Abstract
There is a great deal of interest in a fine-scale population structure in the UK, both as a signature of historical immigration events and because of the effect population structure may have on disease association studies. Although population structure appears to have a minor impact on the current generation of genome-wide association studies, it is likely to have a significant part in the next generation of studies designed to search for rare variants. A powerful way of detecting such structure is to control and document carefully the provenance of the samples involved. In this study, we describe the collection of a cohort of rural UK samples (The People of the British Isles), aimed at providing a well-characterised UK-control population that can be used as a resource by the research community, as well as providing a fine-scale genetic information on the British population. So far, some 4000 samples have been collected, the majority of which fit the criteria of coming from a rural area and having all four grandparents from approximately the same area. Analysis of the first 3865 samples that have been geocoded indicates that 75% have a mean distance between grandparental places of birth of 37.3 km, and that about 70% of grandparental places of birth can be classed as rural. Preliminary genotyping of 1057 samples demonstrates the value of these samples for investigating a fine-scale population structure within the UK, and shows how this can be enhanced by the use of surnames.
Keywords: people of the British Isles, population structure, control population, admixture, surnames
Introduction
During the last 10 years there has been much interest in a fine-scale population structure, particularly in the UK, both as a signature of historical immigration events1, 2, 3, 4, 5, 6 and because of the effect population structure may have on disease association studies,7, 8 although this depends on the magnitude of the associations.9 Fine-scale population structure is principally the outcome of historical movements of people into Britain following the last ice age about 10 000 years ago, with the major subsequent detectable influences likely to be from Anglo-Saxon, Norse and Norman admixture.10 Although population structure appears to have a minor impact on the current generation of genome-wide association studies,9 it is likely to have a significant part in the next generation of studies designed to search for rare variants.11, 12 It is, therefore, important that suitable control population cohorts are available for such studies. In this study we describe the collection and preliminary analysis of a set of carefully chosen samples, to represent the areas of the UK from where they have come.
A powerful way of detecting a fine-scale population structure is to control and document carefully the provenance of the samples involved. This can be carried out by, for example, ensuring that volunteers are chosen for whom all four grandparents were born in the same rural area. This approach should maximise the probability of recruiting individuals whose families have been stable inhabitants of the area for many generations, as most recent migration has been into larger towns and cities. Genotyping a collection of such samples from throughout the UK should then allow identification of high-quality ancestrally informative markers and enable a detailed analysis of population structure. These samples can then be used to assess the impact of population structure on disease and other phenotype association studies, particularly when searching for rare variants. The resulting body of data will also provide an excellent basis for relating population structure to the known history and archaeology of the UK population.
A further way to investigate and refine the genetic signals of population structure is to use surnames when analysing the genetic data.3, 4, 13 The distribution of surnames has been remarkably stable over at least the last 130 years (GB Names Profiler, gbnames.publicprofiler.org14), supporting the notion that the rural British population has been quite sedentary until relatively recently. Although evidence based on studies of testimonials15 suggests that there has been a great deal of movement, this is mainly over short distances. Thus, 75% of reported residential mobility was less than 10 km, with women historically averaging greater distances than men. Classification of surnames into those that have markedly local distributions, in contrast to those with wider, more national distributions, should help to enhance the signals of population structure.
Here we describe the collection of a cohort of samples carefully chosen using the above considerations, and present a preliminary analysis of some genotype and surname data on a small pilot subset of these samples. These are part of a much larger ongoing UK-wide project (The People of the British Isles (PoBI), http://www.peopleofthebritishisles.org), funded by the Wellcome Trust, to set up a well characterised and carefully collected UK-control population as a resource that can be used by the research community. Preliminary data analysis demonstrates that population structure can be detected within the UK even with a limited number of samples and loci, and that the analysis can be enhanced by using information on surnames. Here a population refers to a County or a region of the UK.
Materials and methods
Sample collection
Approximately 4000 rural samples from throughout the UK have so far been collected using the criteria that all four grandparents were born in the same rural area, defined as lying within 60 km linear map distance of each other. For each sample, a self-reported questionnaire was completed. Details requested included place and year of birth of grandparents, parents and the volunteer, place of residence, gender and surname at birth. As approved by the Research Ethics Committee, samples were anonymised upon collection therefore, for research undertaken outside the core research group, surname data and full date of birth will be excluded. During the period of sample collection, consent for genotyping has broadened (see Supplementary Information). The whole project was subjected to the UK standard research ethical consent procedures (Leeds (West) REC – 05/Q1205/35).
A volume of 20μl of blood was collected from each volunteer and peripheral blood lymphocytes (PBLs) were harvested (see Supplementary Information). A number of the stored viable PBLs were subsequently transformed with Epstein Barr virus16 by the European Collection of Cell Cultures and the Avon Longitudinal Study of Parents and Children to check viability and to replenish some depleted DNA stocks, with a success rate of 531/539 (98.5%). DNA was prepared from the 10 ml of blood residue remaining after sterile separation (see Supplementary Information).
Samples
Basic information on numbers, gender and the age distribution of the total sample, and separately, of the sample used for the pilot genotyping is given in Table 1. At the time of this analysis, 3865 of the samples collected have had their birthplaces geocoded by assigning longitude and latitude coordinates. From these coordinates, the mean distance (MD) between the known grandparental birthplaces of each volunteer who gave details of all four grandparents was calculated (see Supplementary Information).
