Abstract
Purpose
Screening of mutations in the fibrillin-1 (FBN1) gene in a Chinese family with autosomal dominant Marfan syndrome (MFS).
Methods
It has been reported that FBN1 mutations account for approximately 90% of Autosomal Dominant MFS. FBN1 mutations were analyzed in a Chinese family of 36 members including 13 MFS patients. The genomic DNAs from blood leukocytes of the patients and their relatives were isolated and the entire coding region of FBN1 was amplified by PCR. The sequence of FBN1 was dertermined with an ABI 3100 Genetic Analyzer.
Results
A previously unreported the missense mutation G214S (caused by a 640 A→G heterozygous change) in FBN1 was identified in the Chinese family. The mutation was associated with the disease phenotype in patients, but not detected in their relatives or in the 100 normal controls.
Conclusions
This is the first report of molecular characterization of FBN1 in the MFS family of Chinese origin. Our results expand the spectrum of FBN1 mutations causing MFS and further confirm the role of FBN1 in the pathogenesis of MFS. Direct sequencing of the mutation in FBN1 may be used for diagnosis of MFS.
Introduction
Marfan syndrome (MFS) is an inherited, autosomal dominant,systemic disorder of connective tissue. The estimated prevalence of MFS is one in 10,000 to 20,000 individuals. It is well known that three major systems: skeletal, ocular, and cardiovascular are affected by the disease. The clinical criteria for MFS include a combination of skeletal manifestations, ectopia lentis, dural ectasia, and dilatation or dissection of the ascending aorta.
The main pathogenesis of MFS is currently thought to be driven by mechanisms due to haploinsufficiency of wild-type fibrillin-1 [1]. Fibrillins are ubiquitous extracellular matrix molecules that assemble into microfibrils [2-4] and target growth factors to the extracellular matrix [5-7]. Fibrillin-1 mutations disrupt microfibril formation, thereby resulting in fibrillin protein abnormalities, and subsequently weakening the connective tissue. More than 1000 mutations have been identified in the gene for fibrillin-1 (FBN1) [8]. Most mutations are unique for a certain MFS family, and only approximately 10% of mutations are recurrent in different families [9].
FBN1 is a 230-kb gene with 65 exons that encodes the structural protein fibrillin-1, located at chromosome 15q-21.1. In this study, we analyzed a four-generation, non-consanguineous Chinese family, which has been diagnosed as MFS, and identified a mutation in the 7th exon of FBN1 (c. 640 A→G) resulting in the substitution of glycine by serine at codon 214 (p. G214S). Our data of FBN1 mutations causing MFS further confirm the role of FBN1 in the pathogenesis of MFS. Obviously this is the first report of molecular characterization in FBN1 in the Chinese MFS family.
Methods
Clinical data and 5 ml of blood samples were collected from a Chinese Zhang family with MFS. The patient underwent complete physical and ophthalmic examinations. The Institutional Review Board approved the project and investigators followed the principles of the Declaration of Helskinki. Informed consent was obtained from each person.
To identify constitutional mutations, blood specimens (8 ml) were collected in EDTA, and genomic DNA was extracted from peripheral blood cells according to a standard protocol (Roche Diagnostics Corporation, Indianapolis, IN). Briefly, all the exons and exon-intron boundaries of FBN1 were amplified by using the standard PCR buffer system with primers listed in Appendix 1. PCR reactions were each performed in a 10 µl volume, containing 1.5 mM MgCl2, 0.4 mM of each primer, 200 µM dNTPs, 1 U Taq DNA polymerase, and 10-20 ng template DNA. Amplification was performed with an initial denaturation for 3 min at 95 °C, followed by 30 cycles of denaturation at 95 °C for 1 min, annealing at 55 °C for 1 min, extension at 72 °C for 1 min, and a final extension at 72 °C for 3 min.
PCR products were purified by using a PCR product purification kit (QIAquick; Qiagen, Valencia, CA). Purified PCR products were sequenced using the BigDye Terminator Cycle Sequencing v3.1 kit (Applied Biosystems, Foster City, CA). Briefly, about 10 ng of template DNA is added to each reaction and using a temperature program which included 25 cycles of denaturation at 97 °C for 30 s, annealing at 50 °C for 15 s, and extension at 60 °C for 4 min. All samples were analyzed in an ABI Prism 310 Genetic Analyzer (Applied Biosystems). The FBN1 CDNA reference sequence with GenBank accession number NC_000015.9 was used (National Center for Biotechnical Information [NCBI], Bethesda, Md).
