated from RAG1-GFP reporter mice with a Gfi1b-expressing retrovirus and compared levels of RAG1-GFP in these cells to bone marrow cells infected with an empty vector. RAG1-GFP levels were lower in early B cells that overexpressed Gfi1b when compared with those infected with the empty vector (Fig. 2 A). This was also the case for Gfi1 (Fig. 2 B).
Figure 2.
Gfi1b overexpression represses Rag expression in primary cells. (A) Flow cytometric analysis of GFP levels in cultured bone marrow cells from RAG1-GFP heterozygous mice infected with a Thy1.1-marked retrovirus overexpressing either Gfi1b or an empty vector. Plots show populations gated according to B220, IgM, and Thy1.1 expression as indicated. Data are representative of five independent experiments. (B) Flow cytometric analysis of RAG1-GFP expression in IL-7–cultured bone marrow cells from RAG1-GFP heterozygous mice infected with a retroviral Gfi1 overexpression construct or an empty vector. This plot is representative of three different experiments. (C) Flow cytometric analysis of IgM expression in IL-7–cultured bone marrow cells from RAG1-GFP heterozygous mice infected with a retroviral Gfi1b overexpression construct or an empty vector. Data are representative of three independent experiments.
If Gfi1b controls the expression levels of genes critical for B cell development, we predict that overexpression of Gfi1b in bone marrow cells would impair their ability to differentiate to the IgM-positive stage. We thus compared the numbers of IgM-positive cells that accumulate in bone marrow cultures infected with the Gfi1b-expressing virus or an empty vector control. Fewer IgM-positive, Gfi1b-overexpressing cells accumulate when compared with cells infected with the empty vector (Fig. 2 C). Whether this effect is directly and exclusively related to Gfi1b’s repression of Rag is a topic of future study.
Targeted disruption of Gfi1b and Gfi1 causes a severe block in B cell development in vivo
Both Gfi1 and Gfi1b are expressed during the early stages of B cell development (Yücel et al., 2004; Vassen et al., 2007). Targeted disruption of Gfi1 in the mouse causes a defect in differentiation from lin−, sca+, c-kit+ (LSK) multipotent progenitors to B220+ B cells (Rathinam and Klein, 2007). Defects in IL-7 receptor signaling also impair B cell development at the pro–B cell stage in Gfi1-deficient mice. Although Gfi1’s effect on B cell development has been well characterized (Rathinam and Klein, 2007), Gfi1b’s role within the B cell compartment has remained elusive because Gfi1b-deficient mice die at day 15 of gestation (Saleque et al., 2002).
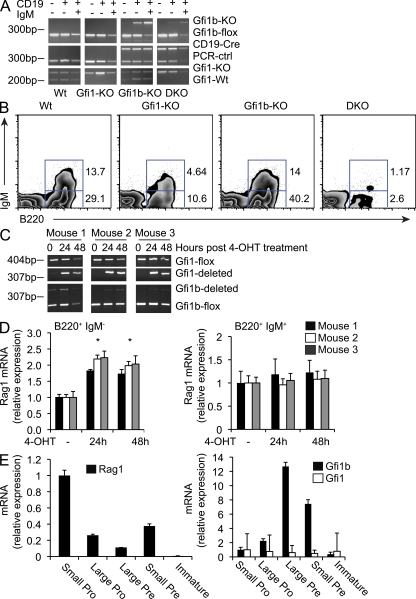
To examine the role of Gfi1b in Rag regulation in vivo, we bred mice carrying a floxed allele of Gfi1b (Khandanpour et al., 2010) with MB-1-Cre mice that express Cre starting at the pro–B cell stage of development (Hobeika et al., 2006). MB-1-Cre–driven deletion of Gfi1b within the B cell compartment did not alter Rag transcript levels or significantly impair B cell development at any of the developmental stages examined (unpublished data), suggesting significant functional redundancy between Gfi1 and Gfi1b as previously demonstrated (Fiolka et al., 2006; van der Meer et al., 2010). This is consistent with our observation that overexpression of Gfi1 in cell lines and in primary cells phenocopies overexpression of Gfi1b (Fig. 1 B and Fig. 2 B) and led us to consider whether Gfi1 could be compensating for the lack of Gfi1b in the B cell–specific Gfi1b KO mouse. To test this idea, we mated a CD19-Cre Gfi1bflox/flox mouse to a Gfi1 KO mouse to obtain B cell–specific Gfi1/Gfi1b double KO animals (DKO; Fig. 3 A). Gfi1/Gfi1b DKO animals were born at less than the expected Mendelian ratios (unpublished data). Flow cytometry revealed that the percentage of B220+IgM− cells in the bone marrow of Gfi1/Gfi1b DKO animals is much lower compared with WT controls or Gfi1b and Gfi1 single KO animals (Fig. 3 B), indicating a very early block in B cell development. The percentage of B220+IgM+ cells is also very low in DKO animals compared with WT and single KO controls (Fig. 3 B). Thus, the defects in B cell development previously described for Gfi1 KO animals (Rathinam and Klein, 2007) are exacerbated when both Gfi1 and Gfi1b are ablated (Fig. 3 B). Although Gfi1 appears to compensate for a deficiency in Gfi1b, we observed that Gfi1b does not fully compensate for a deficiency in Gfi1 during B cell development (Fig. 3 B). This may be a result of the fact that Gfi1 deficiency also results in a hematopoietic stem cell proliferative defect, as well as a defect in the transition from LSK multipotent progenitors to B220+ B cells (Rathinam and Klein, 2007). It is possible that Gfi1b plays less of a role in these critical early points in development, and thus exhibits a weaker developmental phenotype when eliminated. These data point to a partially redundant but crucial role for these transcription factors in B cell development.
Figure 3.
Inactivation of both Gfi1 and Gfi1b results in a severe block in B cell development. (A) Genotyping results for Gfi1+/− (Wt), Gfi1 KO (Gfi1-KO), Gfi1+/− CD19-CRE–induced Gfi1b KO (Gfi1b-KO), and Gfi1/1b DKO mice. PCR was performed on genomic DNA from sorted CD19+IgM+, CD19+IgM−, and CD19−IgM− cells. This data representative of three similar analyses. (B) Flow cytometric analysis of B220 and IgM expression on bone marrow cells from Wt, Gfi1-KO, Gfi1b-KO, or DKO mice as indicated. Although B cell numbers in the DKO samples were variable, this result is representative of three similar analyses. (C) PCR analysis for the presence or absence of the Gfi1 and Gfi1b floxed alleles in bone marrow isolated from Gfi1bflox/flox Gfi1flox/flox ER-Cre mice and treated with tamoxifen (4-OHT) for the indicated time points. This data are representative of two independent analyses. (D) Quantitative RT-PCR analysis of Rag transcripts in IL-7–cultured B220+IgM− (left) and B220+IgM+ (right) bone marrow cells sorted from Gfi1bflox/flox Gfi1flox/flox ER-Cre mice and treated with tamoxifen (4-OHT) or vehicle control. Data are shown for three mice and is representative of three independent experiments. Error bar represents standard deviation of triplicate PCR assays. * indicates a significant difference according to a Student’s t test (P ≤ 0.004 for all mice analyzed). (E) Quantitative RT-PCR analysis of Rag, Gfi1, and Gfi1b transcripts in sorted bone marrow cell fractions from WT mice. Cells were sorted as follows: small pro, IgM−B220+CD43+ forward scatter (FSC) low; large pro, IgM−B220+CD43+ FSC high; large pre, IgM−B220+CD43− FSC high; small pre, IgM−B220+CD43− FSC low; immature B, IgM+IgD−. Error bar represents standard deviation of triplicate PCR assays. This data are representative of three independent experiments.
