Abstract
Cerebral cavernous (or capillary-venous) malformations (CCM) have a prevalence of about 0.1–0.5% in the general population. Genes mutated in CCM encode proteins that modulate junction formation between vascular endothelial cells. Mutations lead to the development of abnormal vascular structures.
In this article, we review the clinical features, molecular and genetic basis of the disease, and management.
Keywords: CCM, molecular mechanism, genetic counselling
Introduction
Cerebral cavernous (or capillary-venous) malformations (CCM; OMIM no. 116 860) are vascular malformations with a prevalence of 0.1–0.5% in the general population, with a familial incidence close to 20%.1, 2, 3 CCM may occur sporadically, but most of the time it has an autosomal dominant inheritance pattern with variable expression and incomplete penetrance.2, 3, 4, 5, 6 At least three genes have been associated with CCM: k-rev interaction trapped protein 1 (KRIT1) (CCM1; OMIM no. 604214), MGC4607 (CCM2; OMIM no. 603284) and programmed cell death 10 (PDCD10) (CCM3; OMIM no. 603285). These genes encode proteins that are involved in junction formation between vascular endothelial cells. Mutations in the CCM genes, which are in general loss-of-function mutations, lead to the development of abnormal vascular structures characterized by thin-walled, dilated blood vessels with gaps between the endothelial cells.1, 7
The underlying genetic mechanism in CCM is partially understood. Second-site genetic mutations have been proposed as one of the possible molecular mechanisms.1, 8
A total of 9% of individuals were symptomatic before age 10 years, 62–72% between 10 and 40 years, and 19% after age 40 years.9, 10 Up to 25% of individuals with CCM remain symptom free throughout their lives.11 This percentage may be an underestimate because many asymptomatic persons go unrecognized. Otten et al12 reported an absence of symptoms in 90% of individuals with CCM ascertained in autopsy. Approximately 50–75% of persons with CCM become symptomatic. Affected individuals most often present with seizures (40–70%), focal neurologic deficits (35–50%), non-specific headaches (10–30%) and cerebral haemorrhage (41%).9, 11, 13, 14 In the most recent study, Denier et al15 found seizures in 55%, focal neurological deficits in 9%, non-specific headaches in 4% and cerebral haemorrhage in 32%.
In most cases, cavernous malformations (or cavernomas) are located within the brain, but in a small proportion of patients with familial CCM, cavernomas may also be observed in the spinal cord, retina, skin or liver.2, 3, 16 Retinal cavernomas occur in about 5% of patients with familial CCM. They are unilateral, generally stable and asymptomatic, and can be diagnosed by routine fundoscopy.2, 17 Cutaneous vascular malformations are seen in 9% of familial CCM patients. Three distinct major phenotypes were identified: hyperkeratotic cutaneous capillary-venous malformations (39%), strongly associated with a KRIT1 mutation. Second, capillary malformations (34%) and finally, venous malformations (21%) mostly seen in patients with a PDCD10 mutation. Patients with a Malcavernin mutation are possibly less prone for cutaneous vascular malformations.2, 16, 18
Molecular and genetic basis of CCMs
Mutated genes and new loci
To date, three genes have been associated with the pathogenesis of CCM, including KRIT1 (also known as CCM1) located on chromosome 7q11.2–21,19, 20 Malcavernin, murine OSM-osmosensing scaffold for MEKK3 (MGC4607, also known as CCM2) on chromosome 7p1319, 21 and PDCD10 (also known as CCM3), originally identified as TF-1 cell apoptosis-related gene-15 (TFAR15) on chromosome 3q26.1 (Table 1).19, 22, 23 In addition, there is at least one further – as yet unspecified – gene that can cause CCM, which has been mapped to chromosome 3q26.3–27.2. Gianfrancesco et al25 reported the zona pellucida-like domain containing 1 gene as possible candidate. This gene is also located on the long arm of chromosome three centromeric of PDCD10.3, 24, 25, 26
Table 1. CCM protein interactions.
| Ligand | Interacted protein | Possible function |
|---|---|---|
| KRIT1 (CCM1) | Rap1a | Tumour suppressor gene |
| ICAP1α | Cell adhesion | |
| Malcavernin | Cell spreading | |
| HEG1 receptor | Direction of endothelial cell migration | |
| Junctional proteins | ||
| (Plus end of) microtubules | ||
| Malcavernin (CCM2) | KRIT1 | Sequester KRIT1 to the cytosol |
| PDCD10 | Lumen formation | |
| Rac1 | Vascular permeability | |
| Kinases involved in the p38 MAPK signalling cascade | Migration | |
| PDCD10 (CCM3) | Malcavernin | Apoptosis |
| STK24 | Direction of endothelial cell migration | |
| STK25 | ||
| MST4 |
Abbreviations: CCM, cerebral cavernous (or capillary-venous) malformations; HEG1, heart of glass 1 receptor; ICAP1α, α isoform of the β1-integrin regulator integrin cytoplasmic adaptor protein 1; KRIT1, k-rev interaction trapped protein 1; PDCD10, programmed cell death 10; STK24/25, serine/threonine protein kinase.