Results
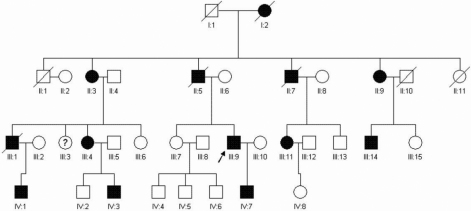
The patients including 8 males and 5 females were from a family in Guangdong Province, China (Figure 1). All of these patients in the family manifested various reduced visual acuities with a bilateral lens dislocation and high myopia. Aortic root dilatation was present in 2 of the 13 patients. One patient had an aortic aneurysm together with either aortic valve stenosis or aortic valve insufficiency. The other patient had a cloverleaf appearance in the cross-section, but the diameter of his aortic root was apparently normal. All patients had facial and skeletal features of MFS including joint laxity, dolichostenomelia, pectus excavatum or pectus carinatum, and scoliosis (Table 1). The cause of death for two patients was cardiovascular malformations.
Figure 1.
The pedigree of the family is shown. Squares and circles indicate males and females, respectively, and the darkened symbols represent the affected members. The patient above the arrow is the proband.
Table 1. Clinical evaluation of affected family members.
| Patient ID | II:3 | II:9 | II:4 | II:9 | II:11 | II:14 | IV:1 | IV:3 | IV:7 |
|---|---|---|---|---|---|---|---|---|---|
| Age |
69 |
59 |
41 |
37 |
32 |
26 |
12 |
9 |
5 |
| Sex |
F |
F |
F |
M |
F |
M |
M |
M |
M |
| Ocular system | |||||||||
| (1) Ectopia lentis |
+ |
+ |
+ |
OEL* |
+ |
+ |
+ |
+ |
+ |
| (2) Myopia |
+ |
+ |
+ |
OEL* |
+ |
+ |
+ |
+ |
+ |
| (3) Strabismus |
+ |
- |
- |
- |
- |
- |
+ |
- |
- |
| (4) Glaucoma |
- |
- |
- |
+ |
- |
- |
- |
- |
- |
| (5) Retinal detachment |
- |
- |
- |
- |
- |
- |
+ |
- |
- |
| Cardiovascular system | |||||||||
| (1) Aortic root dimension (mm) |
31.6 |
29 |
27.5 |
39.2 |
26 |
25.1 |
24.9 |
24 |
20.3 |
| (2) Mitral valve prolapse |
- |
- |
- |
+ |
- |
- |
- |
- |
- |
| (3) aortic aneurysm |
+ |
- |
- |
- |
- |
- |
- |
- |
- |
| Skeletal system | |||||||||
| (1) Height (H;cm) |
168 |
170 |
172 |
192 |
174 |
198 |
171 |
168 |
145 |
| (2) Arm span (AS;cm) |
173 |
175 |
178 |
201 |
180 |
208 |
179 |
177 |
149 |
| (3) AS/H |
1.03 |
1.03 |
1.03 |
1.05 |
1.03 |
1.05 |
1.04 |
1.05 |
1.02 |
| (4) Scoliosis |
- |
- |
- |
+ |
- |
- |
- |
- |
- |
| (5) Arachnodactyly |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
+ |
- |
| (6) Joint hypermobility |
+ |
+ |
- |
- |
- |
- |
- |
- |
- |
| (7) pectus excavatum |
+ |
+ |
- |
- |
- |
- |
- |
- |
- |
| (8) Pectus carinatum |
- |
- |
- |
- |
- |
- |
+ |
+ |
- |
| Other manifestations | |||||||||
| (1) Hyperextensible skin |
+ |
+ |
- |
- |
- |
- |
- |
- |
- |
| (2) Striae |
+ |
+ |
- |
- |
- |
- |
- |
- |
- |
| (3) Hernia | - | - | - | - | - | - | - | - | + |
* Operated for ectopia lentis.