Gfi1 and Gfi1b repress Rag transcription in vivo
To further assess the role of Gfi1 and Gfi1b during B cell development, we bred a Gfi1bflox/flox Gfi1flox/flox mouse to a mouse carrying an ER-Cre transgene at the Rosa26 locus (Ventura et al., 2007). Total bone marrow was isolated from these mice and cultured in the presence of IL-7 with either a vehicle control or with tamoxifen to delete both Gfi1 and Gfi1b. Tamoxifen-treated and control cells were then sorted into B220+IgM− and B220+IgM+ fractions and RNA was purified from each population. PCR assays were used to check for deletion of the floxed alleles (Fig. 3 C). It should be noted that mouse 1 exhibited partial deletion of the floxed Gfi1b allele before tamoxifen treatment; we believe this to be the result of leaky expression of the ER-Cre transgene. We found that transcription of Rag1 increased upon treatment with tamoxifen specifically in the IgM− fraction (Fig. 3 D, left), whereas this was not the case for IgM+ cells (Fig. 3 D, right). Differences in Rag transcription in IgM− cells were statistically significant for each mouse analyzed (calculated P ≤ 0.004 according to a Student’s t test). However, deletion of the floxed alleles for both Gfi1 and Gfi1b was not complete within the timeframe of tamoxifen treatment (Fig. 3 C). This implies that the observed increase in Rag transcription in tamoxifen-treated IgM− cells likely represents an underestimate of the true effect of deleting Gfi1 and Gfi1b. Although a longer period of tamoxifen treatment may have resulted in a more complete deletion of the floxed alleles, we observed an increase in cell death at later time points, thus confounding the interpretation of the data after 48 h of tamoxifen treatment. The increase in Rag transcription in IgM− cells that occurs upon deletion of the floxed alleles of Gfi1 and Gfi1b indicates that these proteins are bona fide repressors of Rag transcription in vivo, and that their repressive activity operates mainly at the pro–and pre–B cells stages of B cell development. The fact that this effect is observed specifically in IgM− cells is consistent with the fact that transcription of Gfi1 and Gfi1b ceases at the immature stage of B cell development (Vassen et al., 2007). Thus, we would not necessarily expect that deletion of Gfi1 and Gfi1b would result in an increase in Rag transcription in cells that have already progressed to the IgM+ stage.
Finally, we found that levels of Rag transcription were inversely correlated with levels of Gfi1b but not Gfi1 transcription in pro– and pre–B cells sorted directly from WT bone marrow (Fig. 3 E). Gfi1b transcript levels were highest in actively dividing cells during the large pre–B cell stage of development where Rag transcription is low. Gfi1b transcript levels decreased at the small pre–B cell stage during which Ig light chain rearrangement takes place, correlating with an increase in Rag transcription (Fig. 3 E). The fact that transcription of Gfi1 does not vary significantly during the early stages of development (Fig. 3 E) may indicate that, of the two proteins, Gfi1b plays the more dominant role, although this has not been conclusively shown in our analysis. The fact that simultaneous deletion of Gfi1 and Gfi1b causes an increase in Rag transcription in pro– and pre–B cells, together with the inverse relationship between transcription of Gfi1b and Rag in cycling versus actively rearranging cells, point to a role for repression of Rag transcription by Gfi1b during early B cell development.
Studies in Abelson cells reveal a dual mechanism of repression of Rag transcription by Gfi1b
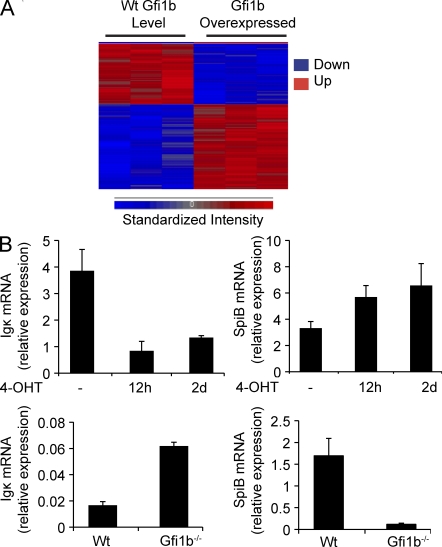
A more global role for Gfi1 and Gfi1b during B cell development is supported by the fact that expression of Gfi1b in Abelson cells results in changes in expression of a large number of genes (Fig. 4). Microarray analysis revealed ∼300 genes, including Rag1 and Rag2, that showed significant differences in transcript levels (P < 0.01) when Gfi1b-ER–expressing cells were exposed to tamoxifen for 12 h, as measured by an ANOVA statistical test (Miller et al., 2002; Fig. 4 A). Several regulated genes, including SpiB, c-Rel, Aiolos, Igβ, Blk, Id2, and Zap70, are critical for B cell development. We validated our microarray results using quantitative real-time RT-PCR (Fig. 4 B). Genes whose transcripts were up-regulated or down-regulated by at least twofold (P < 0.01) are shown in Tables S1 and S2.
Figure 4.
Gfi1b regulates transcription of a large number of genes in AMuLV pro–B cells. (A) Hierarchically clustered heat map of genes whose transcription level is significantly altered (P < 0.01) upon retroviral expression of Gfi1b in RAG1-GFPHigh cells. Each column represents an independent replicate. (B, top) Quantitative RT-PCR analysis of germline Igκ and SpiB transcripts relative to HPRT in RNA from Gfi1b-ER–overexpressing AMuLV cells treated with tamoxifen (4-OHT) as indicated. (B, bottom) Quantitative RT-PCR analysis of these same transcripts relative to HPRT using RNA from wt or Gfi1b−/− AMuLV cells. Error bar represents standard deviation of triplicate PCR assays.
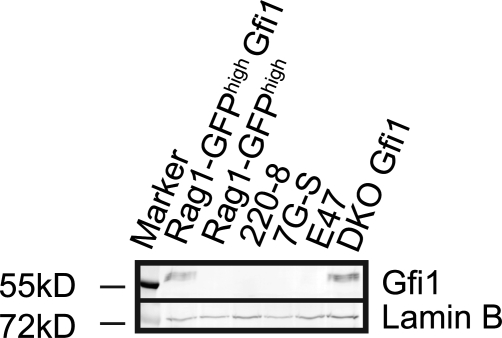
Because of the technical limitation involved in modeling the absence of Gfi1 and Gfi1b proteins in vivo (almost no B cells are produced), we turned to the Abelson cell system to learn more about the role of both proteins in the regulation of Rag expression. We transformed cultured bone marrow from poly-IC–treated, Gfi1bflox/flox × Mx-Cre mice with AMuLV to generate a Gfi1b−/− pro–B cell line (Khandanpour et al., 2010). Gfi1−/− and DKO-transformed cell lines were created in parallel. One peculiarity of the Abelson cell system is that although Gfi1b is expressed normally, Gfi1 is not expressed in WT (Wt) cells (unpublished data), even though both proteins are normally expressed by early B cells in vivo (Yücel et al., 2004). We performed Western blots on a panel of independently generated Abelson lines to address whether the absence of Gfi1 was a peculiarity of our newly generated cell lines, or a general feature of Abelson-transformed cells. A positive control for Gfi1 expression was provided by using two different cell lines transduced with a retrovirus to express Gfi1 (RAG1-GFPhigh Gfi1 and DKO Gfi1). We found that none of the independently generated Abelson cell lines expressed detectable levels of Gfi1 (Fig. 5). Thus, the lack of Gfi1 expression in Abelson cells appears to be a general feature of these transformed cells. These data also indicate that the high level of Rag transcription observed in the RAG1-GFPhigh cell line used in the screen is likely independent of the lack of Gfi1 expression in these cells. Because Gfi1 is not expressed in Abelson cells, we focused our attention on the mechanism of repression of Rag by Gfi1b in this system.
Figure 5.
Anti-Gfi1 Western blot analysis of four independently generated Abelson cell lines and two lines retrovirally transduced with a Gfi1 cDNA as a positive control (RAG1-GFPhigh Gfi1 and DKO Gfi1). Anti-Lamin B was used to control for loading. Data are representative of two independent experiments.
Gfi1b deficiency results in increased Rag expression and recombination
We first verified that deletion of Gfi1b in Abelson cells results in higher levels of endogenous Rag transcription. As in our in vivo studies, we found that Rag1 and Rag2 transcripts were expressed at higher levels in Gfi1b−/− cells compared with their WT (Wt) counterparts in pools of Abelson-transformed mutant cells (Fig. 6 A).
Figure 6.
