Abstract
The trafficking of protein and RNA cargoes between the cytoplasm and the nucleus of eukaryotic cells, which is a major pathway involved in cell regulation, is mediated by nuclear transport sequences in the cargoes and by shuttling transport factors. The latter include receptors (karyopherins) that recognize the cargoes and carry them across the nuclear pore complex (NPC), and the small GTPase Ran, which modulates karyopherin–cargo binding. Nuclear import can be studied in vitro using digitonin-permeabilized cells, which are depleted of shuttling transport factors. Nuclear import can be reconstituted in the permeabilized cells with exogenous cytosol or with purified recombinant transport factors, and can be quantified by light microscopy of fluorescently labeled cargoes or by immunofluorescence staining. Here we describe procedures for in vitro nuclear import in permeabilized mammalian cells, and for the preparation of recombinant transport factors (importin α, importin β, importin 7, transportin, Ran, NTF2) and other reagents commonly used in the assay. This assay provides means to characterize the molecular mechanisms of nuclear import and to study the import requirements of specific cargoes.
Keywords: Nuclear protein import, Digitonin-permeabilized cells, Recombinant protein expression, Recombinant protein purification, Shuttling nuclear transport factors, Karyopherins, Importins, Ran
1 Introduction
In eukaryotic cells, molecules traffic between the cytoplasm and the nucleus through nuclear pore complexes (NPCs), large proteinaceous channels that perfo-rate the nuclear envelope. Whereas small molecules are able to diffuse passively through the NPC, proteins and nucleoprotein complexes larger than ~20–40 kDa require signals and receptors for their nuclear import and export (1). The facilitated transport of receptor–cargo complexes through NPC is saturable and energy dependent, and commonly involves nucleocytoplasmic shuttling receptors of the importin β/karyopherin β family and the small GTPase Ran (2,3).
Nuclear import and export is mediated by nuclear localization sequences (NLSs) and nuclear export sequences (NESs), respectively, in the cargoes. NLSs typically are short amino acid stretches enriched in basic residues, which bind to their cognate karyopherins in the cytoplasm either directly or via adaptors. The resulting import complexes then pass through the NPC by means of multiple interactions between the karyopherins and NPC proteins (nucleoporins) containing Phe-Gly (FG)-repeats. The directionality of nuclear import is regulated by Ran, which exists predominantly in its GTP-bound form in the nucleus, and in its GDP-bound form in the cytoplasm. The nucleocytoplasmic Ran-GTP gradient results from the nuclear localization of Ran guanine nucleotide exchange factor (RanGEF, RCC1), which generates Ran-GTP from Ran-GDP, and from the cytoplasmic localization of RanGTPase-activating protein (RanGAP), which promotes GTP hydrolysis on Ran. When import complexes reach the nucleoplasm, Ran-GTP binds directly to the karyopherins and thereby induces the release of cargoes (4). The Ran-GTP-karyopherin complexes then are translocated back to the cytoplasm, where the Ran-GTP is hydrolyzed by RanGAP and the karyopherins are released for further import. Ran is transported back into the nucleus by NTF2, a receptor for Ran-GDP. Ran and its regulators also play a key role in the directionality of export, since Ran-GTP promotes the binding of NESs to export karyopherins, and the hydrolysis of Ran-GTP by RanGAP in the cytosol promotes the disassembly of export complexes.
An in vitro nuclear import assay involving digitonin-permeabilized cells, developed in our laboratory over 15 years ago, has been widely used to characterize the shuttling factors involved in nuclear import, the import requirements of specific cargoes, and the mechanisms of nuclear transport (5). This method is simple and can be used with virtually any cultured mammalian cells (adherent or non-adherent). Methods for in vitro nuclear assembly and import using extracts from Xenopus eggs also have been developed and extensively used, especially to study nuclear assembly (6). Digitonin is a nonionic detergent that binds cholesterol selectively, and at low concentrations permeabilizes the plasma membrane while leaving the nuclear envelope and endoplasmic reticulum (ER) intact and functional for transport (7). During the process of permeabilization and subsequent washing, most of the shuttling nuclear transport factors are released from the cells, rendering them unable to support nuclear import of exogenous cargoes. However, efficient nuclear import can be restored if cells are reconstituted with exogenous cytosol to provide shuttling nuclear transport factors. Moreover, transport can be reconstituted in permeabilized cells with purified recombinant transport factors instead of cytosol, i.e., karyopherins, Ran, and NTF2. This assay can be used for analyzing endpoints of transport assays (i.e., the level of cargo accumulated in the nucleus at a fixed time) or for real-time imaging of cargo accumulation.