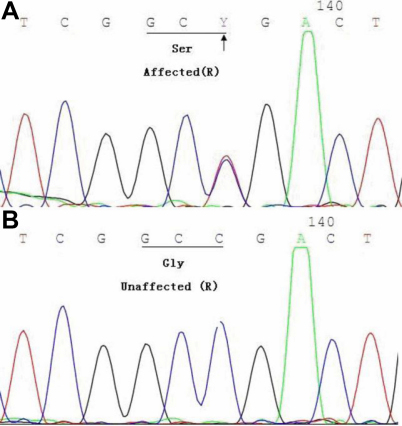
Sequencing of the 65 coding exons of FBN1 in the patients revealed a mutation (c. 640 A→G; Figure 2) which resulted in substitution of glycine by serine (G214S) in exon 7. The mutation was linked to the disease phenotype in all patients, but not found in other unaffected relatives or in the 100 normal controls.
Figure 2.
A novel FBN1 missense mutation in extron 7. The partial nucleotide sequence of FBN1 in is shown. A: The corresponding normal sequence in an unaffected family member. B: A heterozygous change A→G (indicated by the arrow) was identified in an affected family member.
Discussion
MFS is diagnosed using the Ghent criteria, a group of clinical findings that are specific for MFS. The Ghent criteria is through a comprehensive assessment largely based on a combination of major and minor clinical manifestations in the various organ systems and the family history [10]. Although the clinical manifestations of the cardiovascular, ocular, and skeletal systems are important criteria for diagnosis of the disease, a family genetic history can also provide useful information for determining etiology and designing treatment strategy. For example, an individual with these clinical features such as tall, thin body habitus, long limbs, arachnodactyly, pectus deformities, and sometimes scoliosis with family history may be suggestive of a diagnosis of Marfan’s syndrome [11].
Cardiovascular malformations are the most life-threatening presentations of MFS. Affected patients are at risk for aortic dissection and/or severe ocular and orthopedic disorders. Although we have observed the following cardiovascular manifestations of MFS: mitral valve prolapse, calcification of the mitral valve annulus and dilatation or dissection of the descending aorta, the prevalence of cardiovascular manifestations is not very high in this family.
The clinical manifestations of the MFS patients in the family become more evident with age. The most common symptom among them is myopia and 100% of the patients have ectopia lentis. Besides, there was one patient with retinal detachment and two patients underwent IOL surgery.
The pathogenesis of the Marfan’s syndrome has not been fully elucidated. FBN1 mutations are believed to exert a dominant negative effect [12]. In this study we identified a novel FBN1 mutation in patients by genotype-phenotype analysis, which perfectly met the Ghent criteria (p=0.005). The mutation was also detected in those who had ectopia lentis (EL; p<0.0001) [13].
In rare cases, MFS is caused by a mutation in the transforming growth factor beta (TGF-β) receptor 1 or 2 genes [14-16].
The coding sequence of FBN1 is spread over 65 coding exons. FBN1 is comprised mainly of repeated modules such as epidermal-growth-factor like (EGF) domains and transforming growth factor β1 binding protein-like (TB) domains. There are a total of 47 EGF domains on the protein. Among them, 43 of which are of the calcium-binding (cb) type (cb-EGF). There are also 7 TB domains distributed throughout FBN1, separated by variable numbers of the tandem repeated EGF domains [17-19]. Most mutations of FBN1 occur in the EGF domains [20].
FBN1 mutations disrupt microfibril formation, thereby resulting in fibrillin protein abnormalities, and subsequently weakening the connective tissue [21]. Over 601 distinct mutations have been documented in the Marfan mutation database [22]. Two-thirds of the mutations are missense mutations, and the majority of these are cysteine substitutions. Nonsense mutations account for about 10% of all reported mutations. Small insertions, deletions, or duplications represent about 13% among these mutations. Another 13% of the reported mutations are caused by various classes of splicing errors [23].
Interestingly, mutations frequently occur in exons 2, 15, 22, 27, 46, 55, and 62 but much less in exons 7, 41, and 65 [24] although the cause remains to be elucidated. According to Universal Marfan Database – FBN1 (UMD-FBN), only 2 different mutations in exon7 have been reported [25,26].
