Abstract
Dyslexia is a developmental disorder characterised by extensive difficulties in the acquisition of reading or spelling. Genetic influence is estimated at 50–70%. However, the link between genetic variants and phenotypic deficits is largely unknown. Our aim was to investigate a role of genetic variants of FOXP2, a prominent speech and language gene, in dyslexia using imaging genetics. This technique combines functional magnetic resonance imaging (fMRI) and genetics to investigate relevance of genetic variants on brain activation. To our knowledge, this represents the first usage of fMRI-based imaging genetics in dyslexia. In an initial case/control study (n=245) for prioritisation of FOXP2 polymorphisms for later use in imaging genetics, nine SNPs were selected. A non-synonymously coding mutation involved in verbal dyspraxia was also investigated. SNP rs12533005 showed nominally significant association with dyslexia (genotype GG odds ratio recessive model=2.1 (95% confidence interval 1.1–3.9), P=0.016). A correlated SNP was associated with altered expression of FOXP2 in vivo in human hippocampal tissue. Therefore, influence of the rs12533005-G risk variant on brain activity was studied. fMRI revealed a significant main effect for the factor ‘genetic risk' in a temporo-parietal area involved in phonological processing as well as a significant interaction effect between the factors ‘disorder' and ‘genetic risk' in activation of inferior frontal brain areas. Hence, our data may hint at a role of FOXP2 genetic variants in dyslexia-specific brain activation and demonstrate use of imaging genetics in dyslexia research.
Keywords: dyslexia, imaging genetics, FOXP2, fMRI
Introduction
The feasibility to combine functional imaging studies and genetics was demonstrated in several studies.1 Imaging genetics approaches investigate the role of certain genetic variants (eg, single nucleotide polymorphisms – SNPs) on brain activation independently of indirect measurements like behavioural or neuropsychological testing. Promising results were already shown for other neurological disorders, for example, in studies of anxiety,2 attentional processes3 or schizophrenia.4 However, to our knowledge, no functional imaging genetics study has been published on dyslexia so far.
Dyslexia is a specific and severe disorder of reading and spelling, with ∼5% affected schoolchildren in Germany.5 About 50–70% of dyslexia can be explained by genetic influence.6 However, as all the so far identified disease genes account only for a small part of genetic risk, more dyslexia-related genes need to be identified. FOXP2 is a highly relevant candidate gene, situated on chromosome 7q31, close to 7q32, a genomic region in linkage with dyslexia.7 FOXP2 was discovered to be a central gene in language development8, 9, 10 not limited to humans.11, 12, 13, 14 Its functional mechanism could be connected with a downregulation of CNTNAP2, a gene implicated in neuronal recognition and cell adhesion.15 FOXP2's role in language originally became apparent by the discovery of a missense mutation (R553H) leading to developmental verbal dyspraxia.16 However, a mutation screen of FOXP2 in six affected individuals7 found no specific mutations in dyslexia.
Several functional magnetic resonance imaging (fMRI) studies revealed specific differences between dyslexics and normal readers in reading-related brain regions. Consistently, three left hemispheric areas (inferior frontal,17 partly involving Broca's area, dorsal, and ventral temporal cortices, including Wernicke's area) showed significant activation differences in dyslexics in phonological tasks.18, 19 The dorsal and ventral areas show, independently from language,20 less activation in dyslexics compared with normal readers. In older dyslexics, the inferior frontal area is often overactivated. However, this overactivation could also be interpreted as a compensational reading strategy.19
The functional effect of a FOXP2 mutation on inferior frontal and supramarginal brain areas has been demonstrated by Liégeois et al10 for silent and spoken language processing. Therefore our aim was to analyse the role of genetic variants of FOXP2 in the processing of written language in dyslexia. Thus we combined genetics and fMRI using an imaging genetics approach. To select a possibly functionally relevant FOXP2 variant, we initially applied a case/control (n=245) study. Relevance of the most promising variant was investigated in respect to potential functional consequences on brain activation in a reading-related task in fMRI.
Materials and methods
Ethical approval
Our study was approved by the ethics committees of the Universities of Leipzig and Jena as well as the Saxon Ministry of Culture and Sports. Informed and written consent was obtained from subjects' parents.
Study group for genetic epidemiology
This group consisted of 61 dyslexics of German origin and 184 healthy, ethnically matched blood donors as controls. Dyslexics were recruited in the 3rd or 4th grade of special Saxonian dyslexia classes. Age was between 8.8 and 11.4 years, 63% were male. Cases were ascertained in a two-stage approach: first, schools with special dyslexia classes were contacted. Children in those classes were already tested thoroughly at the end of 2nd grade by the local school board with a variety of psychometric tests, including non-verbal intelligence, letter knowledge, phoneme mergence, spelling, memory, mathematical skills as well as reading and listening comprehension. Pursuant to the admission criteria for the special dyslexia classes only children without memory or math problems and a discrepancy between IQ and reading performance of at least 1.25 SDs get access to those classes.