Fluorescently labeled cargoes, which are easily detected by light microscopy, are commonly used in nuclear import assays. In addition, cargo import can be monitored by immunofluorescent staining of cells after the import reaction. Import can be quantified by digital light microscopy to determine the nuclear-localized fluorescence (for adherent cells or for non-adherent cells attached to a slide after the transport reaction) or by flow cytometry to determine the cell-associated fluorescence (for non-adherent cells). The quantification of nuclear import by flow cytometry allows facile analysis of a large number of cells (~10–50,000) and is rapid, although the number of individual samples that can be analyzed is limited. Flow cytometry measures the total fluorescence of cells (both nuclear and cytoplasmic), and is not recommended for nuclear import assays reconstituted with recombinant factors or involving the import inhibitor wheat germ agglutinin (WGA), because nonspecific cytoplasmic binding of cargoes can occur in these cases. Quantification of import by digital light microscopy requires the analysis of at least 50–300 cells (typically three microscope fields with a ×40 objective), since there often is considerable cell-to-cell variability in nuclear transport levels (8). While quantitative analysis of import by light microscopy is laborious if done manually, this method can be automated for high throughput analysis using a computer-controlled microscope with a motorized x-y-z stage together with image analysis software.
Here we describe protocols for analyzing the nuclear import of proteins in digitonin-permeabilized adherent cells reconstituted with cytosol or recombinant transport factors. We provide methods for analyzing several of the karyopherin import pathways that have been characterized, but in principle the protocol can be extended to analysis of any shuttling receptor that can be expressed recombinantly. Moreover, nuclear export can be similarly analyzed in this system, using either exogenous or endogenous cargoes (9, 10). We describe cargoes and recombinant factors that can be used to analyze the import pathways involving importin β (karyopherin β1), importin α/β, importin 7, the importin 7/β heterodimer, and transportin (karyopherin β2).
2 Materials
2.1 Cells
HeLa S3 cells, a clonal derivative of the parent HeLa cell line adapted for continuous growth in suspension (ATCC, Manassas, VA, USA; #CCL-2.2).
HeLa medium: Joklik's modified minimal essential medium (Sigma-Aldrich, St. Louis, MO, USA; #M0518-10L), 10% v/v fetal bovine serum, and 1% w/v penicillin and streptomycin.
NRK (normal rat kidney) cells (ATCC; #CRL-6509).
NRK medium: DMEM medium (4.5 g/L d-glucose, l-glutamine, 110 mg/L sodium pyruvate without sodium carbonate; Gibco-BRL, Invitrogen, Carlsbad, CA, USA; #12800-082), 10% fetal bovine serum, and 1% penicillin/streptomycin.
2.2 Preparation of Cytosol
Phosphate-buffered saline (PBS) (1 L): 10 mM sodium phosphate, pH 7.4, 140 mM NaCl.
Dithiothreitol (DTT): 1 M stock solution in distilled water, store at −20°C.
Wash buffer (500 mL): 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, and 2 mM DTT.
Hypotonic buffer (100 mL): 5 mM HEPES, pH 7.3, 10 mM potassium acetate, 2 mM magnesium acetate, 2 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 μg/mL each of aprotinin, pepstatin, leupeptin.
Digitonin (Calbiochem, EMD Chemicals, Gibbstown, NJ, USA): 10% w/v stock solution in dimethyl sulfoxide (DMSO), store at −20°C.
Trypan Blue solution (0.4% w/v) (Sigma; #T-8154).
Transport buffer (TB): 20 mM HEPES, pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM ethylene glycol-bis (β-aminoethyl ether)-N,N,N′,N;-tetraacetic acid (EGTA), 2 mM DTT, 1 mM PMSF, and 1 μg/mL each of aprotinin, pepstatin, and leupeptin.
Spectra/Por dialysis membranes (6,000–8,000-kDa cut-off) (Spectrum Laboratories, Rancho Dominguez, CA, USA; #132645) and clamps.
2.3 Preparation of FITC-labeled Transport Cargoes
NLS peptide: synthetic peptide containing the SV40 large T antigen wild-type nuclear localization signal (11, 12), preceded by a tri-glycine spacer and an N-terminal cysteine for coupling (CGGGPKKKRKVED).
1% v/v acetic acid.
Sephadex G10 (Sigma; #G10120), chromatography column (10-mL bed volume, 1-cm diameter).
50 mM HEPES, pH 7.0.
Ellmann's buffer: 0.1 M sodium phosphate buffer, pH 7.4, and 5 mM EDTA.
Ellmann's reagent: 1 mM dithiobisnitrobenzoic acid in methanol.
FITC isomer I (Molecular Probes, Invitrogen; #F1906) in dimethylformamide (DMF). Prepare fresh. DMF is toxic and all procedures using this chemical should involve the use of gloves and be done under a chemical fume hood.
Coupling buffer 1: 0.1 M sodium bicarbonate, pH 9.