The new mutation we identified was a missense mutation in exon 7 (640 A→G heterozygous change) of FBN1 in the Chinese family. This mutation was found in all patients, but not in unaffected family members and the 100 unrelated controls, suggesting that the new mutation may be responsible for the pathogenesis of MFS in this family.
Sequences for TB domains found in fibrillin show a high level of amino acid conservation. The TB domain is characterized by eight cysteines predicted to form four intraomolecular disulphide bonds. There are totally 11 reported mutations in the TB domains of human FBN1 which are associated with MFS. Structural analysis implicates that those mutations involving cysteines (C661R, C711Y, C996R, and C1589F) are likely to cause domain misfolding, since one of the four disulfide bonds may be disrupted. Three mutations related to either a frameshift (N1713 and E2105) or the formation of a premature stop codon (Y2113X) cause disruption of the primary sequence of this protein. Point mutations in the TB domains, including A705T, V984I, G1013R, and K1023N have also been reported [27]. Here we report that the new mutation we identified in the TB1 domain may interfere with microfibrillar assembly via producing an unstable mutant protein cleavage products.
Conclusion
We have identified a single missense mutation in FBN1 (c.640 A→G). in a Chinese family with Marfan syndrome. Our results expand the spectrum of FBN1 mutations causing MFS, and further confirm the role of FBN1 in the pathogenesis of MFS. Direct sequencing of the FBN1 mutation could be used for diagnosis of MFS.
Acknowledgments
We are grateful to the patients and their family members for their cooperation in this study. This study was supported by the National Natural Science Foundation (No. 30950007) and Beijing Natural Science Foundation (No. 7102160). The study was approved by the Ethics Committee of the Peking University Third Hospital and conformed to the Declaration of Helsinki.
Appendix 1.
Appendix 1. Primers used to amplify the exons of FBN1. To access the data, click or select the words “Appendix 1.” This will initiate the download of a compressed (pdf) archive that contains the file.
References
- 1.Haneline M, Lewkovich GN. A narrative review of pathophysiological mechanisms associated with cervical artery dissection. J Can Chiropr Assoc. 2007;51:146–57. [PMC free article] [PubMed] [Google Scholar]
- 2.Sakai LY, Keene DR, Engvall E. Fibrillin, a new 350-kD glycoprotein, is a component of extracellular microfibrils. J Cell Biol. 1986;103:2499–509. doi: 10.1083/jcb.103.6.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H, Apfelroth SD, Hu W, Davis EC, Sanguineti C, Bonadio J, Mecham RP, Ramirez F. Structure and expression of fibrillin-2, a novel microfibrillar component preferentially located in elastic matrices. J Cell Biol. 1994;124:855–63. doi: 10.1083/jcb.124.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charbonneau NL, Dzamba BJ, Ono RN, Keene DR, Corson GM, Reinhardt DP, Sakai LY. Fibrillins can co-assemble in fibrils, but fibrillin fibril composition displays cell-specific differences. J Biol Chem. 2003;278:2740–9. doi: 10.1074/jbc.M209201200. [DOI] [PubMed] [Google Scholar]
- 5.Isogai Z, Ono RN, Ushiro S, Keene DR, Chen Y, Mazzieri R, Charbonneau NL, Reinhardt DP, Rifkin DB, Sakai LY. Latent transforming growth factor beta-binding protein 1 interacts with fibrillin and is a microfibril-associated protein. J Biol Chem. 2003;278:2750–7. doi: 10.1074/jbc.M209256200. [DOI] [PubMed] [Google Scholar]