Table 1. Basic information on numbers, gender and the age distribution of the total sample and, separately, of the sample used for the pilot genotyping is given in the top part of the table.
| Overall | Proportion | Pilot | Proportion | |
|---|---|---|---|---|
| Gender | ||||
| M | 1824 | 0.472 | 506 | 0.479 |
| F | 1982 | 0.513 | 497 | 0.470 |
| Unknown | 59 | 0.015 | 54 | 0.051 |
| Total | 3865 | 1057 | ||
| Age (in 2009) | ||||
| <20 | 8 | 0.002 | 0 | 0.000 |
| 20–29 | 82 | 0.021 | 13 | 0.012 |
| 30–39 | 180 | 0.047 | 33 | 0.031 |
| 40–49 | 462 | 0.120 | 66 | 0.062 |
| 50–59 | 688 | 0.178 | 172 | 0.163 |
| 60–69 | 1161 | 0.300 | 295 | 0.279 |
| 70–79 | 915 | 0.237 | 246 | 0.233 |
| 80–89 | 291 | 0.075 | 96 | 0.091 |
| 90–99 | 21 | 0.005 | 12 | 0.011 |
| >100 | 10 | 0.003 | 2 | 0.002 |
| Unknown | 47 | 0.012 | 122a | 0.115 |
| Total | 3865 | 1057 | ||
| MD between grandparental birthplaces | ||||
| Median (km) | 16.05 | 16.31 | ||
| 25% quartile (km) | 2.96 | 3.72 | ||
| 75% quartile (km) | 44.85 | 48.92 | ||
| n | 3646 | 893 | ||
| No. missing | 219 | 65 | ||
| Orkneya | 0 | 99 | ||
The lower part of the table gives the median and 25 and 75% quartiles of the mean distance (MD) between grandparental birthplaces for volunteers who gave information for all four grandparents.
99 of the unknown age group in the pilot data are previously collected Orkney samples.19 These are not included in the overall geocoded data set.
The geocoded place names make it possible to estimate, for any given set of volunteers, what proportion of their grandparents was born in a rural or in an urban area. For this analysis, the extent of UK urban areas was derived from a map layer provided by ESRI (http://www.esri.com). For each sample, the mean geographical position (MGP) of the grandparental birthplaces was mapped using the ArcGIS 9.3 package (http://www.esri.com). To determine whether a MGP was rural, the distance to the fringe of the nearest urban area was calculated based on the straight line to the closest point in the fringe. MGPs were then assigned as rural if they were greater than a defined distance away from the edge of that urban area of a given population size, based on the 2001 census. A range of values of the distances and sizes of urban populations for this definition of rural was investigated.
Use of surnames to subdivide populations
Surnames of the volunteers were routinely collected and this knowledge should allow a more detailed investigation of population structure. Individuals whose surnames are localised to an area are more likely to have ancestry from that area down the male lineage, and should be more representative of the region over a long time period. This should be backed up by the genetics.
Although it is possible to determine a surname's area of origin from contemporary data, historical data sets are advantageous because they are less affected by recent migrations. The digitisation of the 1881 Census of Great Britain (UK Data Archive, http://www.data-archive.ac.uk) provides an invaluable resource for the definition of area of origin. Although it is not the earliest available census, it remains the one that has been digitally encoded (by the Church of Jesus Christ of the Latter-day Saints) to the highest quality. It provides the names and place of enumeration (Parish and Registration District) for 29 million people, with a total of 425 000 unique surnames, ∼49 000 of which occur in more than 20 individual census records. These data have been geocoded to registration district (RD) level (mean population 4900) and linked to a shapefile containing the historical boundary data.17
Some surname distributions are very localised (eg Grahamslaw, Forster or Pedlar, Supplementary Figure 1), while other surnames are much more prevalent throughout the UK (eg Smith or Grey). The distribution of the frequencies of surnames in districts throughout the UK provides an approach to assessing how local a surname is. This can be carried out using the location quotient, which compares the relative frequency of a surname in a given region with the relative frequency of that surname at a more aggregate spatial level,18 for example a county or district versus Great Britain as a whole. It is defined as follows:
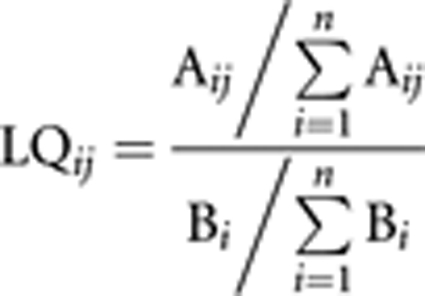 |
where Aij is the count of surname i in registration district (RD) j, Bi is the count of surname i in Great Britain, n is the total number of surnames in Britain and LQij is the location quotient of surname i in region j. LQ values greater that 1 indicate an RD with a higher concentration of the selected name that would be expected if the surname had a uniform distribution throughout the Britain.
The RDs with the three highest LQs for a given surname are taken to define the surname's core locality. In many cases these are contiguous or at least very close to each other, and this is taken to indicate that the surname has a single core. If this is not the case, the surname may either have more than one core or a dispersed distribution.
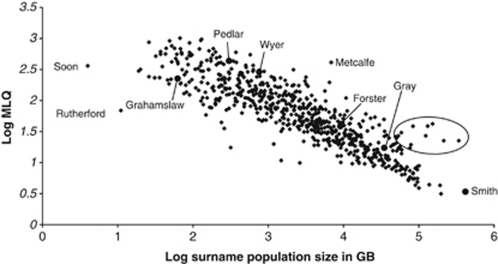
The district with the maximum LQ (MLQ) can be used as a starting point for assigning a surname as local or non-local. In general it appears that surnames with high MLQs tend to be comparatively rare (Figure 1) and are more likely to have a local distribution (eg Pedlar MLQ=323). There are, however, some surnames with relatively lower MLQs that are relatively common but still have, in essence, local distributions (eg Forster MLQ=45). To investigate the effects of using surname localisation on the ability to detect genetic population structure, a range of MLQs was at first used as a cut off to define local versus non-local surnames. These were 19, 45 and 120, respectively, the lower quartile, the median and the upper quartile of the distribution of the highest MLQs for each surname. The definitions of local and non-local were then refined according to whether there were two or more non-adjacent RDs with similarly high LQs, in which case the surname was re-classified as non-local (eg Wyer, MLQ=297). A further refinement was based on whether the MGP of the birthplaces of the four grandparents of a given individual was less than either 83 km (the median of the distribution of the MDs) from the district with the MLQ for the given individual's surname, or less than 120 km from the district (twice the maximum distance between birthplaces of the grandparents of a given individual aimed at when collecting samples). Only if both the MLQ and distance from the MLQ criteria were satisfied, the surname was classified as local (Supplementary Figure 2).