Second, additional tests21, 22, 23 described elsewhere in detail24 were applied to assess reading performance, minimise inclusion of children with attention deficit/hyperactivity (ADD/ADHD) and ensure an IQ≥85. No siblings or twins were included.
SNP selection
We identified tagging SNPs covering common variants with a minor allele frequency (MAF) ≥0. 1 (HapMap data release 24, Haploview-Version 4.0 beta 13 (http://www.broad.mit.edu/mpg/haploview/) and HapMap Public Release #21a, (International HapMap Project http://www.hapmap.org/) applying aggressive tagging, an r2 threshold of 0.8, and a logarithm of the odds threshold for multi-marker tests of 3.0. Nine SNPs were selected: rs12533005, rs10228350, rs10268637, rs4727799, rs17137124, rs7782412, rs12670585, rs936146, and rs10953766. They covered 87% of all common HapMap SNPs located in FOXP2 with a mean r2 of 0.97. Furthermore, we genotyped the mutation R553H, previously found in a large Pakistani family with severe speech and language disorder.8
Genotyping and genetic analysis
DNA extraction and PCR primer design were done as described.24 Primers for single base extension including photo-cleavable sites25 were designed using CalcDalton.26 SNPs were genotyped applying the method ‘GenoSNIP' as described previously.25, 27 Primer sequences are shown in Supplementary Table 1.
Genetic statistics
We analysed SNPs for association with dyslexia applying χ2 statistics in allelic, dominant, and recessive models as well as applying the Cochran–Armitage test. SNPs of cases and of controls were in Hardy–Weinberg Equilibrium (HWE) (P>0.05) indicating no major genotyping errors. Average genotyping rate was 98% for cases and controls. Haploview 4.1 was used to test for haplotype association for haplotypes with a frequency >5%. P-values are shown without correction for multiple testing. The study was powered to detect a difference of at least 15% in the MAF of selected SNPs between cases and controls translating to a minimum detectable odds ratio (OR) of 1.85.28
Prediction of altered gene expression in silico
MatInspector and Genomatix software suite (NCBI 37, ElDorado 12-2010, Genomatix Software GmbH, Munich, Germany), were used to investigate possible loss/gain of transcription factor (TF)-binding sites as described elsewhere.29 Minimum core similarity (score of the highest conserved positions of a matrix match) was 1.00.
Analysis of differential allelic gene expression in human hippocampus
Biopsy samples (n=142) were obtained from patients with chronic pharmacoresistant temporal lobe epilepsy. After quality control, 138 individuals were included in subsequent analyses (63 male, mean age 31.72 years, SD=16.27, age range newborn to 64 years). Fresh frozen human hippocampal segments were prepared as tissue slices under cryostat conditions (Bonn tissue bank). Total DNA and RNA were isolated using the AllPrep DNA/RNA Micro Kit (Qiagen, Hilden, Germany). Quality of total RNA of all samples was checked for degradation (RNA integrity number RIN>7.9) via BioAnalyzer measurements (Agilent Technologies, Waldbronn, Germany). A volume of 50 ng of total RNA were amplified (Illumina TotalPrep 96-RNA Amplification Kit, Ambion/Applied Biosystems, Darmstadt, Germany). Labelled cRNA was hybridised to Illumina human HT-12 Expression v3 BeadChips (Illumina, San Diego, CA, USA). All expression profiles were extracted using GenomeStudio software (Illumina). For genome-wide SNP-genotyping 200 ng of DNA was hybridised to the Illumina Human660W-Quad v1 DNA Analysis BeadChip (Infinium HD Assay Super manual, Illumina). Individuals were defined as outliers and excluded from analysis if their coordinates on at least one of the first two axes of the multi-dimensional scaling analysis on the identical by state matrix of genotyped SNPs was more than six standard deviations away from the mean position of all other individuals. Quality thresholds were as follows: HWE P-value ≥1 × 10−5, MAF≥1%, individual callrate ≥98%, SNP callrate ≥98%, and a false discovery rate of 1% for autosomal heterozygosity. Average genotyping rate was 98%. The sequences of expression probes were re-aligned to UCSC version 18 (hg18, http://genome.ucsc.edu/) allowing only perfect matches. Probes containing either intrinsic polymorphisms or matching to multiple positions in the human genome were excluded from downstream analysis. Remaining probes were normalised using the vsn2 option implemented in the package ‘VSN' for R. For quantitative trait analysis, linear regression of an additive allelic model was performed using the GenABEL package for R (http://www.genabel.org/). Covariates included in the model were gender and age at sampling. To take population stratification into account, we also included the first five components resulting from multidimensional scaling analysis.