PBS: 10 mM sodium phosphate, pH 7.4, 140 mM NaCl.
Sulfosuccidimidyl [N-maleimidomethyl]cyclohexane carboxylate (SMCC) (Pierce; Rockford, IL, USA; 22322): 20 mM in DMSO, prepared immediately before use.
PD-10 column (prepacked G25 column, 10-mL bed volume, Amersham Biosciences, Piscataway, NJ, USA; #17-0851-01).
Fatty acid-free bovine serum albumin (BSA) (Roche, Boehringer Mannheim, Indianapolis, IN, USA; #100062).
Histone H1: Solution of 20 mg of histone H1 in 1 mL of water (Upstate, St. Charles, MO, USA; #14-155).
Coupling buffer 2: 130 mM sodium carbonate, pH 7.0.
2.4 Plasmids, Expression, and Purification Systems
2.4.1 Cargoes: GST-M9 and GST-IBB
Isopropyl-β-Δ-thiogalactopyranoside (IPTG):1 M stock solution in water, aliquot, store at −20°C.
pGEX2T-IBB: this construct contains GST fused to residues 1–65 of human importin α1 (i.e., importin beta binding [IBB] domain) inserted into BamHI-EcoRI sites of pGEX2T (Amersham Biosciences; #27-4801-01) (13).
pGEX5X-M9: this construct contains GST fused to residues 263–306 of human hnRNP A1 (termed the “M9 domain”) inserted into EcoRI-XhoI sites of pGEX5X (Amersham Biosciences) (13).
GST lysis buffer: 5% v/v glycerol, 50 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 1 mM magnesium acetate, 2 mM DTT, 1 mg/mL lysozyme, 10 μg/mL DNase I, 0.1% v/v Triton X-100, and 1 μg/mL each of leupeptin, pepstatin, and aprotinin.
Glutathione Sepharose 4B (Amersham Biosciences; #17-0756-01).
GST washing buffer: 50 mM Tris-HCl, pH 8.0, 0.5 M NaCl, and 1 μg/mL each of leupeptin, pepstatin, and aprotinin.
Reduced glutathione (Sigma; #6529).
GST elution buffer: 50 mM Tris-HCl, pH 8.0, and 15 mM reduced glutathione.
2.4.2 Importin α1
pRSETB-importin α1: this construct contains a 6×-His tag at its N-terminus and an Xpress epitope for detection (pRSETB vector; Invitrogen; #V351-20) and human importin α1 (hSRP1 α, karyopherin α2) sequence inserted into BamH1-Xho1 sites (14).
PMSF: 17.4 mg/mL PMSF in isopropanol (100 mM). Divide the solution in aliquots and store at −20°C. PMSF is toxic and all procedures using this chemical should involve the use of gloves and be done under a chemical fume hood.
Importin α lysis buffer: 5% v/v glycerol, 50 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 1 mM β-mercaptoethanol, 1 mM PMSF, and 1 μg/mL each of leupeptin, pepstatin, and aprotinin.
Talon metal affinity resin (Clontech, Mountain View, CA, USA; #635503).
2.4.3 Importin β
pTYB4-importin β: this construct contains intein/chitin-binding domain at the C-terminus and human importin β sequence inserted into the NcoI-NotI sites (13).
ER2566 cells (New England Biolabs, Ipswich, MA, USA).
Tris (2-carboxyethyl)-phosphine hydrochloride (TCEP) (Pierce, #20490): 0.2 M stock solution in water, store at −20°C.
Importin β lysis buffer: 50 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 2 mM magnesium chloride, 10 mM CHAPS, 0.2 mM TCEP (add fresh), and 1 μg/mL each of leupeptin, pepstatin, and aprotinin.
Chitin beads (New England Biolabs; #S6651L).
Importin β elution buffer: 50 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 2 mM MgCl2, 1 mM CHAPS, 30 mM DTT, and 1 μg/mL each of leupeptin, pepstatin, and aprotinin.
2.4.4 Importin 7
pQE9-importin 7 (RanBP7): this construct contains a 6×-His tag at the N-terminus (pQ9 vector, Qiagen, Valencia, CA, USA; #32915) and the human importin 7 sequence (15).
TG-1 (Zymo Research, Orange, CA, USA; #T3017) and M15 (Qiagen; #34210) cells.
Importin 7 phosphate buffer: 50 mM sodium phosphate, pH 8.0, 300 mM NaCl, and 1 μg/mL each of leupeptin, pepstatin, and aprotinin.
2.4.5 Transportin
pET28c-transportin: this construct contains a 6×-His-tag and a T7 tag at its N-terminus, with a thrombin cleavage site between the two tags (pET28c vector, Novagen, EMD Chemicals, Gibbstown, NJ, USA; #69866) and the human transportin sequence inserted into the BamH1-SalI sites (16).