Figure 1.
Graph of the log (MLQ) of the RD with the highest LQ for each surname (y-axis) against log (surname population size) in the 1881 census (x-axis). There are a number of surnames (circled) with a higher MLQ than might be expected for the surname sample size (Jones, Davies, Evans, Thomas, Hughes, James and Phillips), which are established Welsh surnames. The surnames from Supplementary Figure 1 are also marked.
Genotyping
A total of 1057 of the samples were used in an initial pilot genotyping project, which included cell lines from 99 Orcadian samples previously collected by the laboratory.19 The samples were genotyped with a number of markers that were chosen because they have been used to differentiate populations by many different studies over the years. Specifically, they were: HLA20, 21, 22 (typed at a low-medium resolution, Table 2, Supplementary Table 1), MC1R (R151C (rs1805007) and R160W (rs1805008), the minor alleles of which are associated with red hair23),24, 25 ABO26, 27 (rs7853989, the SNP that differentiates alleles A and B) and the Y chromosome (NRY).1, 2, 28, 29 The six most common NRY halogroups2 were typed (Table 2) as defined by specific SNPs (R1a1 (rs3908), F(xI/J2/R1) (rs2032652), E (rs9306841), I (rs2032597), J2 (rs2032604) and R1(xR1a1) (rs2032624)).
Table 2. Allele and haplotype frequency data.
| NRY | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R1a1 | I | J2 | R1 xR1a1 | F xI/J2/R1 | E | n | ||||||||||
| SW | 0.013 | 0.150 | 0.025 | 0.750 | 0.038 | 0.025 | 80 | |||||||||
| CN | 0.053 | 0.140 | 0.018 | 0.684 | 0.105 | 0.000 | 57 | |||||||||
| E | 0.035 | 0.161 | 0.023 | 0.598 | 0.138 | 0.046 | 87 | |||||||||
| N | 0.022 | 0.202 | 0.033 | 0.656 | 0.071 | 0.016 | 183 | |||||||||
| OR | 0.342 | 0.079 | 0.000 | 0.579 | 0.000 | 0.000 | 38 | |||||||||
| HLA-A | HLA-B | HLA-C | ||||||||||||||
| |
01 |
02 |
03 |
11 |
2n |
07 |
08 |
15 |
35 |
44 |
2n |
03 |
05 |
06 |
07 |
2n |
| SW | 0.169 | 0.307 | 0.156 | 0.055 | 326 | 0.151 | 0.170 | 0.077 | 0.071 | 0.106 | 311 | 0.123 | 0.141 | 0.083 | 0.368 | 326 |
| CN | 0.191 | 0.270 | 0.157 | 0.071 | 267 | 0.139 | 0.147 | 0.053 | 0.090 | 0.109 | 266 | 0.167 | 0.104 | 0.089 | 0.333 | 270 |
| E | 0.172 | 0.304 | 0.175 | 0.052 | 326 | 0.134 | 0.130 | 0.103 | 0.103 | 0.090 | 322 | 0.175 | 0.089 | 0.092 | 0.316 | 326 |
| N | 0.177 | 0.271 | 0.151 | 0.076 | 661 | 0.186 | 0.105 | 0.060 | 0.081 | 0.124 | 651 | 0.131 | 0.122 | 0.087 | 0.366 | 666 |
| OR | 0.183 | 0.291 | 0.091 | 0.080 | 175 | 0.222 | 0.090 | 0.084 | 0.030 | 0.204 | 167 | 0.171 | 0.114 | 0.091 | 0.381 | 176 |
| HLA-DRB1 | HLA-DQB1 | |||||||||||||||
| |
01 |
03 |
04 |
07 |
11 |
13 |
15 |
2n |
02 |
03 |
05 |
06 |
2n |
|
|
|
| SW | 0.106 | 0.156 | 0.240 | 0.100 | 0.065 | 0.109 | 0.122 | 321 | 0.225 | 0.338 | 0.184 | 0.225 | 320 | |||
| CN | 0.105 | 0.165 | 0.173 | 0.102 | 0.094 | 0.109 | 0.132 | 266 | 0.229 | 0.342 | 0.150 | 0.259 | 266 | |||
| E | 0.079 | 0.142 | 0.195 | 0.145 | 0.085 | 0.101 | 0.123 | 318 | 0.280 | 0.341 | 0.137 | 0.213 | 314 | |||
| N | 0.116 | 0.144 | 0.177 | 0.147 | 0.055 | 0.090 | 0.164 | 654 | 0.262 | 0.308 | 0.150 | 0.253 | 652 | |||
| OR | 0.061 | 0.141 | 0.160 | 0.184 | 0.043 | 0.086 | 0.209 | 163 | 0.267 | 0.320 | 0.111 | 0.273 | 172 | |||
| HLA-A, -B and -DRB1 haplotypes | ||||||||||||||||
| |
01-08-03 |
03-07-15 |
02-44-04 |
02-07-15 |
02-15-04 |
29-44-07 |
2n |
|
|
|
|
|
|
|
|
|
| SW | 0.076 | 0.023 | 0.030 | 0.020 | 0.030 | 0.003 | 304 | |||||||||
| CN | 0.088 | 0.023 | 0.012 | 0.012 | 0.008 | 0.008 | 260 | |||||||||
| E | 0.077 | 0.026 | 0.016 | 0.026 | 0.019 | 0.016 | 310 | |||||||||
| N | 0.062 | 0.019 | 0.021 | 0.015 | 0.011 | 0.011 | 623 | |||||||||
| OR | 0.051 | 0.013 | 0.044 | 0.051 | 0.013 | 0.044 | 158 | |||||||||
| MC1R (rs1805007) | MC1R (rs1805008) | ABO (rs7853989) | ||||||||||||||
| |
C |
2n |
C |
2n |
C |
2n |
|
|
|
|
|
|
|
|
|
|
| SW | 0.906 | 340 | 0.945 | 328 | 0.929 | 328 | ||||||||||
| CN | 0.924 | 264 | 0.909 | 264 | 0.893 | 270 | ||||||||||
| E | 0.908 | 326 | 0.932 | 310 | 0.920 | 326 | ||||||||||