For the investigation of the SNP's potential influence on FOXP2 expression, we had to use proxy SNPs as rs12533005 was not included on the Illumina Human660-W Quad array. We identified proxy SNPs by filtering variants correlated with rs12533005 with r2>0.5 according to HapMap release 22 (International HapMap Project http://www.hapmap.org), and an additional effect on gene expression of FOXP2 probes with a nominal P-value ≤0.05. Out of five resulting SNPs, we report data of the variant with strongest effect size (rs10249531).
Study group for fMRI investigation
In an additional, independent sample we investigated children with fMRI. The sample consisted of 19 dyslexics (12 male, mean age 11.46 years, SD=1.04) and 14 controls (11 male, mean age 11.56, SD=0.73). Dyslexics and controls were recruited in Thuringia, a German federal state adjacent to Saxony. Inclusion criteria were an IQ≥85 (non-verbal part of HAWIK-III30), right handedness,31 no attention deficit disorder21, 31), and no history of neurological or psychiatric disorders. Diagnosis of dyslexia was based upon two criteria: (a) discrepancy between non-verbal IQ and reading32/spelling33, 34, 35, 36 ≥1.5 SDs and (b) reading/spelling performance (transformed to IQ scale) <85. Normal performance of controls was controlled by reading/spelling performance (transformed to IQ scale) >85. Controls were matched to dyslexics according to non-verbal IQ and age. Both groups did not differ significantly in IQ and age. In accordance with the additive model applied in differential allelic gene expression analysis, carriers of allele rs12533005-G were classified as subjects with genetic risk, individuals homozygous for rs12533005-C as subjects without genetic risk.
fMRI paradigm
fMRI data were acquired in a Siemens Magnetom Vision 1.5. Tesla MRI Scanner (Siemens, Erlangen, Germany) using a T2-weighted epi-sequence (TE=60 ms, TR=0.6 ms, TA=3496 ms, FOV=192 mm, FA=90°, 64 × 64 matrix) with 32 slices (4 mm slice thickness).
To operationalise phonological as well as low-level visual processing we used a paradigm comprising (1) phonological stimuli (‘rhyming': do the names of two letters rhyme; eg, ‘G' and ‘T') and (2) visual control stimuli (‘slashes': are two slashes inclined in the same direction; eg, ‘/' vs ‘').
The experiment was conducted in a block design. Four scans were acquired per block. For each stimulus type, 11 blocks were acquired, totalling 44 scans per task. Each stimulus block was followed by a baseline-task block (‘fixation': fixation cross for 13984 ms, 21 baseline blocks in total with four scans per block). The block sequence was kept the same between subjects.
Subjects had to indicate their decision via button press, allowing the acquisition of reaction time and error rate (Presentation, Neurobehavioral Systems, http://www.neurobs.com/presentation). The trial setup was fixed to a duration of 3000 ms followed by a 450-ms inter stimulus interval. During each trial the stimuli were presented for a maximum of 3000 ms. The stimulus disappeared after the button press, leaving a blank screen until the end of the 3000 ms interval.
fMRI data analysis
fMRI data were analysed with the SPM8 package (Institute of Neurology, London, UK, http://www.fil.ion.ucl.ac.uk/spm/software/spm8/). Spatial pre-processing included the following: (a) realignment of all functional images to the session's first image and computation of a mean image out of realigned images, (b) normalisation of realigned functional images to the EPI-MNI-template (International Consortium for Brain Mapping template, Montreal Neurological Institute, Montreal, Canada) by estimation of normalisation parameters for the mean image and their application to functional images, and (c) smoothing of images with a Gaussian kernel of 9 mm FWHM.
First-level analyses were computed for each subject using a voxel-by-voxel t-test for the contrasts of (A) rhyming vs fixation as well as (B) rhyming vs slashes. Second-level analyses for the contrasts (A) and (B) were computed via 2 × 2 analysis of variance with factors ‘disorder' (dyslexic or normal reader) and ‘genetic risk' (risk variant rs12533005-G or no risk variant) using a full factorial design. Post-hoc t-tests were done to test intergroup effects for (1) controls vs dyslexics, (2) dyslexics vs controls, (3) no-risk subjects vs risk subjects, and (4) risk subjects vs no-risk subjects. fMRI results are reported on an uncorrected P-level of 0.001 and a cluster level of 10.