Distribution and frequency of gene mutations
Close to 100 mutations (88 germline mutations) have been identified in the KRIT1 gene, representing about 40–53% of the CCM families. Mutations in the MGC4607 gene may account for 15–20% of familial CCM cases.3, 7, 26, 27, 28 The only missense mutation in the MGC4607 gene reported so far, is a leucine to arginine substitution at amino acid 198 (L198R), located in the phosphotyrosine-binding domain (PTB) of Malcavernin.29 Approximately 10–40% of CCM families have been linked to the PDCD10 gene.26, 28 With a single exception, mutations in the PDCD10 gene are either truncating or large genomic deletions of the entire gene. The only known in-frame deletion of PDCD10 is located in exon 5, encompassing amino acids L33–K50, encoding the serine/threonine kinase binding and phosphorylation domain .22, 23, 30 In about 22% of CCM cases with multiple lesions no mutation is detected in the three CCM genes.26 Although de novo mutations have been reported for all three CCM genes, they appear to be more common in the PDCD10 gene.2, 31
The proportion of familial cases has been estimated approximately at 20% in the general population, and estimated to be as high as 50% in Hispanic-American patients of Mexican descent. These families are all apparently related to the same founder mutation (Q455X) in the KRIT1 gene.2, 3, 5
Genotype–phenotype relationship
CCM is an autosomal dominant disorder with a clinical penetrance of 88% in CCM1 families, 100% in CCM2 and 63% in CCM3 families.2, 32
Different explanations have been provided for the molecular pathogenesis of lesion formation in CCM. First, a Knudsonian two-hit mechanism might be involved. According to this mechanism, CCM formation would require a complete loss of the two alleles of a given CCM gene within affected cells. Loss of one of the alleles (first hit) would be the result of a germline mutation, whereas loss of the second allele (or second hit) will occur somatically. In this view, familial CCM exhibits an autosomal dominant mode of inheritance, but is likely recessive at the cellular level.2, 27, 33 On the basis of animal, as well as human studies, evidence grows for the two-hit mechanism. For example, in Ccm heterozygous mice, homozygous knockout for Msh2, penetrance of CCM lesions has been increased. Even so, in surgically resected mature lesions from CCM patients, mutations have been found in both alleles.34 Second, haploinsufficiency may also be an explanation in CCM pathophysiology. In this case, the patient has only a single functional copy of one of the CCM genes, due to mutational inactivation of the other. The single functional copy of the gene, however, does not result in sufficient protein for, for example, an adequate functional junction formation between endothelial cells, which in turn leads to the development of abnormal vascular structures. Third, paradominant inheritance might explain several CCM features. In paradominant inheritance, heterozygous individuals carrying a ‘paradominant' mutation are phenotypically normal, but the trait only becomes manifest when a somatic mutation occurs during embryogenesis, giving rise to loss of heterozygosity and formation of a mutant cell population that is homozygous for the mutation. In addition, a second hit may be caused by environmental factors. The exposure of CCM mutated, presensitized microvascular regions to oxidative stress generated by endothelial nitric oxide synthase uncoupling and reactive oxygen species formation could lead to perivascular astrocytosis.35, 36, 37 The localized nature and the number of lesions (usually a single one in sporadic cases versus multiple lesions in familial cases), as well as the age of first presentation of the phenotype being earlier in familial cases and fits in this type of inheritance. Finally, trans-heterozygosity, in which a patient has synergistic mutations in different genes of the CCM pathway (for example, a germline mutation in the KRIT1 gene with an additional somatic mutation in the MGC4607 or PDCD10 gene), might also explain intrafamilial clinical variability. Indeed, it has been shown that a decrease in the KRIT1, MGC4607 or PDCD10 gene alone caused little or no effect independently, but when combined, resulted in very high incidence of intracranial haemorrhage.1, 3, 27, 38
Biology of CCMs
Protein function and expression pattern
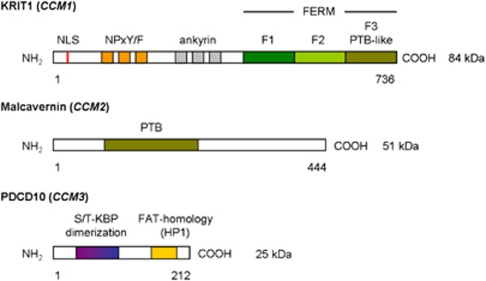
The KRIT1 gene contains 20 exons of which 16 encode a 736 amino acid protein containing three NPxY/F motifs and three ankyrin repeat domains at the N-terminus, and one C-terminal band 4.1 ezrin radixin moesin domain (FERM) found in exons 14–20 (Figure 1).1, 2, 27, 39, 40, 41 The NPxY/F motifs may be involved in dimerization either intramolecular folding of the KRIT1 protein, resulting in a closed and open conformation of KRIT1.41 After that, the first NPxY/F motif interacts with the α-isoform of the β1-integrin regulator integrin cytoplasmic adaptor protein 1 (ICAP1α). ICAP1α is a 200 amino acid protein containing a PTB domain and even as KRIT1 a nuclear localization signal motif in the N-terminus. There is evidence that both KRIT1 and ICAP1α can translocate into the nucleus, where they could cooperate in regulating gene expression. In particular, an open/closed conformation switch regulates KRIT1 nucleocytoplasmic shuttling and molecular interactions.40, 41 The ankyrin repeats in KRIT1 are thought to be involved in protein–protein interaction and have been found in many proteins. No partner interacting with KRIT1 ankyrin repeats has yet been found .27, 39, 41 The FERM domain in KRIT1 is composed of three subdomains, F1–F3, arranged in cloverleaf shape. The F3 subdomain has a PTB-like domain, which recognizes the NPxY/F motif on the cytoplasmic tail of transmembrane receptors. Rap1a is also bind by the FERM domain, suggesting that KRIT1 may function as a scaffold for transmembrane receptors and Rap1a.27, 29, 39, 41
Figure 1.
Functional domains of CCM proteins. Ankyrin, protein–protein interaction domain; FERM, four-point one ezrin radixin moesin domain; NLS, nuclear localization signal; NPxY/F, ICAP1α – Malcavernin binding site; PTB, phosphotyrosine-binding domain; S/T-KBP, serine/threonine kinase binding and phosphorylation domain.