The deletion of Gfi1b results in increased Rag transcription and gene rearrangement. (A) Quantitative RT-PCR analysis of Rag transcripts in Wt and Gfi1b−/− AMuLV cell lines. Levels are expressed relative to HPRT transcripts and consist of the mean of triplicate assays. (B) Flow cytometric analysis of Wt and Gfi1b−/− AMuLV cell lines infected with a rearrangement reporter construct diagrammed above. Upon rearrangement, the IRES-GFP segment is deleted and hCD4 is expressed. Eμ, IgHC enhancer; VHKI, VH gene promoter; triangles, RSSs. Data are representative of two independent experiments. (C) PCR assay for Vκ-Jκ1 coding joints in a series of diluted DNA samples from Wt and Gfi1b−/− AMuLV cell lines. S, splenic DNA; H2O, water-only control. Data are representative of two independent experiments. (D) LM-PCR analysis of DNA breaks at the Jκ1 RSS in Wt and Gfi1b−/− AMuLV cells. The inverted image of an agarose gel analysis of PCR products is shown. Data are representative of two independent experiments. (E, left) Quantitative RT-PCR analysis of Rag1 transcript levels (relative to HPRT) in Wt and Gfi1b−/− AMuLV cell lines treated for the indicated times with STI-571, and, where indicated, washed and cultured for an additional 36h. (E, right) Replot of data shown on the left on a smaller scale to reveal values from the wt cell line. Error bars denote standard deviation of triplicate assays. Data are representative of two independent experiments.
To ascertain whether higher levels of Rag expression in Gfi1b−/− cell lines result in greater Ig gene rearrangement potential, we infected wt or Gfi1b−/− Abelson cells with a V(D)J recombination reporter construct. Upon rearrangement, an ires-GFP sequence surrounded by two RSSs is deleted, bringing a human CD4 gene under control of a heavy chain gene promoter element. Cells having undergone a rearrangement event gain hCD4 expression. We observed higher levels of recombination in cells lacking Gfi1b when compared with Wt cells (Fig. 6 B). In addition, we detected higher levels of V-to-J rearrangements at the endogenous Igκ locus in these same mutant cells (Fig. 6 C), as well as higher levels of Jκ signal end dsDNA breaks, an intermediate in the recombination pathway (Schlissel et al., 1993; Fig. 6 D). We conclude that higher levels of Rag transcripts in cells lacking Gfi1b result in a higher rearrangement potential in these cells, indicating that one role for Gfi1 or Gfi1b proteins in early B cell development may be to prevent deregulated rearrangement during periods of proliferation or after production of a functional BCR.
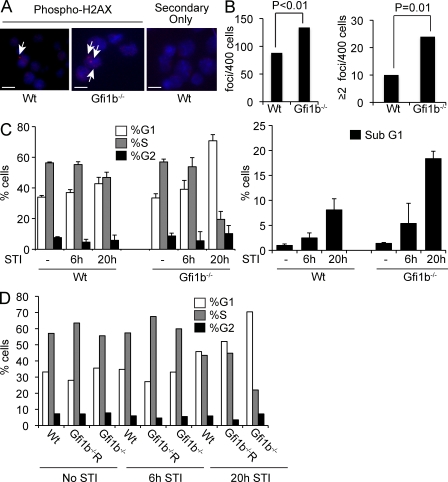
Treatment of AMuLV-transformed cells with STI-571 mimics key aspects of the transition from the large pro–B to the small pre–B stage in B cell development (Muljo and Schlissel, 2003). To understand how loss of Gfi1b might affect this transition, we treated wt and Gfi1b−/− AMuLV-transformed cell lines with STI-571 and monitored Rag transcription. Rag transcription was induced to strikingly high levels in cells deleted for Gfi1b (Fig. 6 E). This led us to hypothesize that a biological role for Gfi1b might be to limit Rag protein levels during rearrangement so that promiscuous DNA cleavage does not occur at rearranging loci, cryptic RSSs, or elsewhere in the genome. To test whether global levels of double-strand breaks are higher in cells deleted for Gfi1b, we performed immunofluorescence experiments using an antibody to phospho-H2AX, a histone variant found in association with double-strand DNA breaks (Ayoub et al., 2009). We found higher numbers of phospho-H2AX foci per cell in Gfi1b−/− cells when compared with wt cells (Fig. 7, A and B). Because Rag proteins generate double-strand breaks, we interpret these data to indicate that Gfi1b’s expression within the B cell compartment may prevent Rag proteins from reaching levels high enough to cause genomic instability.
Figure 7.
Deletion of Gfi1b leads to Rag deregulation. (A) Immunofluorescence of Wt and Gfi1b−/− AMuLV-transformed cells stained with anti–phospho-H2AX antibodies and counterstained with DAPI. Arrows indicate foci of anti–phospho-H2AX staining. Bars, 10 µM. Data are representative of three independent experiments. (B) Quantitation of data from the experiment in A. The total number of H2AX foci/cell was counted (left) for 400 cells. Data were analyzed using Student’s t test. Right, number of cells with two or more foci/cell, n = 400. Data are representative of 3 biological replicates. (C, left) Cell cycle analysis using PI staining in Wt and Gfi1b−/− AMuLV-transformed cell lines treated with STI-571 (STI) for the indicated times. (C, right) Percentage of cells with sub-G1 amounts of DNA indicating apoptosis. Error bar represents standard deviation of flow cytometry cell cycle analysis performed in triplicate. Data are representative of three independent experiments. (D) Cell cycle analysis of PI stained AMuLV cells treated with STI-571 and analyzed by flow cytometry. Gfi1bR, Gfi1b−/− cells reconstituted with a Gfi1b expression vector. Data are representative of two independent experiments.
We tested whether treatment with STI-571 causes perturbations in the cell cycle in Gfi1b−/− cells. We performed PI staining on Gfi1b−/− cells treated with STI-571 and compared the cell cycle profile in these cells to that of identically treated wt cells. Treatment with STI-571 led to a greater number of mutant cells in the G1 phase and fewer in S phase of the cell cycle compared with wt cells (Fig. 7 C). In addition, a much greater fraction of Gfi1b−/− cells had sub-G1 DNA content, indicating increased apoptosis (Fig. 7 C). We verified that these cell cycle effects were specific to Gfi1b by reconstituting a Gfi1b−/− cell line with Gfi1b and treating these cells with STI-571. Cell cycle profiles in the reconstituted cell line were nearly identical to those observed in the wt cell line (Fig. 7 D). We conclude that Gfi1b protects Abelson cells from apoptosis and cell cycle arrest caused by treatment with STI-571.
Gfi1b binds directly to the Rag locus and recruits chromatin modifying complexes to repress Rag in a replication-dependent manner
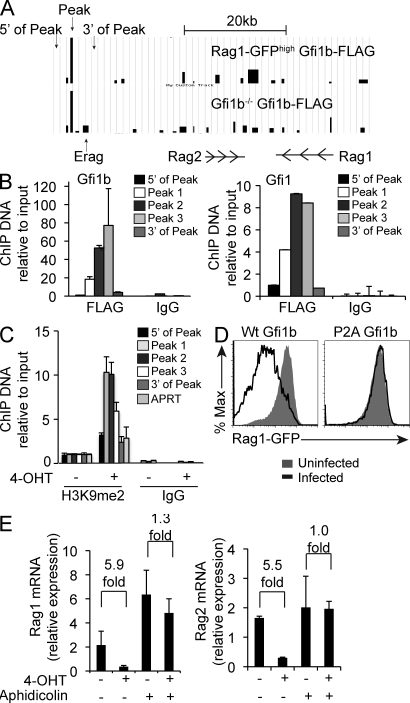
To test whether Gfi1b down-regulates Rag transcription by directly binding to the locus, we performed a chromatin immunoprecipitation (ChIP)–Chip analysis using a FLAG epitope-tagged Gfi1b construct expressed in both the RAG1-GFPhigh cell line as well as a Gfi1b null cell–cell line. We found significant amounts of Gfi1b binding at a region ∼35 kb 5′ of the Rag2 first exon, neighboring the B cell–specific enhancer element Erag (Hsu et al., 2003; Fig. 8 A). A cluster of three AATC core Gfi1b binding sites lies within the peak (chr2 101,399,676-101399,733; Fig. S1). We then performed ChIP experiments using an anti-FLAG or IgG control antibody followed by quantitative real-time PCR with three primer sets within the peak of Gfi1b binding and two primer sets flanking the peak (Fig. 8 A). Larger amounts of Gfi1b bound within the peak when compared with the regions 5′ and 3′ of the peak, confirming the array result (Fig. 8 B, left). Similar results were obtained by performing ChIP using an epitope-tagged Gfi1 construct (Fig. 8 B, right). These results indicate that both Gfi1 and Gfi1b can bind directly to the Rag locus.