Results
Selection of genetic variants for imaging analysis
The following SNPs in FOXP2 were investigated for association with dyslexia: rs12533005 and rs10228350 (intron 1), rs10268637 and rs4727799 (intron 2), rs17137124 (intron 3), rs7782412 and rs12670585 (intron 9), rs936146 (intron 11), rs10953766 (intron 17) and their resulting haplotypes. We also studied the R553H mutation described by Lai et al.8 A summary including the linkage disequilibrium structure of FOXP2 in our population is shown in Supplementary Figure 1.
SNP rs12533005 showed nominal significant association (OR genotype G/G, recessive model: 2.1 (95% confidence interval 1.1–3.9) P=0.016; Cochran–Armitage test P=0.049, for more details see Supplementary Table 2). No effect of age on allele frequency as described elsewhere37 was found. Other tested SNPs and haplotypes did not show significant frequency differences between cases and controls (Supplementary Tables 2 and 3). None of the haplotypes showed stronger association than single marker rs12533005. No sex-specific differences were observed. Haplotype structure in our cohort was comparable to that in HapMap CEU panel. Mutation R553H was not present in any cases or controls. Hence, we selected rs12533005 for further imaging genetic analyses.
Prediction of altered gene expression in silico
Three TF-binding sites for regulators ATBF1 (=ZFHX3), LEF1, and MEL1 (=PRDM16) are predicted in silico to be present for carriers of the risk variant rs12533005-G, but not for rs12533005-C. Matrix similarity (score of the complete matrix match) for ATBF1, LEF1, and MEL1 was 0.80, 0.86, and 1.00, respectively.
Analysis of differential allelic gene expression in human hippocampus
As rs12533005 was not originally included in the analysis of differential allelic expression in human hippocampus, we identified rs10249531 located 41 kb upstream of rs12533005 as most relevant proxy (r2=0.6, D′=1.0). SNP rs10249531-C (correlating with rs12533005-G) was nominally associated with decreased FOXP2 expression (P=0.018), accounting for 4.1% of gene expression variability (Supplementary Figure 2). Within the additive model representing the effect of rs10249531-C on FOXP2 gene expression levels, we found a regression coefficient beta of −0.024. Within our sample, there was no evidence for age-dependent expression differences.
fMRI results
Among analysed SNPs of FOXP2, rs12533005 showed the strongest signs of association with dyslexia. Therefore, the functional relevance of this SNP in language and speech processing was further analysed using fMRI.
Intragroup results
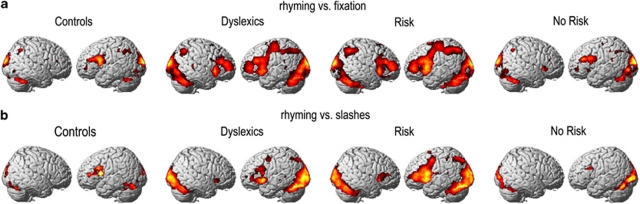
Individuals of the fMRI sample were grouped according to disorder state: (1) dyslexics (n=19), (2) controls (n=14); and according to carriage of rs12533005-G: (3) subjects with genetic risk (n=25), (4) subjects without genetic risk (ie, individuals homozygous for rs12533005-C, n=8). Intragroup results for the contrast (A) rhyming vs fixation for groups (1) and (2) showed in each group activations in the expected (phonological) reading network comprising occipital and inferior temporal brain areas, the angular gyrus, the insula and inferior frontal brain areas (Figure 1). The same areas were found to be activated when groups were defined according to (3) and (4). For contrast (B), rhyming vs slashes, activity in the same network could be observed.
Figure 1.
Intragroup results according to state of disorder and genetic risk. (a) Intragroup results for the contrast rhyming vs fixation, (b) intragroup results for the contrast rhyming vs slashes. Sample sizes: dyslexics n=19, controls n=14, risk (carriers of rs12533005-G) n=25, no risk (non-carriers of rs12533005-G) n=8.
Main effects and interactions
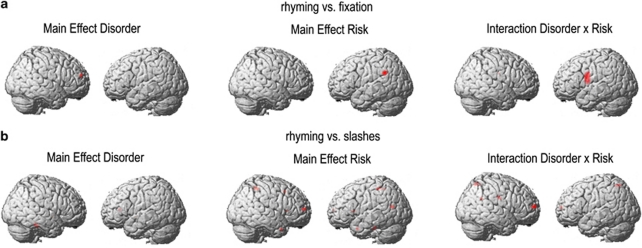
In contrast (A) there was only a single cluster of right middle frontal brain areas associated with the main effect ‘disorder'. For main effect ‘genetic risk' there was an association in a superior temporal cluster bordering to the angular gyrus. For the interaction effect ‘disorder × genetic risk' a left-sided cluster in the Rolandic operculum could be observed.