The MGC4607 gene contains 10 exons encoding Malcavernin, a 444 amino acid protein containing a PTB domain similar to that of ICAP1α. Malcavernin binds KRIT1 by the PTB domain and inhibits in this way nuclear translocation of the KRIT1–ICAP1α complex.2, 7, 21, 41
The PDCD10 gene contains seven exons encoding a 212 amino acid protein containing a dimerization domain at the N-terminus, and a C-terminal focal adhesion targeting-homology domain with a highly conserved HP1 surface.42 Previous studies suggested also the presence of an N-terminal serine/threonine kinase binding and phosphorylation domain, which binds proteins of the germinal centre kinase III family (STK24, STK25, and Mst4).2, 30, 43, 44, 45, 46 The dimerization domain mediates dimerization of PDCD10. The Fat-homology domain is important for stabilization of the expressed PDCD10 protein and interacts with the PTB domain of Malcavernin and paxillin LD motifs.42 PDCD10 also binds Ptdlns(3,4,5)P3 and functions in this way in the (PI3k–)PIP3–PDPK1–Akt signalling pathway.23, 47 He et al suggested that the C-terminus of PDCD10 could be important in the stabilization of VEGFR2 signalling, which is crucial for vascular development.23, 48
Despite the vascular nature of CCM, in situ hybridization studies have shown KRIT1 mRNA and protein expression in astrocytes, neurons, and various epithelial cells. KRIT1 protein was also detected in vascular endothelial cells during early angiogenesis, localized in the cell–cell junctions.3, 49 Guzeloglu-Kayisli et al50 demonstrate that KRIT1 is also present in endothelial cells and cells involved in the formation of the blood-brain barrier, which implicates an important role for KRIT1 in intercellular communication and adherence. MGC4607 mRNA expression has been detected in neurons and astrocytes, as well as in cerebral vessels. PDCD10 mRNA is expressed in neuronal cells at adult stages, but also during embryogenesis.3, 51
Additional, neural expression of KRIT1, MGC4607 and PDCD10 imply that vascular malformations in CCM could also result from a defect in signalling between endothelial and neural cells, but it is still unclear whether the primary defect is of vascular or neuronal origin.3 In spite of this, most research has been focused on endothelial cells.
Histology
The vessel wall in CCM is characterized by less and abnormal junction formation between endothelial cells. After that, the expression of intercellular junction proteins is increased to compensate for the loose of cell contacts. Another characteristic of CCM is the lack of subendothelial support in the vessel wall of CCM made visible by decline in the presence of perivascular supporting cells (pericytes) and deposition of a basal lamina with disorganized collagen bundles. In addition, the formation of microgaps at the interendothelial junction sites was observed using scanning electron microscopy.52, 53 Zhao et al54 suggested that CCM may develop as a result of irregular organization of endothelial cells, as a consequence of an increased proliferation and migration potential of these cells. In line with this hypothesis, increased migratory and proliferatory endothelial cell function would indeed require reduced cell–cell contact and reduced presence of pericytes.
Molecular pathogenesis of CCMs
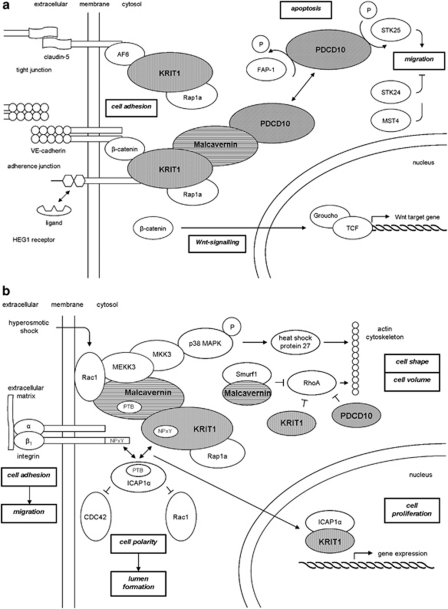
At the molecular level CCM proteins regulate cell–cell adhesion (Figure 2a), cell polarity and most likely cell adhesion to the extracellular matrix (Figure 2b).1, 46, 55
Figure 2.
Integrin signalling pathway. (a) Cell–cell adhesion. HEG1 receptor, heart of glass type 1 receptor; KRIT1, k-rev interaction trapped protein 1; PDCD10, programmed cell death 10; STK24/25, serine/threonine protein kinase; TCF, T-cell factor protein. (—∣) inhibition; ( → ) stimulation; (↔) interaction. (b) Cell-extracellular matrix adhesion. ICAP1α, α isoform of the β1-integrin regulator integrin cytoplasmic adaptor protein 1; KRIT1, k-rev interaction trapped protein 1; PDCD10, programmed cell death 10; PTB, phosphotyrosine-binding domain. (—∣) inhibition; ( → ) stimulation; (↔) interaction.
Cell–cell adhesion
Initiation and maintenance of cell–cell adhesion require the assembly of adherence junctions. The formation of these adherence junctions is stimulated by Rap1a, which forms a complex with KRIT1. Rap1a recruits KRIT1 to the plasma membrane, where it binds to the heart of glass 1 (HEG1) receptor to form a ternary complex of HEG1, KRIT1 and Malcavernin.19, 41, 56 HEG1 is a transmembrane protein, expressed specifically in the endothelium and endocardium. No binding ligand for HEG1 is currently known, although it has been suggested in previous studies that HEG1 may be involved in the Wnt/β-catenin signalling, possibly by binding KRIT1.41, 57, 58, 59 KRIT1 binds β-catenin and stimulate the association of β-catenin with vascular endothelial-cadherin, required for adherence junction formation.60 KRIT1 may also function as a tumour suppressor gene; KRIT1-β-catenin binding prevents β-catenin translocation to the nucleus where displacement of the transcriptional repressor Groucho from T-cell factor proteins by β-catenin would activate Wnt target gene expression.55, 61, 62 However, β-catenin activity in the nucleus is also vital for the blood-brain barrier, as many regulatory proteins involved in its development are under Wnt/β-catenin control.41, 62
Cell polarity
Adherence junctions also promote tight junction assembly. This takes place by the formation of a ternary complex of KRIT1, AF6/afadin and claudin-5.55, 63 Tight junctions may function as a physical barrier along the cell surface. As a consequence of asymmetrical distribution of proteins and lipids across this barrier, cell polarization takes place.41, 64 Cell polarity is important in the process of lumen formation.41 Except tight junction formation, cell polarity is also established through a reshaping of the intracellular cytoskeleton organization. This is regulated by ROCK, a RhoA effector. Crose et al65 showed that Malcavernin regulates RhoA protein level. Malcavernin binding of Smurf1 increases Smurf1-mediated degradation of RhoA. After that, Borikova et al66 also showed that KRIT1 and PDCD10 in addition to Malcavernin are required for the regulation of RhoA protein levels. KRIT1 is a negative regulator of RhoA activity. The functional mechanism of KRIT1 is not yet totally known. In contrast, some aspects of PDCD10 inhibition of RhoA activation has been elucidated, as PDCD10 acts by stabilization of germinal centre kinase III proteins and subsequent activation of moesin, a RhoA inhibitor.67, 68, 69 Loss of the CCM proteins results in an increase of RhoA activity and changes in regulation of ROCK and the cytoskeleton rigidity.