Figure 8.
Gfi1b binds directly to the Rag locus and acts through a chromatin modifying mechanism. (A) α-FLAG ChIP-Chip on RAG1-GFPhigh or Gfi1b−/− cells overexpressing FLAG-tagged Gfi1b analyzed using a custom genomic tiling array. Data are shown for the region upstream and including Rag1 and Rag2 as indicated. Arrows denote the location of primer sets used in B. (B) Quantitative PCR of DNA recovered from ChIP using Gfi1b−/− cells overexpressing FLAG-tagged Gfi1b (left) or Gfi1 (right). IgG is a control IP done in parallel and error bars indicate standard deviation of triplicate assays. (C) Quantitative PCR of H3K9me2 ChIP products in RAG1-GFPhigh Gfi1b-ER cells treated for 3 d in the presence (+) or absence (−) of tamoxifen (4-OHT). Error bar represents standard deviation of triplicate PCR assays. Data are representative of two independent experiments. (D) Flow cytometric analysis of RAG1-GFP levels in RAG1-GFPhigh cells infected with a retrovirus expressing either a wt or P2A mutant Gfi1b protein compared with uninfected cells. Data are representative of two independent experiments. (E) Quantitative RT-PCR of Rag1 and Rag2 transcripts relative to HPRT in RAG1-GFPhigh cells infected with a retroviral Gfi1b-ER construct and treated (+) or not (−) with tamoxifen (4-OHT) for 12 h in the presence (+) or absence (−) of the DNA replication inhibitor aphidicolin. Error bar represents standard deviation of triplicate PCR assays. Numbers indicate fold differences with error bars from triplicate assays.
Target gene repression by Gfi1b is mediated through its association with cofactors that introduce local chromatin modifications (Vassen et al., 2006; Saleque et al., 2007). H3K9 dimethylation is characteristic of de novo silencing (Fuks, 2005), and in other systems, levels of dimethylated H3K9 increase as a result of Gfi1b-mediated recruitment of histone methyltransferases (Vassen et al., 2006; Saleque et al., 2007). We tested whether Gfi1b overexpression altered the level of dimethylated H3K9 at the Gfi1b binding region (GBR) identified by ChIP-Chip. After treatment with tamoxifen, cells infected with the Gfi1b-ER construct had higher levels of H3K9me2 in the GBR (Peak 1–3) of the Rag locus, whereas levels of H3K9me2 were lower in regions 5′ and 3′ of the GBR and at the APRT control locus (Fig. 8 C). Because the SNAG domain of Gfi1b is necessary for its repressor activity (Fiolka et al., 2006), we tested whether mutating this domain would abrogate Gfi1b’s effect on Rag transcription. Mutation of aa 2 in Gfi1b from a proline to an alanine (P2A mutation) does not disrupt its DNA binding activity but does eliminate its association with chromatin modifying cofactors (Fiolka et al., 2006; Saleque et al., 2007). We found that the P2A mutant fails to down-regulate RAG1-GFP when overexpressed (Fig. 8 D). These data imply that Gfi1b’s effect on Rag transcription depends on its association with chromatin-modifying cofactors.
In some systems, changes in patterns of epigenetic gene regulation require DNA replication (Falbo and Shen, 2009). We asked whether treating RAG1-GFPhigh cells with a DNA replication inhibitor would affect Gfi1b’s ability to down-regulate Rag transcription. Although induction of Gfi1b-ER with tamoxifen resulted in decreased Rag mRNA levels, we found that this same treatment had little effect on Rag transcription in the presence of the DNA synthesis inhibitor aphidicolin (Fig. 8 E). As Abelson cells divide approximately every 12 h, the repression of Rag by Gfi1b observed within 12 h of tamoxifen treatment (Fig. 1 C) is consistent with the requirement for replication. Collectively, these data imply that Gfi1b’s effect on Rag transcription is mediated primarily through a replication-dependent chromatin-modifying mechanism.
The Rag locus GBR is necessary for complete down-regulation of Rag expression
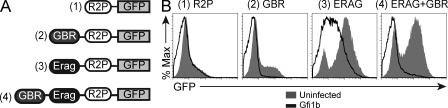
To assess the functional activity of the GBR identified by ChIP-Chip, we cloned a 700-bp fragment encompassing this sequence into a reporter construct containing the Rag2 promoter (R2P) driving GFP and analyzed expression after stable transfection. In addition, we cloned Erag alone and in combination with the GBR upstream of R2P (Fig. 9 A). All reporter constructs were stably integrated into the genome using a selectable drug marker before analyses were performed. The promoter alone supported low levels of GFP expression and inclusion of the GBR upstream of the promoter had little effect on GFP expression. The Erag enhancer substantially increased the fraction of GFP-positive cells when positioned upstream of R2P; however, the presence of the GBR in combination with the Erag enhancer led to a decrease in the fraction of GFP-positive cells (Fig. 9 B). Overexpression of Gfi1b had little effect on cells containing the construct with R2P alone but did decrease GFP expression in the construct also containing the GBR (Fig. 9 B). Overexpression of Gfi1b in cells containing the Erag construct led to a partial reduction in GFP expression, whereas overexpression in cells containing the GBR in combination with Erag led to a complete inactivation of GFP expression (Fig. 9 B). We conclude that binding of Gfi1b to the GBR has a functional effect and that Gfi1b is a direct repressor of Rag transcription. In addition, the data indicate that Gfi1b might act on the Rag locus via an indirect mechanism. We did not see evidence of Gfi1b binding to Erag by ChIP-Chip (Fig. 8 A) or by conventional ChIP (unpublished data), yet Gfi1b can partially inhibit reporter construct activity in the presence of Erag alone (Fig. 9 B). In light of this indirect effect, we hypothesized that Gfi1b might also regulate expression of a positively acting factor that binds to Erag. Because Foxo1 binds to Erag and directly activates Rag transcription during B cell development (Amin and Schlissel, 2008; Dengler et al., 2008), we went on to test whether Gfi1b has an indirect effect on Rag transcription through regulation of Foxo1.
Figure 9.
The Gfi binding region within the RAG locus has a functional effect on RAG expression. (A) Reporter constructs used to assay RAG locus elements for effects on promoter activity. Erag, B cell–specific Rag locus enhancer; GBR, Gfi binding region; GFP, green fluorescent protein. (B) Flow cytometric analysis of GFP expression in AMuLV pro–B cells with stably integrated reporter constructs wherein GFP is driven by the indicated Rag locus DNA binding elements. Cells were analyzed in the absence or presence of a retroviral Gfi1b overexpression construct. Data are representative of two independent experiments.