In contrast (B) there were three clusters associated with the main effect ‘disorder': one left-sided cluster in the white matter close to the insula, one cluster in the grey matter close to the anterior commissure and the third in the right cerebellum. For the main effect ‘genetic risk' various small clusters within the right precuneus, the left and right nucleus caudatus, right-sided superior frontal, the left and right temporal pole, left-sided postcentral, and, finally, in the left-sided fusiform gyrus could be observed. For the interaction effect ‘disorder' × ‘genetic risk' clusters in the left and right precuneus as well as in the right superior medial gyrus could be observed. Main effects and interactions for contrasts (A) and (B) are depicted in Figure 2.
Figure 2.
Main effects and interactions according to state of disorder and genetic risk. (a) Results of the F-test for the contrast rhyming vs fixation, (b) results of the F-test for the contrast rhyming vs letters. Disorder: contrast dyslexics vs controls. Risk: contrast carriers of rs12533005-G vs non-carriers. Sample sizes: dyslexics n=19, controls n=14, risk (carriers of rs12533005-G) n=25, no risk (non-carriers of rs12533005-G) n=8.
Post-hoc t-tests
Post-hoc t-tests were applied to investigate the direction of main effects and interactions. The post-hoc t-tests for the contrast (A) for controls vs dyslexics revealed no significant effects, whereas the t-test for dyslexics vs controls showed a small overactivation in the right-sided fusiform gyrus. The t-test of no genetic risk vs genetic risk (ie, non-carriage vs carriage of rs12533005-G) showed a prominent overactivation in the supramarginal and angular gyri as well as three small clusters of overactivation in (I) an area bordering on the inferior frontal gyrus, (II) the superior occipital gyrus, and (III) the lingual gyrus for subjects with no genetic risk. The t-test of genetic risk vs no genetic risk did not show any significant effects.
Post-hoc t-tests for contrast (B) for controls vs dyslexics showed two clusters in the white matter of the left and right middle frontal, the t-test for dyslexics vs controls showed two small clusters left sided in the lingual, fusiform, and middle occipital gyrus. The t-tests for non-genetic risk vs genetic risk and vice versa showed no significant clusters.
Discussion
Imaging genetics links genetic variants and their functional relevance in brain activation. To our knowledge we report for the first time results of fMRI-based imaging genetics in dyslexia. We initially performed a genetic association analysis of FOXP2 variants to prioritise markers for use in fMRI analysis. We selected FOXP2-tagging SNPs and the coding-variant R553H, a mutation initially found to be associated with severe speech and language disorder in a Pakistani family by Lai et al.8 No other coding variants of FOXP2 were investigated as no non-synonymously coding SNPs were reported in dbSNP (Build 130) and no non-synonymously coding SNPs in FOXP2 were found in dyslexics in a previous study.7 The coding mutation R553H was not found in any individual in our study, strengthening the hypothesis that R553H is a rare mutation present only in some sparse families and not of relevance for diseases in the general population.38 Of studied variants, only SNP rs12533005 showed nominal significant association with dyslexia (P=0.016). As FOXP2 haplotypes did not show stronger association than single marker rs12533005, this variant was chosen for imaging genetics analysis.
As rs12533005 is an intronic SNP, it does not change the protein sequence of FOXP2. It may be a marker for a yet unknown functional variant or it may modify regulation of gene expression. Notably, according to the PupaSuite database,39 rs12533005 is classified to be situated in a conserved region. Three TF-binding sites (ATBF1, LEF1, and MEL1) are predicted to be present in carriers of risk variant rs12533005-G, but not for rs12533005-C. MEL1 and ATBF1 have a role in regulatory transcription processes in the mammalian central nervous system.40 Positive MEL1 regulation indicates a multistep regulatory network aimed at expression of specific neuronal repressors.41 ATBF1 acts as a repressor of gene expression by downregulating AT-rich enhancer elements42, 43 and LEF1 seems to be involved especially in downregulation of E-cadherin, which is important for cellular polarity and adhesion processes during embryonic development in general44 as well as embryonic brain morphogenesis.45
To verify the results of our in silico analysis we examined differential allelic expression of FOXP2 rs12533005 in 138 human resected hippocampi. In this data set, the most relevant proxy for rs12533005 – which was not contained on the Illumina SNP array used by us – was rs10249531. We found significant reduction of FOXP2 gene expression for rs10249531-C, which is associated with rs12533005-G (Supplementary Figure 2). While there is no perfect linkage disequilibrium between the two SNPs, our data suggest differential allelic expression of FOXP2 depending on rs1253305 alleles. This supports our in silico prediction for a repressor binding site generated by rs12533005. We note that hippocampal tissues were derived from epileptic patients. However, we believe that the identified differential allelic expression of FOXP2 is not connected to a specific epilepsy-related process as neither the FOXP2 locus nor FOXP2 expression levels are reported to be associated with epilepsy. Based on these promising data we examined the role of rs12533005 in reading-related brain activation.