- 6.Gregory KE, Ono RN, Charbonneau NL, Kuo CL, Keene DR, Bächinger HP, Sakai LY. The prodomain of BMP-7 targets the BMP-7 complex to the extracellular matrix. J Biol Chem. 2005;280:27970–80. doi: 10.1074/jbc.M504270200. [DOI] [PubMed] [Google Scholar]
- 7.Sengle G, Charbonneau NL, Ono RN, Sasaki T, Alvarez J, Keene DR, Bächinger HP, Sakai LY. Targeting of bone morphogenetic protein growth factor complexes to fibrillin. J Biol Chem. 2008;283:13874–88. doi: 10.1074/jbc.M707820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faivre L, Collod-Beroud G, Loeys BL, Child A, Binquet C, Gautier E, Callewaert B, Arbustini E, Mayer K, Arslan-Kirchner M, Kiotsekoglou A, Comeglio P, Marziliano N, Dietz HC, Halliday D, Beroud C, Bonithon-Kopp C, Claustres M, Muti C, Plauchu H, Robinson PN, Adès LC, Biggin A, Benetts B, Brett M, Holman KJ, De Backer J, Coucke P, Francke U, De Paepe A, Jondeau G, Boileau C. Effect of mutation type and location on clinical outcome in 1,013 probands with Marfan syndrome or related phenotypes and FBN1 mutations: an international study. Am J Hum Genet. 2007;81:454–66. doi: 10.1086/520125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collod-Béroud G, Le Bourdelles S, Ades L, Ala-Kokko L, Booms P, Boxer M, Child A, Comeglio P, De Paepe A, Hyland JC, Holman K, Kaitila I, Loeys B, Matyas G, Nuytinck L, Peltonen L, Rantamaki T, Robinson P, Steinmann B, Junien C, Béroud C, Boileau C. Update of the UMD-FBN1 mutation database and creation of an FBN1 polymorphism database. Hum Mutat. 2003;22:199–208. doi: 10.1002/humu.10249. [DOI] [PubMed] [Google Scholar]
- 10.Gelb BD. Marfan's syndrome and related disorders–more tightly connected than we thought. N Engl J Med. 2006;355:841–4. doi: 10.1056/NEJMe068122. [DOI] [PubMed] [Google Scholar]
- 11.Dean JC. Marfan syndrome: clinical diagnosis and management. Eur J Hum Genet. 2007;15:724–33. doi: 10.1038/sj.ejhg.5201851. [DOI] [PubMed] [Google Scholar]
- 12.Erentug V, Polat A, Bozbuga NU, Polat E, Erdogan HB, Kirali K, Guler M, Akinci E, Yakut C. Cardiovascular reoperations in Marfan syndrome. J Card Surg. 2006;21:455–7. doi: 10.1111/j.1540-8191.2006.00276.x. [DOI] [PubMed] [Google Scholar]
- 13.Robinson PN, Godfrey M. The molecular genetics of Marfan syndrome and related microfibrillopathies. J Med Genet. 2000;37:9–25. doi: 10.1136/jmg.37.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizuguchi T, Collod-Beroud G, Akiyama T, Abifadel M, Harada N, Morisaki T, Allard D, Varret M, Claustres M, Morisaki H, Ihara M, Kinoshita A, Yoshiura K, Junien C, Kajii T, Jondeau G, Ohta T, Kishino T, Furukawa Y, Nakamura Y, Niikawa N, Boileau C, Matsumoto N. Heterozygous TGFBR2 mutations in Marfan syndrome. Nat Genet. 2004;36:855–60. doi: 10.1038/ng1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh KK, Rommel K, Mishra A, Karck M, Haverich A, Schmidtke J, Arslan-Kirchner M. TGFBR1 and TGFBR2 mutations in patients with features of Marfan syndrome and Loeys Dietz syndrome. Hum Mutat. 2006;27:770–7. doi: 10.1002/humu.20354. [DOI] [PubMed] [Google Scholar]
- 16.Sakai H, Visser R, Ikegawa S, Ito E, Numabe H, Watanabe Y, Mikami H, Kondoh T, Kitoh H, Sugiyama R, Okamoto N, Ogata T, Fodde R, Mizuno S, Takamura K, Egashira M, Sasaki N, Watanabe S, Nishimaki S, Takada F, Nagai T, Okada Y, Aoka Y, Yasuda K, Iwasa M, Kogaki S, Harada N, Mizuguchi T, Matsumoto N. Comprehensive genetic analysis of relevant four genes in 49 patients with Marfan syndrome or Marfan-related phenotypes. Am J Med Genet A. 2006;140:1719–25. doi: 10.1002/ajmg.a.31353. [DOI] [PubMed] [Google Scholar]