Gfi1b represses Foxo1, a positive regulator of Rag expression
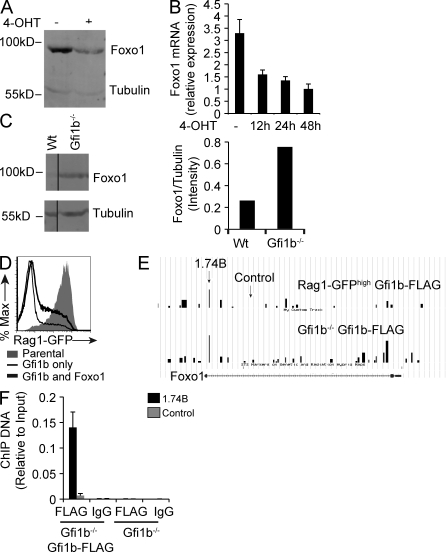
We assayed Foxo1 mRNA and protein levels in AMuLV-transformed pro–B cells overexpressing Gfi1b, as compared with control pro–B cells, and found that levels of Foxo1 protein and mRNA are decreased in cells overexpressing Gfi1b (Fig. 10, A and B). Conversely, levels of Foxo1 are higher in mutant pro–B cells lacking Gfi1b (Fig. 10 C). To test whether Gfi1b’s effect on Rag transcription is solely mediated through Foxo1, we sorted Gfi1b-overexpressing Rag reporter cells and asked whether Foxo1 overexpression could restore Rag transcripts to a high level. We found that overexpression of Foxo1 led to a partial rescue of Rag transcription (Fig. 10 D), indicating that Gfi1b likely uses multiple pathways to down-regulate Rag transcription in early B cells. ChIP-Chip analysis at the Foxo1 locus revealed a peak of Gfi1b binding at a site 3′ to the first exon of Foxo1 (Fig. 10 E), and these results were confirmed using conventional ChIP (Fig. 10 F). Thus, Gfi1b’s partial repression of GFP expression in the Erag reporter construct (Fig. 9 B) might be mediated by repression of Foxo1. Because both Erag and the GBR are necessary for complete down-regulation of reporter construct activity (Fig. 9 B), we conclude that Gfi1b regulates Rag expression via a dual mechanism: by direct binding to the locus, and by repression of the trans-activating Foxo1.
Figure 10.
Gfi1b’s effect on Rag transcription is mediated in part through Foxo1. (A) Anti-Foxo1 Western blot analysis of whole cell extracts from RAG1-GFPhigh cell lines infected with a Gfi1b-ER retroviral overexpression construct and treated (+) or not (−) with tamoxifen (4-OHT) for 12 h. Tubulin was used as a loading control. Data are representative of three independent experiments. (B) Quantitative RT- PCR assay for Foxo1 transcripts in RNA from Gfi1b-ER–overexpressing RAG1-GFPhigh cells treated or not (−) with tamoxifen (4-OHT) for the indicated times. Error bars denote standard deviation of triplicate assays. (C, left) Anti-Foxo1 Western blot using whole cell extracts from the indicated AMuLV cell lines and anti-tubulin as a loading control. (C, right) Quantitation of this Western blot showing the ratio of Foxo1 to tubulin signals. Data are representative of three independent experiments. (D) Flow cytometric analysis of RAG1-GFP expression in RAG1-GFPhigh parental cells, or RAG1-GFPhigh cells overexpressing Gfi1b alone, or Gfi1b and Foxo1. Data are representative of two independent experiments. (E) α-FLAG ChIP-Chip analysis of RAG1-GFPhigh and Gfi1b−/− cells overexpressing FLAG-tagged Gfi1b analyzed using a custom genomic tiling array. Data are shown for the Foxo1 gene locus. Arrows indicate location of primer sets used in F. (F) Quantitative PCR analysis of the indicated ChIP samples conducted on Gfi1b−/− cells overexpressing FLAG-tagged Gfi1b or on untransduced Gfi1b−/− cells using either the 1.74B or control primer sets. Error bar represents standard deviation of triplicate PCR assays. Data are representative of two independent experiments.
DISCUSSION
Mouse models in which GFP has been knocked into either the Gfi1 or Gfi1b locus have elucidated where these genes are expressed within the hematopoietic system. Gfi1 is highly expressed in early B cells and in T cells, where its activity peaks at the pre-TCR stage. Gfi1 expression is absent in mature B and T cells but is reinduced when T cells become activated (Yücel et al., 2004). Gfi1 is also expressed in hematopoietic stem cells, common lymphoid progenitors, and in monocytes, granulocytes, and their progenitors. It is absent in common myeloid progenitors, as well as in megakaryocytes, erythroid cells, and their progenitors (Zeng et al., 2004). Although expressed in early B and T cells, Gfi1b is not induced upon T cell activation. Unlike Gfi1, it is not expressed within the macrophage/granulocyte lineages. Instead, it is expressed in erythroid and megakaryocyte cells and their monocyte progenitors (MEP; Vassen et al., 2007).
We have identified a new role for the transcriptional repressors Gfi1 and Gfi1b as potent negative regulators of Rag expression in B-lineage cells (Figs. 1, 2, and 3). Our data indicate that Gfi1b represses Rag transcription in pro– and pre–B cells in vivo, and that transcription of this protein inversely correlates with levels of Rag transcription in early B lineage cells (Fig. 3). Using the Abelson system, we further demonstrate that Gfi1 and Gfi1b bind directly to a region upstream of Erag, and that binding of these proteins is followed by changes in chromatin structure at the Rag locus (Fig. 8).
Our efforts to define distinct roles for Gfi1 and Gfi1b in the regulation of Rag transcription have been complicated by several factors. Although it has been proposed that target genes may exist that are specific for Gfi1 and not Gfi1b (Fiolka et al., 2006), both proteins have many overlapping targets and exhibit functional redundancy (Fiolka et al., 2006; van der Meer et al., 2010). In addition, Gfi1 and Gfi1b both auto- and cross-regulate one another’s expression (Doan et al., 2004; Yücel et al., 2004; Huang et al., 2005; Vassen et al., 2005; Laurent et al., 2009). This may explain why the single deletion of either Gfi1 or Gfi1b in vivo does not alter the level of Rag transcription in developing B cells (unpublished data). However, deletion of both proteins simultaneously results in an increase in Rag transcription in IgM− progenitor B cells, indicating that Gfi1 and Gfi1b are bona fide repressors of Rag transcription in vivo. Although it is clear from the single KO studies that Gfi1 can fully compensate for the absence of Gfi1b, transcriptional profiling of the two proteins reveals that transcription of Gfi1b, and not Gfi1, is inversely correlated with levels of Rag transcription (Fig. 3 E). This may point to a more dominant role for Gfi1b in controlling Rag transcription in vivo.
Rag-mediated double-strand DNA breaks can lead to senescence, apoptosis, or chromosomal abnormalities, such as large deletions or translocations. Some translocations activate oncogenes and result in the development of leukemias and lymphomas (Franco et al., 2006; Schlissel et al., 2006). We find that cells lacking Gfi1b have a buildup of signal ends (Fig. 6 D) and accumulate multiple DNA breaks per cell (Fig. 7 A), and that STI-571 treatment of these cells results in a more rapid G1 arrest (Fig. 7 C). We suggest that Gfi family members may be responsible for keeping recombinase levels in check during gene rearrangement to prevent the promiscuous generation of DNA double-strand breaks. They may also be required to repress Rag after heavy chain gene rearrangement so that early B cells can progress forward in development without risking additional DNA damage, particularly in those undergoing DNA replication. This is in agreement with our finding that Gfi1b transcription peaks in cycling large pre–B cells (Fig. 3 D) and points to a role for this protein in repressing Rag specifically in early proliferating B cells.
Gfi1b and Gfi1 are encoded by genes on separate chromosomes but have very similar structures. These include an N-terminal SNAG domain that mediates association with chromatin modifying proteins, and a zinc finger C-terminal domain that allows the proteins to bind directly to DNA. The intermediate region between these domains is variable (Grimes et al., 1996; Zweidler-Mckay et al., 1996; Lee et al., 2010). Association of Gfi1 and Gfi1b with chromatin modifiers through their SNAG domains allows them to reversibly repress their targets (Saleque et al., 2007). Accordingly, we demonstrate that the integrity of the SNAG domain is required for Gfi1b-mediated repression of Rag expression and that Gfi1 and Gfi1b interact with chromatin at the Rag locus (Fig. 8), leading to an increase in the level of dimethylated H3K9 within this region. This is consistent with a recent study, reporting that Gfi1b binds to the Rag locus in a stem cell line (Wilson et al., 2010). In transcription reporter constructs containing R2P and the Erag enhancer, inclusion of the GBR is necessary for complete down-regulation, yet Gfi1b is able to mediate partial down-regulation when Erag alone is included in these constructs (Fig. 9). These data support the existence of two modes of Rag down-regulation by Gfi1b: a direct mechanism mediated by binding to the GBR within the Rag locus, and an indirect mechanism mediated by its repression of Foxo1, an activator of Erag (Fig. 10). The interplay between Foxo1 and Gfi1b may coordinate when and where the Rag proteins are expressed. One model is that Foxo1 is induced before the initiation of gene rearrangement and that this event is followed by induction of Gfi1b as part of a feedback loop, limiting recombinase levels during rearrangement by repressing Foxo1 expression. Further repression of Rag by these Gfi proteins may follow gene rearrangement and promote developmental progression, allowing the cells to replicate without risking DNA damage.