Using fMRI we found for the contrast of the ‘rhyming' vs ‘fixation' task a significant main effect of activation difference for the factor ‘risk' (ie, comparison of subjects being carriers and non-carriers of the risk variant rs12533005-G). Subsequent one-tailed post-hoc t-tests indicate that the main effect for the factor ‘risk' can be explained by an overactivation of the non-carriers of the risk-variant in two temporo-parietal brain areas: the angular and the supramarginal gyrus. This finding is in line with former fMRI research:18 the angular as well as the supramarginal gyrus are significantly involved in phonological language processing and showed a higher activation in normal readers compared with dyslexics.18 Thus our observation is consistent with the interpretation that these temporo-parietal brain areas show a functional deficit in carriers of the putative risk variant compared with non-carriers. Therefore our results point to a possible role of SNP rs12533005 in reading-related brain activation in the dorsal temporal cortex.
The interaction of factors ‘disorder' × ‘genetic risk' showed significant activation differences in the Rolandic operculum, a brain region strongly involved in motoric speech production. Post-hoc t-tests revealed for the comparison of non-risk carriers vs risk carriers a small cluster of overactivations in an area next to the inferior frontal gyrus. The interaction effect ‘disorder' × ‘genetic risk' can probably be explained by stronger activation of non-risk carriers in these inferior frontal areas. This result is in line with earlier findings of an effect of a FOXP2 mutation on the function of inferior frontal brain areas.10 Yet, due to the incomplete overlap of the cluster localisation between interaction effect and the post-hoc t-test result, our findings from interaction analysis may require further investigation.
Based on our results, it could be speculated that in presence of the risk variant rs12533005-G repressor binding sites are created, leading to decreased FOXP2 expression. As very tightly regulated FOXP2 expression in the developing brain seems to be required for development of speech and linguistic functions,46 repression might result in neuronal dysfunction giving a possible explanation for our fMRI findings.
We aimed to increase power to detect true positives by integrating genetic and functional data. Yet, given the only moderate sample sizes of the studied populations and no multiple testing correction, our findings need replication.
Our results hint on a possible role of FOXP2 variants in German dyslexics. The functional link may be found in an influence of FOXP2 on the function of the left hemispheric brain area involved in spoken (phonological memory) as well as written language (grapheme-phoneme correspondence and the mental lexicon).
Although replication of our results in an independent, larger cohort is definitely needed, our results further corroborate the role of FOXP2 in speech and language development, hint on the relevance of genetic variants of FOXP2 for reading and spelling, and demonstrate the application of imaging genetics in the investigation of dyslexia.
Acknowledgments
The work presented in this paper was made possible by funding from the German Federal Ministry of Education and Research (BMBF, PtJ-Bio, 0315883) to AW and HK, 01KN0702 to PA, and 01GS08144 to SC and AB, the Interdisciplinary Center for Clinical Research Jena (T.P. 1.2/B 307-04004) and a habilitation grant from the Hochschul- und Wissenschaftsprogramm/Thuringia (personal grant for CL). HK and PA were funded by means of the European Union, by the European Regional Development Fund (ERFD), the European Social Fund and by means of the Free State of Saxony (LIFE Center, University of Leipzig). We thank Nazanin Karbalai for statistical support.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Mattay VS, Goldberg TE. Imaging genetic influences in human brain function. Curr Opin Neurobiol. 2004;14:239–247. doi: 10.1016/j.conb.2004.03.014. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Mattay VS, Tessitore A, et al. Serotonin transporter genetic variation and the response of the human amygdala. Science. 2002;297:400–403. doi: 10.1126/science.1071829. [DOI] [PubMed] [Google Scholar]