Cell adhesion to the extracellular matrix
β1-integrin, essential for the control of the intracellular cytoskeleton organization, regulates endothelial cell adhesion to the extracellular matrix.39, 70 It is proposed that β1-integrin signal to CDC42 and Rac1.41, 71, 72 Both are required for the induction of vacuole and lumen formation in vascular endothelial cells.39, 43, 73, 74 In addition, β1-integrin also promotes blood vessel maturation by stimulating the adhesion of mural cells to endothelial cells.41 β1-integrin function is inhibited by binding of ICAP1α. KRIT1 competes with β1-integrin for binding to ICAP1α, suggesting that KRIT1 may regulate the ICAP1α inhibitory effect on β1-integrin.7, 27, 28, 29, 39, 41
Cell adhesion to the extracellular matrix induces formation of focal adhesion sites in which plaque proteins, such as vinculin and paxillin, provide a bridge between β-integrins and the actin cytoskeleton. Subsequent activation of signalling cascades, regulated by focal adhesion kinase, promote actin cytoskeleton plasticity.75 Malcavernin has been shown to be capable to regulate actin cytoskeleton plasticity. In response to hyperosmotic shock, restoration of cell volume and cell shape is regulated by the p38 MAPK signalling cascade, controlled by Malcavernin. Malcavernin acts as a scaffold protein for Rac1 and the upstream kinases MEKK3 and MKK3. The p38 MAPK signalling pathway leads to the activation of heat shock protein 27, which in turn activates actin polymerization and stabilization.29, 41, 55, 73, 76
Clinical management of CCMs
Genetic counselling and molecular diagnosis
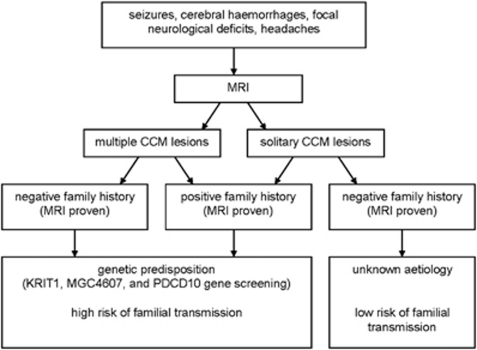
To estimate the genetic risk of CCM, three key points are essential (Figure 3):3
a detailed three-generation family tree with specific enquiry about seizures, cerebral haemorrhages, focal neurological deficits and (recurrent) headaches.
MRI of the brain to differentiate between solitary or multiple CCM lesions.
age of onset.
Figure 3.
Scheme for work-up of CCM patients at clinical presentation. Most patients (50–80%) with CCM are sporadic without a known family history of CCM. Solitary CCM lesions may be found in 8–19% of familial cases and roughly 75% of sporadic cases. Multiple lesions are indicative of familial forms of CCM.27
Genetic testing for KRIT1, MGC4607 and PDCD10 can confirm the clinical diagnosis in patients, and enables predictive and prenatal testing.
The yield of mutation screening in CCM depends on family history. If only a single lesion can be detected, familial transmission is extremely rare. In contrast, sporadic cases with multiple cerebral lesions are most likely to have a genetic cause and need to be considered as familial cases. In these cases, genetic screening of all three CCM genes is indicated. The sensitivity of this screen is estimated to be 57% therefore, the patient should be aware that a negative test does not exclude a genetic cause.2, 3 The explanation for a negative test may be a somatic mosaicism of a de novo mutation during gestation, which is not always detectable in DNA extracted from peripheral mononuclear blood cells. Also additional mutations outside the CCM coding exons may account for altered transcription of CCM associated proteins and fail to be detected by conventional gene mapping techniques.2
In familial cases, sensitivity of genetic screening of all three CCM genes in a CCM proband with an affected relative is 96%. Once the mutation has been identified in a proband, sensitivity of screening of the relatives of this particular patient is 100%.2 Genetic counselling is important to help patients and relatives to come to an informed choice.