Our microarray data indicate that Gfi1b controls several B cell–specific genes (Fig. 4). In addition, chromatin IP experiments indicate that Gfi1b binds to several genes that are important for early B cell development (unpublished data). In light of these data, it is perhaps not surprising that knocking out both proteins causes a severe block in B cell development (Fig. 3). Elucidating the transcriptional pathways by which these factors control developmental progression is an exciting area for future study.
It remains to be seen whether Gfi1b plays a role in preventing Rag expression in non–B lineage cells. Gfi1b is expressed in several other cell types (Vassen et al., 2007) and has been found to play an essential role in development of the erythroid and megakaryocytic lineages (Saleque et al., 2002). Gfi1 proteins also play a role in transcriptional priming that occurs in early hematopoietic progenitor cells. Although high levels of Gfi1 support neutrophil development, lower levels of Gfi1 favor macrophage development (Laslo et al., 2006). We are currently investigating whether repression of Rag transcription by Gfi1 and Gfi1b plays an important role in developmental progression in either the myeloid or erythroid lineages.
MATERIALS AND METHODS
Cell culture.
Cells were cultured in RPMI 1640 medium supplemented with 10% (vol/vol) FCS, 2 mM l-glutamine, 100 g/ml penicillin, 100 g/ml streptomycin, and 50 mM 2-mercaptoethanol and were grown at 37°C in 5% CO2.
Retroviral production and infection.
Retrovirus was harvested from the EcoPack2 packaging cell line (Takara Bio Inc.). EcoPack2 cells were transfected with retroviral plasmid resuspended in Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol and viral supernatant was collected and filtered 48 h after transfection. AMuLV-transformed RAG1-GFP pro–B cells were infected by resuspension of the cells in viral supernatant containing 4 g/ml polybrene (Sigma-Aldrich) and cultured overnight. Cells were then expanded into normal media. Primary cells were infected as in (Amin and Schlissel, 2008). Cells were labeled with anti-IgM (II/41), anti-B220 (RA2-6B2), and anti-CD43 (S7), plus anti-Thy-1.1 (OX-7). Cells were analyzed 3–4 d after retroviral infection. Lentiviral constructs were cotransfected with VSVG and pMD2G into 293T cells using Lipofectamine 2000 as described, and virus was harvested and used to infect cell lines as described.
In vitro deletion of Gfi1 and Gfi1b.
Total bone marrow was isolated from Gfi1bflox/flox Gfi1flox/flox ER-Cre transgenic mice and fractionated using histopaque to eliminate red blood cells. Cells were separated into two pools and cultured with either tamoxifen or a vehicle control for 1 or 2 d in the presence of IL-7 at a concentration of 2 ng/ml. Finally, RNA was extracted from sorted B220+IgM− or IgM+ cells and analyzed by RT-PCR for Rag1 expression. All animal experimentation was approved by the University of California, Berkeley Animal Care and Use Committee (protocol R253, approved March 18, 2011).
Expression analysis in WT mice.
Bone marrow was isolated and pooled from eight WT BALB/c mice. After elimination of red blood cells, CD19+ cells were isolated using CD19 MicroBeads (Miltenyi Biotec) on a MACS column (Miltenyi Biotec) and then further sorted into small pro, large pro, large pre, small pre, and immature fractions using anti-IgM (II/41), anti-B220 (RA2-6B2), and anti-CD43 (S7), as well as forward scatter.
Generation of AMuLV-transformed KO cell lines.
MxCre tg Gfi1fl/fl or Gfi1bfl/fl (or compound mutants) were injected with pIpC (Sigma-Aldrich) at a dose of 500 µg per injection every other day for a total of five injections. Bone marrow was infected as described in Retroviral production and infection with the AMuLV previously described in Rosenberg et al. (1975). Cells were cultured for 4–12 wk in standard RPMI until transformed cells grew out.
Gene expression analysis.
RNA was isolated by lysing cells in TRIzol reagent (Invitrogen), followed by chloroform extraction. Reverse transcription was performed using MoMLV-RT (Invitrogen) according to the manufacturer’s instructions. Quantitative real-time PCR was performed using JumpStart Taq polymerase (Sigma-Aldrich) according to the manufacturer’s protocol and fluorescent labeling with EvaGreen (Biotium). PCR cycling conditions were 95°C for 4 min, followed by 45 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min. Primer sequences are given in Table S3.
Rag reporter studies.
AMuLV pro–B cells were electroporated with 4 µg of linearized DNA using the Lonza nucleofector kit V (VCA-1003) and the program T-001 on the Lonza Nucleofector device (AAD-1001). Cells were selected for 2 wk in G418 r 1MG/ML to select for those cells that had integrated the constructs.
Expression plasmids.
All retroviral plasmids were based on the MSCV retroviral vector and were modified to contain an IRES in-frame with a surface marker protein (Thy-1.1, or human CD4) to mark retrovirus-infected cells. The cDNA was cloned upstream of the IRES sequence.
The Gfi1b-ER fusion construct was created by PCR amplification of the ER hormone-binding domain and amplification of the ORF of Gfi1b from the cDNA library. Pfu TurboUltra (Agilent Technologies) was used for PCR according to the manufacturer’s protocol, and fragments were cloned into the MSCV retroviral vector upstream of IRES thy1.1. Constructs were subsequently verified by DNA sequence analysis. The position 2 proline to alanine mutation (P2A)–mutated Gfi1b construct was created by PCR amplification with a primer containing the mutation. The Gfi1 cDNA was PCR amplified with Pfu, as described, and cloned upstream of the IRES Thy1.1 within the MSCV retroviral vector.
The rearrangement reporter construct was created by Pfu-mediated PCR of the Eμ heavy chain enhancer and a VHKI promoter (Bates et al., 2007) and subsequent insertion into the pMX-delCJ (Bredemeyer et al., 2006) rearrangement reporter. The ires was inserted upstream of GFP using appropriate restriction sites. The Eμ, VHKI, ires-GFP, RSS 12, RSS23, and hCD4 fragments were excised in a single unit and inserted into the pLV-UT-tTR-KRAB lentiviral vector obtained from Addgene.
MSCV-Foxo1-ires-hCD4 was described previously (Amin and Schlissel, 2008). The Rag reporter constructs were provided by T. Kuo (University of California, Berkeley, Berkeley, CA). PCR with Pfu TurboUltra (Agilent Technologies) was used to amplify the 700-bp GBR and Erag and these were inserted upstream of R2P using appropriate restriction sites.
Drugs.
Tamoxifen (EMD) was used at a concentration of 1 µM. STI-571 (Novartis) was used at 2.5 µM. Aphidicolin (Sigma-Aldrich) was used at a concentration of 4 µM. G418 was used at 1 mg/ml.
ChIP and ChIP-chip.
Chromatin immunoprecipitation was performed as described in Yu et al. (2000). In brief, 100 million cells were fixed with formaldehyde, sonicated, incubated with an anti-H3K9me2(Abcam), anti-FLAG, or IgG control antibody (Sigma-Aldrich), collected using magnetic protein G beads (Invitrogen) three times with low salt buffer, once with high salt buffer, and once with LiCl buffer as described in Yu et al. (2000). DNA–protein cross-links were reversed, and DNA was precipitated and subjected to quantitative real-time PCR using primers listed in Table S3. For ChIP-chip, DNA was fragmented and ligated using the Whole Genome Amplification kit (Sigma-Aldrich). Samples were labeled and hybridized to a custom genome tiling array generated by Nimblegen.
Rearrangement and LM-PCR.
Genomic DNA was isolated and subjected to 30 cycles of PCR using the VκS and Jκ1 primers listed in Table S3. For LM-PCR, broken ends were ligated to the BW linker and then amplified with the BW-H and k05 primers (see Table S3) as described in Schlissel et al. (1993). Ligated DNA was subjected to 12 cycles of PCR with the following cycling conditions: 94°C for 1 min and 66°C for 2.5 min. BW-H and ko3 primers (see Table S3) were used to amplify 2 µl of DNA from the first reaction for 30 cycles under the same cycling conditions.