- Fan J, Fossella J, Sommer T, Wu Y, Posner MI. Mapping the genetic variation of executive attention onto brain activity. Proc Natl Acad Sci USA. 2003;100:7406–7411. doi: 10.1073/pnas.0732088100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Goldberg TE, Kolachana BS, et al. Effect of COMT Val108/158 Met genotype on frontal lobe function and risk for schizophrenia. Proc Natl Acad Sci USA. 2001;98:6917–6922. doi: 10.1073/pnas.111134598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte-Körne G, Remschmidt H. Lese-Rechtschreibstörung (Legasthenie) - Symptomatik, Diagnostik, Ursachen, Verlauf und Behandlung. Deutsches Ärzteblatt. 2003;100:396–408. [Google Scholar]
- Harlaar N, Spinath FM, Dale PS, Plomin R. Genetic influences on early word recognition abilities and disabilities: a study of 7-year-old twins. J Child Psychol Psychiatry. 2005;46:373–384. doi: 10.1111/j.1469-7610.2004.00358.x. [DOI] [PubMed] [Google Scholar]
- Kaminen N, Hannula-Jouppi K, Kestila M, et al. A genome scan for developmental dyslexia confirms linkage to chromosome 2p11 and suggests a new locus on 7q32. J Med Genet. 2003;40:340–345. doi: 10.1136/jmg.40.5.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- Belton E, Salmond CH, Watkins KE, Vargha-Khadem F, Gadian DG. Bilateral brain abnormalities associated with dominantly inherited verbal and orofacial dyspraxia. Hum Brain Mapp. 2003;18:194–200. doi: 10.1002/hbm.10093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Baldeweg T, Connelly A, et al. Language fMRI abnormalities associated with FOXP2 gene mutation. Nat Neurosci. 2003;6:1230–1237. doi: 10.1038/nn1138. [DOI] [PubMed] [Google Scholar]
- Fee MS, Scharff C. The songbird as a model for the generation and learning of complex sequential behaviors. ILAR J. 2010;51:362–377. doi: 10.1093/ilar.51.4.362. [DOI] [PubMed] [Google Scholar]
- Fisher SE, Scharff C. FOXP2 as a molecular window into speech and language. Trends Genet. 2009;25:166–177. doi: 10.1016/j.tig.2009.03.002. [DOI] [PubMed] [Google Scholar]
- Frayer DW, Fiore I, Lalueza-Fox C, Radovcic J, Bondioli L. Right handed Neandertals: Vindija and beyond. J Anthropol Sci. 2010;88:113–127. [PubMed] [Google Scholar]
- Enard W, Gehre S, Hammerschmidt K, et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell. 2009;137:961–971. doi: 10.1016/j.cell.2009.03.041. [DOI] [PubMed] [Google Scholar]
- Vernes SC, Newbury DF, Abrahams BS, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359:2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargha-Khadem F, Watkins KE, Price CJ, et al. Neural basis of an inherited speech and language disorder. Proc Natl Acad Sci USA. 1998;95:12695–12700. doi: 10.1073/pnas.95.21.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaywitz SE, Shaywitz BA, Pugh KR, et al. Functional disruption in the organization of the brain for reading in dyslexia. Proc Natl Acad Sci USA. 1998;95:2636–2641. doi: 10.1073/pnas.95.5.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligges C, Blanz B. [Survey of fMRI results regarding a phonological deficit in children and adults with dyslexia: fundamental deficit or indication of compensation?] Z Kinder Jugendpsychiatr Psychother. 2007;35:107–115. doi: 10.1024/1422-4917.35.2.107. [DOI] [PubMed] [Google Scholar]
- Maisog JM, Einbinder ER, Flowers DL, Turkeltaub PE, Eden GF. A meta-analysis of functional neuroimaging studies of dyslexia. Ann NY Acad Sci. 2008;1145:237–259. doi: 10.1196/annals.1416.024. [DOI] [PubMed] [Google Scholar]
- Paulesu E, Demonet JF, Fazio F, et al. Dyslexia: cultural diversity and biological unity. Science. 2001;291:2165–2167. doi: 10.1126/science.1057179. [DOI] [PubMed] [Google Scholar]
- Brickenkamp R, Zillmer E. The d2 test of attention. Göttingen: Hogrefe; 2002. [Google Scholar]
- Marx H. Knuspels Leseaufgaben (KNUSPEL-L) Göttingen: Hogrefe; 1998. [Google Scholar]
- Weiss RH. Grundintelligenztest Skala 2. Göttingen: Hogrefe; 1998. [Google Scholar]
- Wilcke A, Weissfuss J, Kirsten H, et al. The role of gene DCDC2 in German dyslexics. Ann Dyslexia. 2009;59:1–11. doi: 10.1007/s11881-008-0020-7. [DOI] [PubMed] [Google Scholar]