When mutation screening is negative, predictive testing of relatives is not an option, which precludes the need for a magnetic resonance imaging (MRI). When mutation screening is positive, an additional MRI would be recommended. Although the sensitivity of MRI is very high, MRI as an initial screening test does not exclude a predisposition for CCM, as the disease may be in its latent phase, devoid of CNS lesions.2, 27
Predictive testing of minors should not be performed, given the possible psychological and socio-economic consequences of genetic testing, late onset, and reduced penetrance.2
Prenatal diagnosis and pregnancy
Prenatal diagnosis or pre-implantation genetic diagnosis is technically feasible in known familial mutations. Decisions about termination of pregnancy in case of familial mutation detection in a foetus might be difficult, because of reduced penetrance and late onset of symptoms. There is no contra-indication for pregnancy and normal delivery in patients with identified small lesions, without recent clinical signs of haemorrhage. Large lesions or recent symptomatic haemorrhages are a relative contraindication for pregnancy. In case of pregnancy, caesarean section should then be considered.2, 27
Clinical management of CCMs
Clinical monitoring of CCM depends on the presence of clinical manifestations. In asymptomatic individuals with an increased risk of CCM, a MRI analysis every 1 or 2 years should be considered. In our hospital MRI will be performed in carriers or at-risk persons. Only if neurological problems arise or increase, MRI will be repeated. The indication for surgery should be discussed individually with the patient in an experienced neurosurgical centre. Thereby, patients clinical course in combination with MRI characteristics of the CCM lesion, such as localization, size or new haemorrhage, are important factors for the decision of surgery. In case of deep-seated or brainstem lesions, surgery is associated with a morbidity rate of 30–70% and a mortality rate of 2%. Stereotactic radiosurgery for these lesions remains controversial.77, 78, 79
Medical treatment consists of inhibition of RhoA by simvastatin, or its effector protein ROCK by fasudil. Also cyclic adenosine monophosphate-elevating drugs should be considered. All of them stabilize CCM lesions by improving vascular integrity.66, 79, 80, 81, 82 Preventing progression of CCM lesions could be reached by sorafenib, an anti-angiogenic drug, targeting VEGF receptors and ERK signalling, which is enhanced in the endothelium of CCM lesions.83, 84 Treatment with antiplatelet drugs should be avoided, whereas anticoagulation with coumadin derivatives is contra-indicated .2, 19, 53
Prognosis of CCMs
The long-term prognosis of familial CCM is not well known, but the available data suggest that it is quite favourable after (surgical) treatment.
MRI identified new lesions appear at a rate of 0.2–0.4 lesions per patient year. The new onset seizure rate is 2.4% per patient year and the haemorrhage rate is 3.1%.2, 3, 10, 27, 85
Conclusions
The pathogenesis of CCM remains to date incompletely clarified. One theory is a perturbed relationship between adhesion and migration of endothelial precursor cells during the formation of the primary vascular plexus. Initiation, guidance and termination of migration are precisely regulated by interaction with the extracellular matrix and neighbouring cells. Adhesion and migration are linked by the CCM pathway proteins. CCM complex components function as bridging molecules between junctional and cytoplasmic proteins. Loss-of-function of one of the CCM proteins leads to a decrease in adhesion. This theory is mainly based on research performed in endothelial cells. Additional studies to the effect of interaction between neural and endothelial cells are necessary, as it remains unclear whether the primary defect of CCM is of vascular or neuronal origin.
The authors declare no conflict of interest.
References
- Gore AV, Lampugnani MG, Dye L, et al. Combinatorial interaction between CCM pathway genes precipitates hemorrhagic stroke. Dis Model Mech. 2008;1:275–281. doi: 10.1242/dmm.000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labauge P, Denier C, Bergametti F, et al. Genetics of cavernous angiomas. Lancet Neurol. 2007;6:237–244. doi: 10.1016/S1474-4422(07)70053-4. [DOI] [PubMed] [Google Scholar]
- Revencu N, Vikkula M. Cerebral cavernous malformation: new molecular and clinical insights. J Med Genet. 2006;43:716–721. doi: 10.1136/jmg.2006.041079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashti SR, Hoffer A, Hu YC, et al. Molecular genetics of familial cerebral cavernous malformations. Neurosurg Focus. 2006;21:e2. doi: 10.3171/foc.2006.21.1.3. [DOI] [PubMed] [Google Scholar]
- Rigamonti D, Hadley MN, Drayer BP, et al. Cerebral cavernous malformations. Incidence and familial occurrence. N Engl J Med. 1988;319:343–347. doi: 10.1056/NEJM198808113190605. [DOI] [PubMed] [Google Scholar]
- Felbor U, Sure U, Grimm T, et al. Genetics of cerebral cavernous angioma. Zentralbl Neurochir. 2006;67:110–116. doi: 10.1055/s-2006-933537. [DOI] [PubMed] [Google Scholar]
- Brouillard P, Vikkula M. Genetic causes of vascular malformations. Hum Mol Genet. 2007;16 (Spec. no. 2:R140–R149. doi: 10.1093/hmg/ddm211. [DOI] [PubMed] [Google Scholar]
- Gault J, Shenkar R, Recksiek P, et al. Biallelic somatic and germ line CCM1 truncating mutations in a cerebral cavernous malformation lesion. Stroke. 2005;36:872–874. doi: 10.1161/01.STR.0000157586.20479.fd. [DOI] [PubMed] [Google Scholar]
- Brunereau L, Labauge P, Tournier-Lasserve E, et al. Familial form of intracranial cavernous angioma: MR imaging findings in 51 families. Radiology. 2000;214:209–216. doi: 10.1148/radiology.214.1.r00ja19209. [DOI] [PubMed] [Google Scholar]