Flow cytometry.
Single-cell suspensions depleted of red blood cells were prepared from mice or from cultured cells and were incubated for at least 10 min with Fc receptor–blocking antibody (2.42G; purified from a hybridoma supernatant) and then were labeled with fluorochrome- or biotin-conjugated antibodies by standard techniques. A FC500 or an Elite XL flow cytometer (Beckman Coulter) was used for flow cytometry; a MoFlo high-speed cell sorter (Dako) was used for sorting. Data were analyzed with FlowJo software (Tree Star) and, with the exception of cell cycle analyses, dead cells were gated out using forward and side scatter for all analyses. All antibodies were obtained from eBioscience, except anti-CD43 and anti-Thy-1.1 (both from BD).
Immunoblot.
AMuLV-transformed pro–B cells were lysed in RIPA buffer (Gilbert et al., 2002), analyzed by Bradford, centrifuged to clear insoluble material, and boiled for 10 min. For immunoblot of Gfi1, nuclei were isolated and the cytosolic fraction removed before boiling. Lysate was separated by 8 or 10% SDS-PAGE and then transferred to nitrocellulose membranes. Membranes were blocked in 5% milk and labeled with primary and secondary antibodies according to the manufacturer’s instructions. Membranes were analyzed with the Odyssey Infrared Imaging System (LI-COR Biosciences). Anti-Foxo1 (9462) was obtained from Cell Signaling Technologies, and anti-Gfi1b (sc-8559), anti-Actin (sc-1615), and anti-Lamin B (sc-M20) were obtained from Santa Cruz Biotechnology, Inc. Anti-FLAG (F1804) and anti-tubulin (T3526) antibodies were obtained from Sigma-Aldrich. Anti-Gfi1 was obtained from R&D Systems. Infrared dye–conjugated secondary antibodies were from Invitrogen.
Immunofluorescence.
Cells were affixed to frosted X slides using a cytospin. Cells were then fixed with 4% paraformaldehyde and blocked before staining with anti-H2AX (Abcam). Cells were washed and stained with DAPI and anti–rabbit infrared dye–conjugated secondary antibody (Invitrogen). Cells were visualized with a Eclipse microscope (E800; Nikon).
Primers.
Primer sequences are listed in Table S3.
Microarray.
RAG1-GFPhigh cells infected with a Gfi1b-ER fusion construct were treated for 12 h with tamoxifen or a vehicle control. mRNA was isolated using Trizol reagent (Invitrogen), followed by cleanup with the RNeasy kit (QIAGEN), and submitted for analysis to the UCSF core facility. The microarray accession no. is GSE33709.
Online supplemental material.
Fig. S1 shows a cluster of three AATC sites that are found in the GBR 5′ of Rag2. Table S1 shows identities of genes whose transcripts are down-regulated by at least twofold (P < 0.01) upon Gfi1b expression in Abelson cells. Table S2 shows identities of genes whose transcripts are up-regulated by at least twofold (P < 0.01) upon Gfi1b expression in Abelson cells. Table S3 shows sequences of primers used in this study.
Acknowledgments
The authors would like to thank Hector Nolla for cell sorting, Dan Huang and Jason Yu for technical assistance, and Christian Vettermann for experimental input, technical assistance, and critical reading of the manuscript.
We thank all members of the Schlissel laboratory for advice and constructive criticisms. This work was supported by a grant to M.S. Schlissel from the National Institutes of Health (HL48702). T. Möröy was supported by a grant from the Canadian Institutes of Health Research (MOP-84238) and a Canada Research Chair (Tier 1).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AMuLV
- Abelson murine leukemia virus
- ChIP
- chromatin immunoprecipitation
- DKO
- double KO
- ER
- estrogen receptor
- GBR
- Gfi1b binding region
- R2P
- Rag2 promoter
- RSS
- recombination signal sequence
References
- Amin R.H., Schlissel M.S. 2008. Foxo1 directly regulates the transcription of recombination-activating genes during B cell development. Nat. Immunol. 9:613–622 10.1038/ni.1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoub N., Jeyasekharan A.D., Bernal J.A., Venkitaraman A.R. 2009. Paving the way for H2AX phosphorylation: chromatin changes in the DNA damage response. Cell Cycle. 8:1494–1500 10.4161/cc.8.10.8501 [DOI] [PubMed] [Google Scholar]
- Bain G., Maandag E.C., Izon D.J., Amsen D., Kruisbeek A.M., Weintraub B.C., Krop I., Schlissel M.S., Feeney A.J., van Roon M., et al. 1994. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 79:885–892 10.1016/0092-8674(94)90077-9 [DOI] [PubMed] [Google Scholar]
- Bates J.G., Cado D., Nolla H., Schlissel M.S. 2007. Chromosomal position of a VH gene segment determines its activation and inactivation as a substrate for V(D)J recombination. J. Exp. Med. 204:3247–3256 10.1084/jem.20071787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer A.L., Sharma G.G., Huang C.Y., Helmink B.A., Walker L.M., Khor K.C., Nuskey B., Sullivan K.E., Pandita T.K., Bassing C.H., Sleckman B.P. 2006. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature. 442:466–470 10.1038/nature04866 [DOI] [PubMed] [Google Scholar]
- Dengler H.S., Baracho G.V., Omori S.A., Bruckner S., Arden K.C., Castrillon D.H., DePinho R.A., Rickert R.C. 2008. Distinct functions for the transcription factor Foxo1 at various stages of B cell differentiation. Nat. Immunol. 9:1388–1398 10.1038/ni.1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan L.L., Porter S.D., Duan Z., Flubacher M.M., Montoya D., Tsichlis P.N., Horwitz M., Gilks C.B., Grimes H.L. 2004. Targeted transcriptional repression of Gfi1 by GFI1 and GFI1B in lymphoid cells. Nucleic Acids Res. 32:2508–2519 10.1093/nar/gkh570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falbo K.B., Shen X. 2009. Histone modifications during DNA replication. Mol. Cells. 28:149–154 10.1007/s10059-009-0127-7 [DOI] [PubMed] [Google Scholar]
- Fiolka K., Hertzano R., Vassen L., Zeng H., Hermesh O., Avraham K.B., Dührsen U., Möröy T. 2006. Gfi1 and Gfi1b act equivalently in haematopoiesis, but have distinct, non-overlapping functions in inner ear development. EMBO Rep. 7:326–333 10.1038/sj.embor.7400618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco S., Alt F.W., Manis J.P. 2006. Pathways that suppress programmed DNA breaks from progressing to chromosomal breaks and translocations. DNA Repair (Amst.). 5:1030–1041 10.1016/j.dnarep.2006.05.024 [DOI] [PubMed] [Google Scholar]
- Fuks F. 2005. DNA methylation and histone modifications: teaming up to silence genes. Curr. Opin. Genet. Dev. 15:490–495 10.1016/j.gde.2005.08.002 [DOI] [PubMed] [Google Scholar]
- Gilbert C., Rollet-Labelle E., Caon A.C., Naccache P.H. 2002. Immunoblotting and sequential lysis protocols for the analysis of tyrosine phosphorylation-dependent signaling. J. Immunol. Methods. 271:185–201 10.1016/S0022-1759(02)00347-2 [DOI] [PubMed] [Google Scholar]
- Grimes H.L., Chan T.O., Zweidler-McKay P.A., Tong B., Tsichlis P.N. 1996. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol. 16:6263–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobeika E., Thiemann S., Storch B., Jumaa H., Nielsen P.J., Pelanda R., Reth M. 2006. Testing gene function early in the B cell lineage in mb1-cre mice. Proc. Natl. Acad. Sci. USA. 103:13789–13794 10.1073/pnas.0605944103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu L.Y., Lauring J., Liang H.E., Greenbaum S., Cado D., Zhuang Y., Schlissel M.S. 2003. A conserved transcriptional enhancer regulates RAG gene expression in developing B cells. Immunity. 19:105–117 10.1016/S1074-7613(03)00181-X [DOI] [PubMed] [Google Scholar]