- 17.Handford PA. Fibrillin-1, a calcium binding protein of extracellular matrix. Biochim Biophys Acta. 2000;1498:84–90. doi: 10.1016/s0167-4889(00)00085-9. [DOI] [PubMed] [Google Scholar]
- 18.Corson GM, Chalberg SC, Dietz HC, Charbonneau NL, Sakai LY. Fibrillin binds calcium and is coded by cDNAs that reveal a multidomain structure and alternatively spliced exons at the 5′ end. Genomics. 1993;17:476–84. doi: 10.1006/geno.1993.1350. [DOI] [PubMed] [Google Scholar]
- 19.Pereira L, D'Alessio M, Ramirez F, Lynch JR, Sykes B, Pangilinan T, Bonadio J. Genomic organization of the sequence coding for fibrillin, the defective gene product in Marfan syndrome. Hum Mol Genet. 1993;2:961–8. doi: 10.1093/hmg/2.7.961. [DOI] [PubMed] [Google Scholar]
- 20.Dietz HC, Saraiva JM, Pyeritz RE, Cutting GR, Francomano CA. Clustering of fibrillin (FBN1) missense mutations in Marfan syndrome patients at cysteine residues in EGF-like domains. Hum Mutat. 1992;1:366–74. doi: 10.1002/humu.1380010504. [DOI] [PubMed] [Google Scholar]
- 21.Cañadas V, Vilacosta I, Bruna I, Fuster V. Marfan syndrome. Part 1: pathophysiology and diagnosis. Nat Rev Cardiol. 2010;7:256–65. doi: 10.1038/nrcardio.2010.30. [DOI] [PubMed] [Google Scholar]
- 22.Robinson PN, Arteaga-Solis E, Baldock C, Collod-Béroud G, Booms P, De Paepe A, Dietz HC, Guo G, Handford PA, Judge DP, Kielty CM, Loeys B, Milewicz DM, Ney A, Ramirez F, Reinhardt DP, Tiedemann K, Whiteman P, Godfrey M. The molecular genetics of Marfan syndrome and related disorders. J Med Genet. 2006;43:769–87. doi: 10.1136/jmg.2005.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faivre L, Collod-Beroud G, Loeys BL, Child A, Binquet C, Gautier E, Callewaert B, Arbustini E, Mayer K, Arslan-Kirchner M, Kiotsekoglou A, Comeglio P, Marziliano N, Dietz HC, Halliday D, Beroud C, Bonithon-Kopp C, Claustres M, Muti C, Plauchu H, Robinson PN, Adès LC, Biggin A, Benetts B, Brett M, Holman KJ, De Backer J, Coucke P, Francke U, De Paepe A, Jondeau G, Boileau C. Effect of mutation type and location on clinical outcome in 1,013 probands with Marfan syndrome or related phenotypes and FBN1 mutations: an international study. Am J Hum Genet. 2007;81:454–66. doi: 10.1086/520125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faivre L, Collod-Beroud G, Child A, Callewaert B, Loeys BL, Binquet C, Gautier E, Arbustini E, Mayer K, Arslan-Kirchner M, Stheneur C, Kiotsekoglou A, Comeglio P, Marziliano N, Halliday D, Beroud C, Bonithon-Kopp C, Claustres M, Plauchu H, Robinson PN, Adès L, De Backer J, Coucke P, Francke U, De Paepe A, Boileau C, Jondeau G. Contribution of molecular analyses in diagnosing Marfan syndrome and type I fibrillinopathies: an international study of 1009 probands. J Med Genet. 2008;45:384–90. doi: 10.1136/jmg.2007.056382. [DOI] [PubMed] [Google Scholar]
- 25.Schrijver I, Liu W, Odom R, Brenn T, Oefner P, Furthmayr H, Francke U. Premature termination mutations in FBN1: distinct effects on differential allelic expression and on protein and clinical phenotypes. Am J Hum Genet. 2002;71:223–37. doi: 10.1086/341581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu WO, Oefner PJ, Qian C, Odom RS, Francke U. Denaturing HPLC-identified novel FBN1 mutations, polymorphisms, and sequence variants in Marfan syndrome and related connective tissue disorders. Genet Test. 1997-1998;1:237–42. doi: 10.1089/gte.1997.1.237. [DOI] [PubMed] [Google Scholar]
- 27.Yuan X, Downing AK, Knott V, Handford PA. Solution structure of the transforming growth factor beta-binding protein-like module, domain associated with matrix fibrils. EMBO J. 1997;16:6659–66. doi: 10.1093/emboj/16.22.6659. [DOI] [PMC free article] [PubMed] [Google Scholar]