- Labauge P, Brunereau L, Laberge S, et al. Prospective follow-up of 33 asymptomatic patients with familial cerebral cavernous malformations. Neurology. 2001;57:1825–1828. doi: 10.1212/wnl.57.10.1825. [DOI] [PubMed] [Google Scholar]
- Siegel AM. Familial cavernous angioma: an unknown, known disease. Acta Neurol Scand. 1998;98:369–371. doi: 10.1111/j.1600-0404.1998.tb07315.x. [DOI] [PubMed] [Google Scholar]
- Otten P, Pizzolato GP, Rilliet B, et al. [131 cases of cavernous angioma (cavernomas) of the CNS, discovered by retrospective analysis of 24,535 autopsies] Neurochirurgie 19893582–83.128–131. [PubMed] [Google Scholar]
- Zabramski JM, Wascher TM, Spetzler RF, et al. The natural history of familial cavernous malformations: results of an ongoing study. J Neurosurg. 1994;80:422–432. doi: 10.3171/jns.1994.80.3.0422. [DOI] [PubMed] [Google Scholar]
- Rigamonti D, Drayer BP, Johnson PC, et al. The MRI appearance of cavernous malformations (angiomas) J Neurosurg. 1987;67:518–524. doi: 10.3171/jns.1987.67.4.0518. [DOI] [PubMed] [Google Scholar]
- Denier C, Labauge P, Brunereau L, et al. Clinical features of cerebral cavernous malformations patients with KRIT1 mutations. Ann Neurol. 2004;55:213–220. doi: 10.1002/ana.10804. [DOI] [PubMed] [Google Scholar]
- Eerola I, Plate KH, Spiegel R, et al. KRIT1 is mutated in hyperkeratotic cutaneous capillary-venous malformation associated with cerebral capillary malformation. Hum Mol Genet. 2000;9:1351–1355. doi: 10.1093/hmg/9.9.1351. [DOI] [PubMed] [Google Scholar]
- Labauge P, Krivosic V, Denier C, et al. Frequency of retinal cavernomas in 60 patients with familial cerebral cavernomas: a clinical and genetic study. Arch Ophthalmol. 2006;124:885–886. doi: 10.1001/archopht.124.6.885. [DOI] [PubMed] [Google Scholar]
- Sirvente J, Enjolras O, Wassef M, et al. Frequency and phenotypes of cutaneous malformations in a consecutive series of 417 patients with familial cerebral cavernous malformations. J Eur Acad Dermatol Venereol. 2009;23:1066–1072. doi: 10.1111/j.1468-3083.2009.03263.x. [DOI] [PubMed] [Google Scholar]
- Kleaveland B, Zheng X, Liu JJ, et al. Regulation of cardiovascular development and integrity by the heart of glass-cerebral cavernous malformation protein pathway. Nat Med. 2009;15:169–176. doi: 10.1038/nm.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laberge-le Couteulx S, Jung HH, Labauge P, et al. Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nat Genet. 1999;23:189–193. doi: 10.1038/13815. [DOI] [PubMed] [Google Scholar]
- Liquori CL, Berg MJ, Siegel AM, et al. Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am J Hum Genet. 2003;73:1459–1464. doi: 10.1086/380314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergametti F, Denier C, Labauge P, et al. Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am J Hum Genet. 2005;76:42–51. doi: 10.1086/426952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CF, Horst JA, Malone MH, et al. Defining the functional domain of programmed cell death 10 through its interactions with phosphatidylinositol-3,4,5,-trisposphate. PLoS One. 2010;5:e11740. doi: 10.1371/journal.pone.0011740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liquori CL, Berg MJ, Squitieri F, et al. Low frequency of PDCD10 mutations in a panel of CCM3 probands: potential for a fourth CCM locus. Hum Mutat. 2006;27:118. doi: 10.1002/humu.9389. [DOI] [PubMed] [Google Scholar]
- Gianfrancesco F, Esposito T, Penco S, et al. ZPLD1 gene is disrupted in a patient with balanced translocation that exhibits cerebral cavernous malformations. Neuroscience. 2008;155:345–349. doi: 10.1016/j.neuroscience.2008.05.030. [DOI] [PubMed] [Google Scholar]
- Riant F, Bergametti F, Ayrignac X, et al. Recent insights into cerebral cavernous malformations: the molecular genetics of CCM. FEBS J. 2010;277:1070–1075. doi: 10.1111/j.1742-4658.2009.07535.x. [DOI] [PubMed] [Google Scholar]
- Gault J, Sarin H, Awadallah NA, et al. Pathobiology of human cerebrovascular malformations: basic mechanisms and clinical relevance Neurosurgery 2004551–16.discussion 16–17. [PubMed] [Google Scholar]
- Wang QK. Update on the molecular genetics of vascular anomalies. Lymphat Res Biol. 2005;3:226–233. doi: 10.1089/lrb.2005.3.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer NW, Zawistowski JS, Marchuk DA. Genetics of cerebral cavernous malformations. Curr Neurol Neurosci Rep. 2005;5:391–396. doi: 10.1007/s11910-005-0063-7. [DOI] [PubMed] [Google Scholar]
- Voss K, Stahl S, Hogan BM, et al. Functional analyses of human and zebrafish 18-amino acid in-frame deletion pave the way for domain mapping of the cerebral cavernous malformation 3 protein. Hum Mutat. 2009;30:1003–1011. doi: 10.1002/humu.20996. [DOI] [PubMed] [Google Scholar]
- Denier C, Labauge P, Bergametti F, et al. Genotype-phenotype correlations in cerebral cavernous malformations patients. Ann Neurol. 2006;60:550–556. doi: 10.1002/ana.20947. [DOI] [PubMed] [Google Scholar]
- Craig HD, Gunel M, Cepeda O, et al. Multilocus linkage identifies two new loci for a mendelian form of stroke, cerebral cavernous malformation, at 7p15-13 and 3q25.2-27. Hum Mol Genet. 1998;7:1851–1858. doi: 10.1093/hmg/7.12.1851. [DOI] [PubMed] [Google Scholar]
- Akers AL, Johnson E, Steinberg GK, et al. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum Mol Genet. 2009;18:919–930. doi: 10.1093/hmg/ddn430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald DA, Shenkar R, Shi C, et al. A novel mouse model of cerebral cavernous malformations based on the two-hit mutation hypothesis recapitulates the human disease. Hum Mol Genet. 2011;20:211–222. doi: 10.1093/hmg/ddq433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goitre L, Balzac F, Degani S, et al. KRIT1 regulates the homeostasis of intracellular reactive oxygen species. PLoS One. 2010;5:e11786. doi: 10.1371/journal.pone.0011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belik J, Jerkic M, McIntyre BAS, et al. Age-dependent endothelial nitric oxide synthase uncoupling in pulmonary arteries of endoglin heterozygous mice. Am J Physiol Lung Cell Mol Physiol. 2009;297:L1170–L1178. doi: 10.1152/ajplung.00168.2009. [DOI] [PubMed] [Google Scholar]