- Wenzel T, Elssner T, Fahr K, et al. Genosnip: SNP genotyping by MALDI-TOF MS using photocleavable oligonucleotides. Nucleosides Nucleotides Nucleic Acids. 2003;22:1579–1581. doi: 10.1081/NCN-120023038. [DOI] [PubMed] [Google Scholar]
- Kirsten H, Dienst S, Emmrich F, Ahnert P. CalcDalton: a tool for multiplex genotyping primer design for single-base extension reactions using cleavable primers. Biotechniques. 2006;40:158–162. doi: 10.2144/000112115. [DOI] [PubMed] [Google Scholar]
- Kirsten H, Teupser D, Weissfuss J, et al. Robustness of single-base extension against mismatches at the site of primer attachment in a clinical assay. J Mol Med. 2007;85:361–369. doi: 10.1007/s00109-006-0129-2. [DOI] [PubMed] [Google Scholar]
- Gordon D, Haynes C, Blumenfeld J, Finch SJ. PAWE-3D: visualizing power for association with error in case-control genetic studies of complex traits. Bioinformatics. 2005;21:3935–3937. doi: 10.1093/bioinformatics/bti643. [DOI] [PubMed] [Google Scholar]
- Tasheva ES, Klocke B, Conrad GW. Analysis of transcriptional regulation of the small leucine rich proteoglycans. Mol Vis. 2004;10:758–772. [PubMed] [Google Scholar]
- Tewes U, Rossmann P, Schallberger U. Hamburg-Wechsler-Intelligenz-Test für Kinder III (HAWIK III) Bern: Verlag Hans Huber; 2002. [Google Scholar]
- Zimmermann P, Gondan M, Fimm B. KiTAP Kinderversion der Testbatterie zur Aufmerksamkeitsprüfung. Herzogenrath: Vera Fimm - Psychologische Testsysteme; 2002. [Google Scholar]
- Grissemann H, Baumberger W. ZLVT 4-6 Zürcher Leseverständnistest für das 4. bis 6. Schuljahr. Bern: Verlag Hans Huber; 2000. [Google Scholar]
- Grund M, Haug G, Naumann CL.Diagnostischer Rechtschreibtest für 4. Klassen (DRT 4)2nd edn.Göttingen: Beltz; 2004 [Google Scholar]
- Grund M, Haug G, Naumann CL. Diagnostischer Rechtschreibtest für 5. Klassen (DRT 5) Göttingen: Beltz; 2004. [Google Scholar]
- Müller R. Diagnostischer Rechtschreibtest für 2. Klassen (DRT 2) Weinheim: Beltz; 1990. [Google Scholar]
- Rathenow P, Laupenmühle D, Vöge J.Westermann Rechtschreibtest 6+2nd edn.Braunschweig: Westermann; 1981 [Google Scholar]
- Bickeboller H, Campion D, Brice A, et al. Apolipoprotein E and Alzheimer disease: genotype-specific risks by age and sex. Am J Hum Genet. 1997;60:439–446. [PMC free article] [PubMed] [Google Scholar]
- Sanjuan J, Tolosa A, Gonzalez JC, et al. FOXP2 polymorphisms in patients with schizophrenia. Schizophr Res. 2005;73:253–256. doi: 10.1016/j.schres.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Conde L, Vaquerizas JM, Dopazo H, et al. PupaSuite: finding functional single nucleotide polymorphisms for large-scale genotyping purposes. Nucleic Acids Res. 2006;34:W621–W625. doi: 10.1093/nar/gkl071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura Y, Tam T, Ido A, et al. Cloning and characterization of an ATBF1 isoform that expresses in a neuronal differentiation-dependent manner. J Biol Chem. 1995;270:26840–26848. doi: 10.1074/jbc.270.45.26840. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, et al. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansregret L, Nepveu A. The multiple roles of CUX1: insights from mouse models and cell-based assays. Gene. 2008;412:84–94. doi: 10.1016/j.gene.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Yasuda H, Mizuno A, Tamaoki T, Morinaga T. ATBF1, a multiple-homeodomain zinc finger protein, selectively down-regulates AT-rich elements of the human alpha-fetoprotein gene. Mol Cell Biol. 1994;14:1395–1401. doi: 10.1128/mcb.14.2.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamora C, DasGupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 2003;422:317–322. doi: 10.1038/nature01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura K, Takeichi M. Local and transient expression of E-cadherin involved in mouse embryonic brain morphogenesis. Development. 1992;116:1011–1019. doi: 10.1242/dev.116.4.1011. [DOI] [PubMed] [Google Scholar]
- Lai CS, Gerrelli D, Monaco AP, Fisher SE, Copp AJ. FOXP2 expression during brain development coincides with adult sites of pathology in a severe speech and language disorder. Brain. 2003;126:2455–2462. doi: 10.1093/brain/awg247. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.