| N | 0.910 | 652 | 0.934 | 664 | 0.914 | 660 | ||||||||||
| OR | 0.887 | 194 | 0.906 | 192 | 0.828 | 192 | ||||||||||
The NRY haplogroups are those that are the most common in Europe, while the HLA alleles (low, allele group, resolution) are those that have a frequency of >7.5% in at least one region. The estimated frequencies of the six most common HLA-A, -B -and DRB1 haplotypes are also shown. Only the major allele frequencies are presented for the MC1R and ABO SNPs. Populations are grouped into regions as defined in the main text. The regions are: SW (Cornwall, Devon and Pembrokeshire), CN (Oxfordshire and the Forest of Dean), E (Sussex, Kent, Norfolk and Lincolnshire), N (Cumbria, Yorkshire and the North East) and OR (Orkney).
Assessment of allele frequency differences and calculation of FST
To conduct a meaningful analysis of population structure with the limited genotyping we have so far carried out on the pilot samples, these were pooled into groups based mainly on geographical association, but also to some extent using historical and archaeological criteria.10 We recognise that these distinctions are somewhat arbitrary and their effect will be investigated in more detail in the future work. Cornwall, Devon and Pembrokeshire were pooled to represent the South/West (SW) and the area that could be considered the closest surrogate to the Ancient British. Kent, Norfolk and Lincolnshire were pooled to represent the East (E) and the area most directly influenced by the Anglo-Saxon invasions. Cumbria, Yorkshire and the North East were pooled broadly to represent the North of England (N); Oxfordshire and the Forest of Dean were combined to represent the Central region of England (CN); and Orkney was kept separate from the others, largely because of the known substantial Norse Viking influence in Orkney. The aim was to achieve a grouping that, a priori and given the limitations of the sample size, would be most likely to reveal differences in a regional fine-scale population structure.
Fisher's exact test was used to assess allele frequency differences using 2 × 2 tables of allele counts to split the data in three ways (see Supplementary Information) and FST was calculated using Weir and Cockerham's method.30
Admixture
To investigate further signals of a fine-scale population structure within the UK, point estimates of admixture were calculated using a maximum likelihood approach31 (see Supplementary Information). Autosomal admixture was estimated using the six most common HLA-A, -B and -DRB1 haplotypes, together with only those HLA alleles not represented on any of those six haplotypes, and the MC1R and ABO SNPs.
Results
Sampling
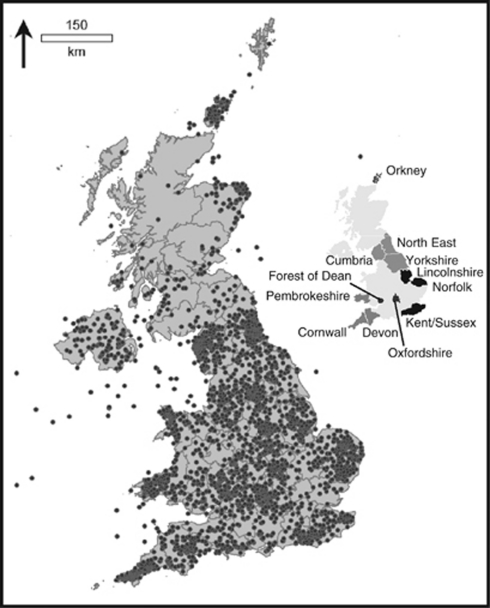
For the 3865 of the samples that have been geocoded the distances between birthplaces could be accurately and consistently calculated. Of these, 958 were genotyped for this study. The distribution in England and Wales of the MGP of each individual's grandparents birthplace is shown in Figure 2. The data on distances between grandparental birthplaces, given in Table 1, show that the median of the MD between grandparental birthplaces for all the geocoded samples is 16.05 km (quartiles 2.96 and 44.85 km), while it is slightly larger for the genotyped samples (16.31 km, (3.72 and 48.92 km)). The overall distribution of these distances is skewed towards the lower values (Supplementary Figure 3). The individuals who did not know where all their grandparents were born, and the 99 genotyped Orkney samples for whom this information was not available, are excluded from these calculations. Overall, 219 out of the 3865 geocoded samples were excluded from further analysis using distance information.
Figure 2.
Distribution of MGP of grandparental birthplaces of the 3646 volunteers for whom there was information for all four grandparents. Dots mark the MPG for individual volunteers. The populations from which samples were taken for the genotyping are marked on the inset map.