- Louvi A, Chen L, Two AM, et al. Loss of cerebral cavernous malformation 3 (Ccm3) in neuroglia leads to CCM and vascular pathology. PNAS. 2011;108:3737–3742. doi: 10.1073/pnas.1012617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer. 2001;1:157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- Marchuk DA, Srinivasan S, Squire TL, et al. Vascular morphogenesis: tales of two syndromes. Hum Mol Genet. 2003;12 (Spec. no. 1:R97–112. doi: 10.1093/hmg/ddg103. [DOI] [PubMed] [Google Scholar]
- Francalanci F, Avolio M, De Luca E, et al. Structural and functional differences between KRIT1A and KRIT1B isoforms: a framework for understanding CCM pathogenesis. Exp Cell Res. 2009;315:285–303. doi: 10.1016/j.yexcr.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Faurobert E, Albiges-Rizo C. Recent insights into cerebral cavernous malformations: a complex jigsaw puzzle under construction. FEBS J. 2010;277:1084–1096. doi: 10.1111/j.1742-4658.2009.07537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang R, Zhang H, et al. Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity. J Biol Chem. 2010;285:24099–24107. doi: 10.1074/jbc.M110.128470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Wu YM, Hsu CY, et al. Caspase activation of mammalian sterile 20-like kinase 3 (Mst3). Nuclear translocation and induction of apoptosis. J Biol Chem. 2002;277:34367–34374. doi: 10.1074/jbc.M202468200. [DOI] [PubMed] [Google Scholar]
- Lu TJ, Lai WY, Huang CY, et al. Inhibition of cell migration by autophosphorylated mammalian sterile 20-like kinase 3 (MST3) involves paxillin and protein-tyrosine phosphatase-PEST. J Biol Chem. 2006;281:38405–38417. doi: 10.1074/jbc.M605035200. [DOI] [PubMed] [Google Scholar]
- Ma X, Zhao H, Shan J, et al. PDCD10 interacts with Ste20-related kinase MST4 to promote cell growth and transformation via modulation of the ERK pathway. Mol Biol Cell. 2007;18:1965–1978. doi: 10.1091/mbc.E06-07-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss K, Stahl S, Schleider E, et al. CCM3 interacts with CCM2 indicating common pathogenesis for cerebral cavernous malformations. Neurogenetics. 2007;8:249–256. doi: 10.1007/s10048-007-0098-9. [DOI] [PubMed] [Google Scholar]
- Schleider E, Stahl S, Wüstehube J, et al. Evidence for anti-angiogenic and pro-survival functions of the cerebral cavernous malformation protein 3. Neurogenetics. 2011;12:83–86. doi: 10.1007/s10048-010-0261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhang H, Yu L, et al. Stabilization of VEGFR2 signalling by cerebral cavernous malformation 3 is critical for vascular development. Sci Signal. 2010;3:ra26. doi: 10.1126/scisignal.2000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denier C, Gasc JM, Chapon F, et al. Krit1/cerebral cavernous malformation 1 mRNA is preferentially expressed in neurons and epithelial cells in embryo and adult. Mech Dev. 2002;117:363–367. doi: 10.1016/s0925-4773(02)00209-5. [DOI] [PubMed] [Google Scholar]
- Guzeloglu-Kayisli O, Amankulor NM, Voorhees J, et al. Krit1/cerebral cavernous malformation 1 protein localizes to vascular endothelium, astrocytes, and pyramidal cells of the adult human cerebral cortex. Neurosurgery. 2004;54:943–949. doi: 10.1227/01.neu.0000114512.59624.a5. [DOI] [PubMed] [Google Scholar]
- Petit N, Blecon A, Denier C, et al. Patterns of expression of the three cerebral cavernous malformation (CCM) genes during embryonic and postnatal brain development. Gene Expr Patterns. 2006;6:495–503. doi: 10.1016/j.modgep.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Wong JH, Awad IA, Kim JH. Ultrastructural pathological features of cerebrovascular malformations: a preliminary report. Neurosurgery. 2000;46:1454–1459. doi: 10.1097/00006123-200006000-00027. [DOI] [PubMed] [Google Scholar]
- Burkhardt JK, Schmidt D, Schoenauer R, et al. Upregulation of transmembrane endothelial junction proteins in human cerebral cavernous malformations. Neurosurg Focus. 2010;29:E3. doi: 10.3171/2010.6.FOCUS10125. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Tan YZ, Zhou LF, et al. Morphological observation and in vitro angiogenesis assay of endothelial cells isolated from human cerebral cavernous malformations. Stroke. 2007;38:1313–1319. doi: 10.1161/01.STR.0000259914.21997.89. [DOI] [PubMed] [Google Scholar]
- Dejana E, Tournier-Lasserve E, Weinstein BM. The control of vascular integrity by endothelial cell junctions: molecular basis and pathological implications. Dev Cell. 2009;16:209–221. doi: 10.1016/j.devcel.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Whitehead KJ, Plummer NW, Adams JA, et al. Ccm1 is required for arterial morphogenesis: implications for the etiology of human cavernous malformations. Development. 2004;131:1437–1448. doi: 10.1242/dev.01036. [DOI] [PubMed] [Google Scholar]
- Huang L, Ren J, Chen D, et al. MUC1 cytoplasmic domain coactivates Wnt target gene transcription and confers transformation. Cancer Biol Ther. 2003;2:702–706. [PubMed] [Google Scholar]
- Lang T, Hansson GC, Samuelsson T. An inventory of mucin genes in the chicken genome shows that the mucin domain of Muc13 is encoded by multiple exons and that ovomucin is part of a locus of related gel-forming mucins. BMC Genomics. 2006;7:197. doi: 10.1186/1471-2164-7-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal U, Venkatraman J, Niranjali D, et al. Interaction of MUC1 with beta-catenin modulates the Wnt target Gene cyclinD1 in H. pylori-induced gastric cancer. Mol Carcinog. 2007;46:807–817. doi: 10.1002/mc.20311. [DOI] [PubMed] [Google Scholar]