- Hu H., Wang B., Borde M., Nardone J., Maika S., Allred L., Tucker P.W., Rao A. 2006. Foxp1 is an essential transcriptional regulator of B cell development. Nat. Immunol. 7:819–826 10.1038/ni1358 [DOI] [PubMed] [Google Scholar]
- Huang D.Y., Kuo Y.Y., Chang Z.F. 2005. GATA-1 mediates auto-regulation of Gfi-1B transcription in K562 cells. Nucleic Acids Res. 33:5331–5342 10.1093/nar/gki838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandanpour C., Thiede C., Valk P.J., Sharif-Askari E., Nückel H., Lohmann D., Horsthemke B., Siffert W., Neubauer A., Grzeschik K.H., et al. 2010. A variant allele of Growth Factor Independence 1 (GFI1) is associated with acute myeloid leukemia. Blood. 115:2462–2472 10.1182/blood-2009-08-239822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwata N., Igarashi H., Ohmura T., Aizawa S., Sakaguchi N. 1999. Cutting edge: absence of expression of RAG1 in peritoneal B-1 cells detected by knocking into RAG1 locus with green fluorescent protein gene. J. Immunol. 163:6355–6359 [PubMed] [Google Scholar]
- Laslo P., Spooner C.J., Warmflash A., Lancki D.W., Lee H.J., Sciammas R., Gantner B.N., Dinner A.R., Singh H. 2006. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 126:755–766 10.1016/j.cell.2006.06.052 [DOI] [PubMed] [Google Scholar]
- Laurent B., Randrianarison-Huetz V., Kadri Z., Roméo P.H., Porteu F., Duménil D. 2009. Gfi-1B promoter remains associated with active chromatin marks throughout erythroid differentiation of human primary progenitor cells. Stem Cells. 27:2153–2162 10.1002/stem.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Doddapaneni K., Hogue A., McGhee L., Meyers S., Wu Z. 2010. Solution structure of Gfi-1 zinc domain bound to consensus DNA. J. Mol. Biol. 397:1055–1066 10.1016/j.jmb.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Metzger D., Clifford J., Chiba H., Chambon P. 1995. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl. Acad. Sci. USA. 92:6991–6995 10.1073/pnas.92.15.6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller L.D., Long P.M., Wong L., Mukherjee S., McShane L.M., Liu E.T. 2002. Optimal gene expression analysis by microarrays. Cancer Cell. 2:353–361 10.1016/S1535-6108(02)00181-2 [DOI] [PubMed] [Google Scholar]
- Muljo S.A., Schlissel M.S. 2003. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat. Immunol. 4:31–37 10.1038/ni870 [DOI] [PubMed] [Google Scholar]
- Rathinam C., Klein C. 2007. Transcriptional repressor Gfi1 integrates cytokine-receptor signals controlling B-cell differentiation. PLoS ONE. 2:e306 10.1371/journal.pone.0000306 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Rosenberg N., Baltimore D., Scher C.D. 1975. In vitro transformation of lymphoid cells by Abelson murine leukemia virus. Proc. Natl. Acad. Sci. USA. 72:1932–1936 10.1073/pnas.72.5.1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleque S., Cameron S., Orkin S.H. 2002. The zinc-finger proto-oncogene Gfi-1b is essential for development of the erythroid and megakaryocytic lineages. Genes Dev. 16:301–306 10.1101/gad.959102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleque S., Kim J., Rooke H.M., Orkin S.H. 2007. Epigenetic regulation of hematopoietic differentiation by Gfi-1 and Gfi-1b is mediated by the cofactors CoREST and LSD1. Mol. Cell. 27:562–572 10.1016/j.molcel.2007.06.039 [DOI] [PubMed] [Google Scholar]
- Schlissel M.S. 2003. Regulating antigen-receptor gene assembly. Nat. Rev. Immunol. 3:890–899 10.1038/nri1225 [DOI] [PubMed] [Google Scholar]
- Schlissel M., Constantinescu A., Morrow T., Baxter M., Peng A. 1993. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 7:2520–2532 10.1101/gad.7.12b.2520 [DOI] [PubMed] [Google Scholar]
- Schlissel M.S., Kaffer C.R., Curry J.D. 2006. Leukemia and lymphoma: a cost of doing business for adaptive immunity. Genes Dev. 20:1539–1544 10.1101/gad.1446506 [DOI] [PubMed] [Google Scholar]
- Sukumar S., Schlissel M.S. 2011. Receptor editing as a mechanism of B cell tolerance. J. Immunol. 186:1301–1302 10.4049/jimmunol.1090129 [DOI] [PubMed] [Google Scholar]
- Tonegawa S. 1983. Somatic generation of antibody diversity. Nature. 302:575–581 10.1038/302575a0 [DOI] [PubMed] [Google Scholar]
- van der Meer L.T., Jansen J.H., van der Reijden B.A. 2010. Gfi1 and Gfi1b: key regulators of hematopoiesis. Leukemia. 24:1834–1843 10.1038/leu.2010.195 [DOI] [PubMed] [Google Scholar]
- Vassen L., Fiolka K., Mahlmann S., Möröy T. 2005. Direct transcriptional repression of the genes encoding the zinc-finger proteins Gfi1b and Gfi1 by Gfi1b. Nucleic Acids Res. 33:987–998 10.1093/nar/gki243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassen L., Fiolka K., Möröy T. 2006. Gfi1b alters histone methylation at target gene promoters and sites of gamma-satellite containing heterochromatin. EMBO J. 25:2409–2419 10.1038/sj.emboj.7601124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassen L., Okayama T., Möröy T. 2007. Gfi1b:green fluorescent protein knock-in mice reveal a dynamic expression pattern of Gfi1b during hematopoiesis that is largely complementary to Gfi1. Blood. 109:2356–2364 10.1182/blood-2006-06-030031 [DOI] [PubMed] [Google Scholar]
- Ventura A., Kirsch D.G., McLaughlin M.E., Tuveson D.A., Grimm J., Lintault L., Newman J., Reczek E.E., Weissleder R., Jacks T. 2007. Restoration of p53 function leads to tumour regression in vivo. Nature. 445:661–665 10.1038/nature05541 [DOI] [PubMed] [Google Scholar]
- Wilson N.K., Foster S.D., Wang X., Knezevic K., Schütte J., Kaimakis P., Chilarska P.M., Kinston S., Ouwehand W.H., Dzierzak E., et al. 2010. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell. 7:532–544 10.1016/j.stem.2010.07.016 [DOI] [PubMed] [Google Scholar]
- Yannoutsos N., Barreto V., Misulovin Z., Gazumyan A., Yu W., Rajewsky N., Peixoto B.R., Eisenreich T., Nussenzweig M.C. 2004. A cis element in the recombination activating gene locus regulates gene expression by counteracting a distant silencer. Nat. Immunol. 5:443–450 10.1038/ni1053 [DOI] [PubMed] [Google Scholar]
- Yu W., Misulovin Z., Suh H., Hardy R.R., Jankovic M., Yannoutsos N., Nussenzweig M.C. 1999. Coordinate regulation of RAG1 and RAG2 by cell type-specific DNA elements 5′ of RAG2. Science. 285:1080–1084 10.1126/science.285.5430.1080 [DOI] [PubMed] [Google Scholar]
- Yu J., Angelin-Duclos C., Greenwood J., Liao J., Calame K. 2000. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell. Biol. 20:2592–2603 10.1128/MCB.20.7.2592-2603.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yücel R., Kosan C., Heyd F., Möröy T. 2004. Gfi1:green fluorescent protein knock-in mutant reveals differential expression and autoregulation of the growth factor independence 1 (Gfi1) gene during lymphocyte development. J. Biol. Chem. 279:40906–40917 10.1074/jbc.M400808200 [DOI] [PubMed] [Google Scholar]
- Zeng H., Yücel R., Kosan C., Klein-Hitpass L., Möröy T. 2004. Transcription factor Gfi1 regulates self-renewal and engraftment of hematopoietic stem cells. EMBO J. 23:4116–4125 10.1038/sj.emboj.7600419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweidler-Mckay P.A., Grimes H.L., Flubacher M.M., Tsichlis P.N. 1996. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol. 16:4024–4034 [DOI] [PMC free article] [PubMed] [Google Scholar]