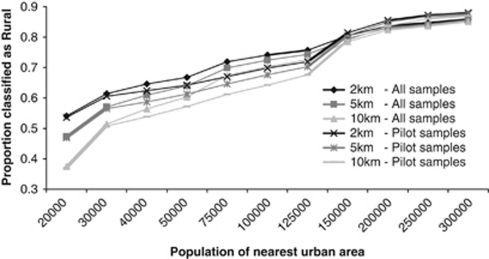
Using the approaches discussed in the methods section for the definition of rural versus urban, the proportion of grandparents from the 3865 geocoded samples who were born in rural areas ranges from 0.375 (assuming the stringent criterion that people born within 10 km of small towns of 20 000 people (as of 2001), such as Penzance, or any towns larger than this, count as urban) to 0.859 (assuming the much less stringent criterion that only those born within 2 km of large cities of 300 000 or more, such as Southampton, count as urban, Supplementary Table 2). Choosing a definitive cut off population size for the distinction between rural and urban is difficult, but from Figure 3, (Supplementary Table 2) plotting the proportion of rural samples against population size for different distances, there seems to be a definite discontinuity at around population size 125 000 (eg Doncaster). Choosing this size as the threshold that distinguishes rural from urban gives estimates of the proportion of rural volunteers, for all geocoded samples, which range from 0.726 to 0.757, depending on the distance from the urban area. In the geocoded samples, there are 683 (4.5%) grandparental birthplaces that were given simply as a county and 365 (2.4%) that were unknown. The corresponding numbers for the genotyped data are 120 (3.1%) and 94 (2.5%).
Figure 3.
Percentage of volunteers with all four grandparents classed as rural according to their distance (2, 5 or 10 km) from an urban area (y axis) of a given population size (x-axis). Estimates are made for all the geocoded samples (all samples) and those genotyped (pilot samples).
Local classification by surname
Surnames of individuals in the pilot set were classified as local using a combination of five different MLQ thresholds and two different thresholds for distances between the MGPs and the district with the MLQ for the individual's surname (Table 3). The proportion of surnames classified as local ranged from 0.034 (Cumbria and Yorkshire with a threshold LQ of 300) to 0.767 (Cornwall with a threshold MLQ of 19). Cornwall and Kent/Sussex generally had, respectively, the highest and second highest proportions of local surnames, and Norfolk and Lincolnshire generally have the next highest proportions of local surnames. Eight hundred and forty five of the geocoded samples, 824 of which had been successfully genotyped, were used for the local classification of surnames.
Table 3. Proportion of surnames classified as local depending on different exclusion criteria.
| Proportion | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Population | MLQ>19, dist<83 km | MLQ>19, dist<120 km | MLQ 19 | MLQ>45, dist<83 km | MLQ>45, dist<120 km | MLQ 45 | MLQ>120, dist<83 km | MLQ>120, dist<120 km | MLQ 120 | MLQ 200 | MLQ 300 |
| Cornwall | 0.550 | 0.583 | 0.767 | 0.467 | 0.483 | 0.533 | 0.417 | 0.433 | 0.467 | 0.267 | 0.217 |
| Cumbria | 0.345 | 0.397 | 0.552 | 0.293 | 0.293 | 0.328 | 0.190 | 0.190 | 0.190 | 0.086 | 0.034 |
| Devon | 0.316 | 0.354 | 0.684 | 0.316 | 0.342 | 0.456 | 0.253 | 0.266 | 0.316 | 0.152 | 0.076 |
| Forest of Dean | 0.164 | 0.299 | 0.478 | 0.149 | 0.209 | 0.239 | 0.090 | 0.134 | 0.149 | 0.119 | 0.045 |
| Kent/Sussex | 0.469 | 0.469 | 0.653 | 0.429 | 0.429 | 0.490 | 0.388 | 0.367 | 0.408 | 0.204 | 0.122 |
| Lincolnshire | 0.367 | 0.433 | 0.667 | 0.367 | 0.400 | 0.567 | 0.267 | 0.267 | 0.333 | 0.133 | 0.067 |
| North East | 0.324 | 0.382 | 0.588 | 0.309 | 0.338 | 0.485 | 0.096 | 0.103 | 0.154 | 0.088 | 0.044 |
| Norfolk | 0.430 | 0.440 | 0.700 | 0.400 | 0.410 | 0.520 | 0.230 | 0.240 | 0.270 | 0.150 | 0.120 |
| Pembrokeshire | 0.436 | 0.487 | 0.590 | 0.231 | 0.256 | 0.359 | 0.103 | 0.103 | 0.128 | 0.051 | 0.051 |
| Oxfordshire | 0.278 | 0.316 | 0.582 | 0.241 | 0.266 | 0.380 | 0.190 | 0.203 | 0.266 | 0.165 | 0.101 |
| Yorkshire | 0.372 | 0.414 | 0.621 | 0.248 | 0.269 | 0.379 | 0.090 | 0.103 | 0.138 | 0.083 | 0.034 |
| All populations | 0.363 | 0.411 | 0.625 | 0.309 | 0.333 | 0.431 | 0.186 | 0.200 | 0.236 | 0.131 | 0.077 |
The two main criteria were a minimum location quotient (LQ) of the district with the highest LQ (MLQ) and maximum distance of the mean grandparental place of birth (MGP) from that district for each sample. When no distance is given, the distance constraint was not used. A number of samples were further excluded because of observed multiple peaks or broad geographic surname distributions (see Supplementary Table 3). These exclusions are incorporated into the proportions here.