- Glading AJ, Ginsberg MH. Rap1 and its effector KRIT1/CCM1 regulate beta-catenin signalling. Dis Model Mech. 2010;3:73–83. doi: 10.1242/dmm.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H. Wnt/beta-catenin signalling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- Liebner S, Corada M, Bangsow T, et al. Wnt/beta-catenin signalling controls development of the blood-brain barrier. J Cell Biol. 2008;183:409–417. doi: 10.1083/jcb.200806024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Mariscal L, Tapia R, Chamorro D. Crosstalk of tight junction components with signalling pathways. Biochim Biophys Acta. 2008;1778:729–756. doi: 10.1016/j.bbamem.2007.08.018. [DOI] [PubMed] [Google Scholar]
- Lampugnani MG, Orsenigo F, Rudini N, et al. CCM1 regulates vascular-lumen organization by inducing endothelial polarity. J Cell Sci. 2010;123:1073–1080. doi: 10.1242/jcs.059329. [DOI] [PubMed] [Google Scholar]
- Crose LE, Hilder TL, Sciaky N, et al. Cerebral cavernous malformation 2 protein promotes smad ubiquitin regulatory factor 1-mediated RhoA degradation in endothelial cells. J Biol Chem. 2009;284:13301–13305. doi: 10.1074/jbc.C900009200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borikova AL, Dibble CF, Sciaky N, et al. Rho kinase inhibition rescues the endothelial cell cerebral cavernous malformation phenotype. J Biol Chem. 2010;285:11760–11764. doi: 10.1074/jbc.C109.097220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Xu C, Di Lorenzo A, et al. CCM3 signalling through sterile 20-like kinases plays an essential role during zebrafish cardiovascular development and cerebral cavernous malformations. J Clin Invest. 2010;120:2795–2804. doi: 10.1172/JCI39679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisinger C, Short B, De Corte V, et al. YSK1 is activated by the Golgi matrix protein GM130 and plays a role in cell migration through its substrate 14-3-3 {zeta} J Cell Biol. 2004;164:1009–1020. doi: 10.1083/jcb.200310061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M, Fraile M, Pires A, et al. CCM3/PDCD10 stabilizes GCKIII proteins to promote Golgi assembly and cell orientation. J Cell Sc. 2010;123:1274–1284. doi: 10.1242/jcs.061341. [DOI] [PubMed] [Google Scholar]
- Rupp PA, Little CD. Integrins in vascular development. Circ Res. 2001;89:566–572. doi: 10.1161/hh1901.097747. [DOI] [PubMed] [Google Scholar]
- Bayless KJ, Davis GE. The Cdc42 and Rac1 GTPases are required for capillary lumen formation in three-dimensional extracellular matrices. J Cell Sci. 2002;115:1123–1136. doi: 10.1242/jcs.115.6.1123. [DOI] [PubMed] [Google Scholar]
- Iruela-Arispe ML, Davis GE. Cellular and molecular mechanisms of vascular lumen formation. Developmental Cell. 2009;16:222–231. doi: 10.1016/j.devcel.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead KJ, Chan AC, Navankasattusas S, et al. The cerebral cavernous malformation signalling pathway promotes vascular integrity via Rho GTPases. Nat Med. 2009;15:177–184. doi: 10.1038/nm.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Rigamonti D, Badr A, et al. Ccm1 regulates microvascular morphogenesis during angiogenesis. J Vasc Res. 2011;48:130–140. doi: 10.1159/000316851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harburger DS, Calderwood DA. Integrin signalling at a glance. J Cell Sci. 2009;122:159–163. doi: 10.1242/jcs.018093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgett JS, Ding J, Guh-Siesel L, et al. Essential role for p38alpha mitogen-activated protein kinase in placental angiogenesis. PNAS. 2000;97:10454–10459. doi: 10.1073/pnas.180316397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham M, Gross BA, Bendok BR, et al. Radiosurgery for angiographically occult vascular malformations. Neurosurg Focus. 2009;26:E16. doi: 10.3171/2009.2.FOCUS0923. [DOI] [PubMed] [Google Scholar]
- Gross BA, Batjer HH, Awad IA, et al. Brainstem cavernous malformations. Neurosurgery. 2009;64:E805–E818. doi: 10.1227/01.NEU.0000343668.44288.18. [DOI] [PubMed] [Google Scholar]
- Yadla S, Jabbour PM, Shenkar R, et al. Cerebral cavernous malformations as a disease of vascular permeability: from bench to bedside with caution. Neurosurg Focus. 2010;29:E4. doi: 10.3171/2010.5.FOCUS10121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisht KM, Whitehead KJ, Niazi T, et al. The pathogenetic features of cerebral cavernous malformations: a comprehensive review with therapeutic implications. Neurosurg Focus. 2010;29:E2. doi: 10.3171/2010.6.FOCUS10135. [DOI] [PubMed] [Google Scholar]
- Stockton RA, Shenkar R, Awad IA, et al. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J Exp Med. 2010;207:881–896. doi: 10.1084/jem.20091258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara S, Sakurai A, Sano H, et al. Cyclic AMP potentiates vascular endothelial cadherin-mediated cell-cell contact to enhance endothelial barrier function through an Epac-Rap1 signalling pathway. Mol Cell Biol. 2005;25:136–146. doi: 10.1128/MCB.25.1.136-146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm S, Carter C, Lynch M, et al. Discovery and development of sorafenib: A multikinase inhibitor for treating cancer. Nat Rev Drug Discov. 2006;5:835–844. doi: 10.1038/nrd2130. [DOI] [PubMed] [Google Scholar]
- Wüstehube J, Bartol A, Liebler SS, et al. Cerebral cavernous malformation protein CCM1 inhibits sprouting angiogenesis by activating DELTA-NOTCH signalling. PNAS. 2010;107:12640–12645. doi: 10.1073/pnas.1000132107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarity JL, Wetzel M, Clatterbuck RE, et al. The natural history of cavernous malformations: a prospective study of 68 patients. Neurosurgery. 1999;44:1166–1173. [PubMed] [Google Scholar]