Figure 1 shows, for each surname, a plot of the MLQ against the surname population size as given in the 1881 UK census. There are a few obvious outliers from the general distribution, which indicates that there are few surnames with higher MLQs than would be expected from their abundance, with MLQs ranging from 23 to 42. These surnames are almost exclusively established Welsh surnames (Jones, Davies, Evans, Thomas, Hughes, James and Phillips), surnames that are distinctive, but at a scale that is region specific. There are also some surnames that were not classified as local despite having a high MLQ. This is either because they had a multi-centre distribution or the average grandparental birthplace was further than 83 or 120 km from the district with the MLQ. The proportion excluded from the local classification for these reasons ranged from 0 (several populations for which high MLQ thresholds were used) to 0.385 (Pembrokeshire, MLQ>19, Supplementary Table 3).
Genotypes
In all, 1019 of the pilot samples were successfully genotyped and the genotype data for the loci typed are given, by region, in Table 2 (Supplementary Figure 4). Only HLA alleles with a frequency greater than 7.5% in at least one population are shown here. The full HLA allele data set is given in Supplementary Table 1. All autosomal loci were in Hardy–Weinberg equilibrium.
Evidence for population structure
Pairwise FST values, calculated separately for each marker, showed no obvious consistent patterns, apart from the suggestion at three loci (HLA-B, rs7853989 and NRY) that the Orcadian samples appear to be significantly different from the rest (Supplementary Table 4). As may be expected from a marker with a lower effective population size, FST values calculated using the NRY data were greater than those for the autosomal markers.
The aim of dividing the samples into those with local as opposed to non-local surnames was to see whether this would accentuate regional divergence, and therefore reveal a greater extent of population substructure. The procedures described in the methods section for distinguishing between local and non-local surnames enable a hierarchical classification of the samples based on a combination of MLQ values and distance constraints. This ranges, as described above, from no constraint (no splitting between local and non-local) to the maximum locality constraint of an MLQ>120 and distance <83 km, with lower LQ cut offs and the lesser distance cut offs lying somewhere between these two extremes. Pairwise FST values calculated from different degrees of locally defined surname samples still did not reveal any consistent patterns (Supplementary Table 5).
Given that the FST analysis was clearly not powerful enough to detect population structure in our pilot sample, we decided to see whether an analysis of population admixture might be more revealing. For this, we first assumed that the central population was a simple mixture between two source populations, namely the South West, a surrogate for the Ancient British and the Eastern, a surrogate for the Anglo-Saxons. Using only local samples of each of the population groups to estimate the admixture, by the maximum likelihood procedure, the autosomal data with the most stringent thresholds (MLQ>120, distance <83 km) suggested that most of the contribution was from the Eastern population (0.945 East (0.895–0.995), Table 4). When only non-local samples are used for the analysis, there was a substantial contribution from both source populations (0.630 East (95% CI 0.591–0.669), Table 4). Using a much lower stringency (MLQ>19, distance120 km), the estimates suggested that there was again a major contribution from the Eastern population (0.900, 0.829–0.971) and again, when non-local samples are used, there was a substantial contribution from both source populations (0.525, 0.482–0.568). The NRY sample sizes were too small to allow analysis of subdivided data. Using all the available male samples, the Eastern contribution to the Central population was still substantially greater than the Western contribution, although the confidence intervals were very large (0.620, 0.000–1.000). At face value these data suggest first of all that there is measurable population substructure in contrast to the FST calculations. Second, they suggest a very substantial contribution to the central population from the East, putatively the Anglo-Saxons. Intriguingly, the difference between the autosomal and NRY analysis suggests that the male Eastern contribution may be less than the female. However, the NRY CIs are large.
Table 4. Maximum likelihood admixture estimates for the most stringent and the least stringent criteria used to define local and non-local surnames.
| ‘Admixed' population | Parental populations | Local (L) or non-local (N) | Proportion East | −95% CI | +95% CI |
|---|---|---|---|---|---|
| LQ>120, distance<83 kma | |||||
| CN | West vs East | L | 0.945 | 0.895 | 0.995 |
| CN | West vs East | N | 0.630 | 0.591 | 0.669 |
| OR | West vs East | L | 0.550 | 0.488 | 0.614 |
| OR | West vs East | N | 0.695 | 0.630 | 0.760 |
| LQ>19, distance<120 kmb | |||||
| CN | West vs East | L | 0.900 | 0.829 | 0.971 |
| CN | West vs East | N | 0.525 | 0.482 | 0.568 |
| OR | West vs East | L | 0.360 | 0.265 | 0.455 |
| OR | West vs East | N | 0.815 | 0.761 | 0.869 |
|
‘Admixed' population |
Parental populations |
Local (L) or non-local (N) |
%Norse |
−95% CI |
+95% CI |
| LQ>120, distance<83 kma | |||||
| OR | Norse vs West | L | 0.375 | 0.331 | 0.419 |
| OR | Norse vs West | N | 0.405 | 0.357 | 0.453 |
| LQ>19, distance<120 kmb | |||||
| OR | Norse vs West | L | 0.315 | 0.266 | 0.364 |
| OR | Norse vs West | N | 0.420 | 0.375 | 0.465 |
The contributions of the putative ancestral populations (East, West and Norse) to the putative admixed population (Central (CN) or Orkney (OR)) were estimated for either the local surnames (L) alone or only the non-local (N) surnames. For the Orkney anaylsis, all Orcadian samples were compared with either local or non-local stratified PoBI samples.
Most stringent criteria
Least stringent criteria
The Orcadian population is thought to be a mixture of Norse Vikings and, mostly, the Ancient British.1, 28 Because our Norse population surrogate was based on limited published Norwegian data, we used only a subset of the autosomal data (HLA-A, -B, -C, -DQB1, MC1R and rs7853989) for the admixture analysis. The source populations were the South Western set, as before as a proxy for Ancient British ancestry, and published Norwegian (or Swedish) data as a proxy for Norse Viking ancestry. The estimate of Norse ancestry was 0.375 (0.331–0.419) for the local surnames, rising just slightly to 0.405 (0.357–0.453) when non-local surnames were used at the highest stringency. These estimates were 0.315 (0.266–0.364) and 0.420 (0.375–0.465) at a lower stringency. The NRY estimate of Norse ancestry was 1.000 (0.139–1.000), again with a very wide CI.
We repeated the analysis on the Orcadian samples using the Eastern set instead of Norway. This comparison showed a lower admixture from the East for the local than the non-local samples, especially using the less stringent criteria. This may well be because the non-local samples are ‘contaminated' with some Viking admixture, although possibly mainly from the Danish Vikings, who must have been very closely related to the Anglo-Saxons as they came from essentially the same geographical area. Using the most stringent criteria for local, the estimates of admixture from West versus East and Norse versus West match remarkably well, suggesting in both cases a nearly 50% contribution from Ancient British to Orcadian ancestry, with a likely higher Norse contribution from males than females. There can be no doubt that the admixture analysis is much more sensitive for the detection of population structure in these rather closely related populations, and that the use of local surnames, does affect the analysis and helps to create a finer population subdivision.
Discussion
The PoBI samples represent a very carefully recruited set of rural volunteers with the intention that they can be used as a standard UK-control population. The main advantage of the samples is that the provenance of the four grandparents is known, reaching further into the past than by simply using the volunteer's place of birth. This greatly improves the chance that the volunteers are locally representative samples and avoids recent admixture events as far as possible.
The most challenging aspect of this project has been to collect samples from volunteers who fit the stringent selection criteria. A number of methods were used to recruit the volunteers through a collaboration with 10 groups spread throughout the country, and it took a full 5 years to collect the current 4000 PoBI samples. This is largely due to the fact that, from our experience, a small proportion of people (probably less that 5% of the population in general) fit the criteria. Indeed, the age range of the samples, with the majority being over 60, suggests that there is likely to have been more movement in recent years, and hence in the future, fewer people will fit these criteria. It should, however, be borne in mind that this bias in the ages will also, to some extent, be dictated by availability of volunteers to attend events because of restrictions caused by work and some self-selection of volunteers with an interest in family history. Our volunteers are older than the population average (the average age for starting genealogical research is likely to be 40 (psych.fullerton.edu/genealogy/#elderly)), but the older age distribution has the advantage of giving a greater time depth to the set of samples. Number of individuals who fit the criteria will continue to decline as a result of an increasingly mobile global community and hence now may well be the last opportunity to collect such samples with relative ease.
The majority of the samples collected did fit the criteria required. Analysis of the first 3865 samples that have been geocoded indicates that 75% have an MD between grandparental birthplaces of 37.3 km (Table 1), and about 70% of grandparental birthplaces could be classed as rural, although this does depend on the criteria used. These figures emphasise the quality of the samples collected, which gives the potential for a finer-scale analysis of the UK population that can be carried out using other available control sample collections.
Preliminary genotyping of 1057 samples, using nine loci, demonstrates the value of these samples for investigating a fine-scale population structure within the UK. The use of traditional methods such as pairwise estimation of FST, PCA and STRUCTURE (PCA and STRUCTURE were both applied but showed no patterns) failed to detect any structure in this pilot project, probably because the sample sizes and numbers of loci used are too small to detect such differences. Instead, we have used an admixture analysis, based on historical priors, to investigate whether a fine-scale structure in the UK could be detected in these samples and to see if partitioning the samples by surnames, an important asset of our PoBI cohort, enhances the power to detect structure. Simple point admixture estimates, based on linear combinations of contributions from ancestral populations, did reveal the expected population structure. This was more finely dissected using the surname data to further stratify the samples by local and non-local surnames. In particular, for both the high and low stringencies, there is a significant difference between admixture estimates for the local versus non-local surnames in both the CN (Central) and Orkney populations when the Eastern and Western populations are used as parental populations (Table 4).
The project has now collected about 4000 samples that are available for further analysis. Just under 3000 of the samples have recently been genotyped as replication controls for WTCCC2 on both the Illumina 1.2M and Affymetrix v6.0 whole-genome SNP platforms and these data will facilitate a more detailed investigation of UK population structure. These genotype data should be further enhanced by using surnames to improve the sample localisation, together with a careful geocoding analysis using the detailed knowledge of the grandparental birthplaces. A total of 100 of the samples, split between regions likely to be most representative of the Ancient Britons and the subsequent Anglo-Saxon and Norse Viking incursions (Cornwall, Kent, Orkney and West Scotland), are being sequenced as part of the 1000 Genomes Project.12 These will also be available to the research community.
We believe that our method of selecting volunteers is a powerful way to collect a set of samples that can be used for high-quality analysis of a fine-scale population structure in the UK. Subsequent localisation using surnames can sharpen the results of the structure analysis. Even with limited data, an appropriate admixture analysis can give a much more refined result than use of FST, or PCA and similar structure detection analyses.
Acknowledgments
This project was funded by the Wellcome Trust, and we would like to acknowledge Lon Cardon and David Bicknell for their help in putting the grant application together. Sorrel May and Nick Godwin helped to kick start the large-scale recruitment of volunteers, while Chris Abell, Tanefa Apekey, Carolina Bonilla, Julie Burton, John Burns, Joy Hadfield, Christopher Hand, Amanda Howe, Pat Jonas, Ciaran Kilkelly, Donald Lehman, Julie Lewis, Louise Lynagh, Shan Owens, Jan Tawn, Malcolm Taylor and Stan Urbaniak were heavily involved in recruiting. This project was supported by the Peninsula NIHR Clinical Research Facility (University of Exeter) and the Wellcome Trust Clinical Research Facility (Manchester). Finally, we would like to thank all the volunteers themselves and the many friends and colleagues who helped to recruit them.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Wilson JF, Weiss DA, Richards M, Thomas MG, Bradman N, Goldstein DB. Genetic evidence for different male and female roles during cultural transitions in the British Isles. Proc Natl Acad Sci USA. 2001;98:5078–5083. doi: 10.1073/pnas.071036898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelli C, Redhead N, Abernethy J, et al. A Y chromosome census of the British Isles. Curr Biol. 2003;13:979–984. doi: 10.1016/s0960-9822(03)00373-7. [DOI] [PubMed] [Google Scholar]
- King TE, Ballereau SJ, Schurer KE, Jobling MA. Genetic signatures of coancestry within surnames. Curr Biol. 2006;16:384–388. doi: 10.1016/j.cub.2005.12.048. [DOI] [PubMed] [Google Scholar]
- Bowden GR, Balaresque P, King TE, et al. Excavating past population structures by surname-based sampling: the genetic legacy of the Vikings in northwest England. Mol Biol Evol. 2008;25:301–309. doi: 10.1093/molbev/msm255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Dushlaine CT, Morris D, Moskvina V, et al. Population structure and genome-wide patterns of variation in Ireland and Britain. Eur J Hum Genet. 2010;18:1248–1254. doi: 10.1038/ejhg.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Bell JI. Association study designs for complex diseases. Nat Rev Genet. 2001;2:91–99. doi: 10.1038/35052543. [DOI] [PubMed] [Google Scholar]
- Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles D. The Tribes of Britain: Who Are We and Where Do We Come From? London: Weidenfeld and Nicolson; 2005. [Google Scholar]
- Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1000 Genomes Project Consortium A map of human genome variation from population-scale sequencing. Nature. 2010;467:1062–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TE, Jobling MA. Founders, drift and infidelity: the relationship between Y chromosome diversity and patrilineal surnames. Mol Biol Evol. 2009;26:1093–1102. doi: 10.1093/molbev/msp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheshire J, Mateos P, Longley PA.Creating a regional geography of Britain through the spatial analysis of surnames Geoforumdoi: 10.1016/j.geoforum.2011.02.001in press). [DOI]
- Whyte ID.Migration in early-modern Scotland and Englandin Pooley CG, Whyte ID (eds):: Migrants Emigrants and Immigrants New York: Routledge; 1991 [Google Scholar]
- Sly WS, Gurbax SS, Kennett R, Bodmer WF, Bodmer JG. Permanent lymphoid lines from genetically marked lymphocytes: success with lymphocytes recovered from frozen storage. Tissue Antigens. 1976;7:165–172. doi: 10.1111/j.1399-0039.1976.tb01047.x. [DOI] [PubMed] [Google Scholar]
- Woollard M, Allen M.1881 Census for England, Wales, The Channel Islands and the Isle of Man: Introductory user Guide V 0.4Distributed by the History Data Service, UK Data Archive.1991
- Burt J, Barber G, Rigby D.Elementary Statistics for Geographers3rd ednLondon: The Guilford Press; 2009 [Google Scholar]
- Bodmer JG, De Luca M, Moses JH, Heyes JM, Marsh SGE. HLA Class I and II study in an Orcadian population. Hum Immunology. 1996;47:51. [Google Scholar]
- Bodmer WF, Bodmer JG. Evolution and function of the HLA system. Br Med Bull. 1978;34:309–316. doi: 10.1093/oxfordjournals.bmb.a071518. [DOI] [PubMed] [Google Scholar]
- Solberg OD, Mack SJ, Lancaster AK, et al. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum Immunol. 2008;69:443–464. doi: 10.1016/j.humimm.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evseeva I, Nicodemus KK, Bonilla C, Tonks S, Bodmer WF. Linkage disequilibrium and age of HLA region SNPs in relation to classic HLA gene alleles within Europe. E J Hum Gen. 2010;18:924–932. doi: 10.1038/ejhg.2010.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees JL. The melanocortin 1 receptor (MC1R): more than just red hair. Pigment Cell Res. 2000;13:135–140. doi: 10.1034/j.1600-0749.2000.130303.x. [DOI] [PubMed] [Google Scholar]
- Harding RM, Healy E, Ray AJC, et al. Evidence for variable selective pressures at MC1R. Am J Hum Genet. 2000;66:1351–1361. doi: 10.1086/302863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstenblith MR, Goldstein AM, Fargnoli MC, Peris K, Landi MT. Comprehensive evaluation of allele frequency differences of MC1R variants across populations. Hum Mutat. 2007;28:495–505. doi: 10.1002/humu.20476. [DOI] [PubMed] [Google Scholar]
- Mourant AE, Kopeć AC, Domaniewska-Sobczak K. The Distribution of Human Blood Groups. London: Oxford University Press; 1976. [Google Scholar]
- Garratty G, Glynn SA, McEntire R, Retrovirus Epidemiology Donor Study ABO and Rh(D) phenotype frequencies of different racial/ethnic groups in the United States. Transfusion. 2004;44:703–706. doi: 10.1111/j.1537-2995.2004.03338.x. [DOI] [PubMed] [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, et al. The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci USA. 2001;98:10244–10249. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y Chromosome Consortium A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Winney B, Litton C, Parkin D, Feare C. The subspecific origin of the inland breeding colonies of the cormorant Phalacrocorax carbo in Britain. Heredity. 2001;86:45–53. doi: 10.1046/j.1365-2540.2001.00807.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.