BL21 (DE3) (Stratagene, Agilent, Santa Clara, CA, USA; #200131).
Transportin lysis buffer: 50 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 1 mM magnesium acetate, 5% v/v glycerol, 2 mM CHAPS, 1 mg/mL lysozyme, 10 μg/mL DNase I, and 1 μg/mL each of leupeptin, pepstatin, and aprotinin.
Transportin washing buffer: 50 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 1 mM magnesium acetate, 1 mM CHAPS, and 1 μg/mL each of leupeptin, pepstatin, and aprotinin.
Transportin elution buffer: 50 mM Tris-HCl, pH 6.8, 0.5 M NaCl, 1 mM magnesium acetate, and 1 μg/mL each of leupeptin, pepstatin, and aprotinin.
2.4.6 Ran
Ran vector: Human Ran is in modified pET11d (Novagen; #69439-3). The pET11d was digested by Nco1 and BamH1 such that there is no purification tag. The NcoI site was introduced at the start codon of Ran by PCR and the fragment was inserted into NcoI-BamHI sites of the modified pET11d (17).
Ran lysis buffer: 50 mM Tris pH 8.0, 75 mM NaCl, 1 mM MgCl2, 1 mM DTT, 1 mM PMSF, 1 mg/mL lysozyme, 10 μg/mL Dnase I, and 1 μg/mL each of leupeptin, pepstatin, aprotinin.
DE52: Pre-swollen microgranular DEAE cellulose (Whatman, Maidstone, Kent, UK).
Transport buffer (TB): 20 mM HEPES pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 2 mM DTT, 1 mM PMSF, and 1 μg/mL each of aprotinin, pepstatin, leupeptin.
Hiload 26/60 Superdex 200 prep grade (Amersham Biosciences).
SP buffer: 50 mM Tris pH 6.8, 10 mM KCl, 1 mM MgCl2, 1 mM DTT, and 1 μg/mL each of aprotinin, pepstatin, and leupeptin.
PD-10 column (prepacked G25 column, 10-mL bed volume, Amersham Biosciences).
SP-SepharoseT Fast Flow (Amersham Biosciences).
2.4.7 NTF2
BL21 (DE3) pLysS (Stratagene; #200132).
pET23b-NTF2: This plasmid contains human NTF2. The pET23b vector (Novagen; #69746-3) was modified to include a stop codon at the HincII site, thereby eliminating translation of the histidine tag (18).
TNE buffer: 50 mM Tris-HCl pH 8.0, 100 mM NaCl, and 10 mM EDTA.
S200 HR: Superdex 200 10/300 GL (Amersham Biosciences).
Q Sepharose Fast Flow (Amersham Biosciences).
Q buffer: 50 mM HEPES pH 7.4, 1 mM MgCl2, 1 mM DTT, and 1 μg/mL each of aprotinin, pepstatin, and leupeptin.
2.5 Nuclear Import Assay
Ten-well microscope slides (ICN Biomedicals, Irvine, CA, USA; #6041805).
Hydrophobic slide marker for staining procedure (Research Products International, Mt. Prospect, IL, USA; #195505).
Transfer pipet (Fisher Scientific, Pittsburgh, PA, USA; #13-711-5A).
Square cell culture plate (Fisher Scientific; #0875711A) that can accommodate three ten-well slides.
Two metal blocks, one prechilled on ice and the other prewarmed to 30°C.
Humidified chamber at 30°C. We use a plastic box covered by aluminum foil, which contains the prewarmed metal block on moistened absorbent paper. Temperature is maintained at 30°C using an incubator or water bath.
Cytosol from HeLa S3 cells: 10 mg/mL cytosol in TB (see Section 3.2).
Fluorescent transport cargoes (see Section 3.3) or unlabeled transport cargoes (see Section 3.4). The cargoes are in TB and stored as aliquots at −80°C.
Transport buffer (TB): 20 mM HEPES pH 7.4, 110 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 2 mM DTT, and 1 μg/mL of each leupeptin, pepstatin, and aprotinin. The TB solution is prepared freshly using a 20-fold concentrated stock solution containing HEPES, potassium acetate, and magnesium acetate (stored at 4°C), a solution stock of 0.2 M EGTA, pH 7.4 (stored at 4°C), and 1 M stock solutions of DTT and protease inhibitors (stored at −20°C).
Digitonin (Calbiochem; #300410): Make a 10% w/v stock in DMSO and store at −20°C.
Adenosine triphosphate (ATP) (Sigma): 200 mM stock in 20 mM HEPES, pH 7.4, and 100 mM magnesium acetate. Dissolve ATP in 100 mM magnesium acetate, adjust the pH to ~7.4 with 10 M NaOH, and add HEPES to 20 mM from a 1 M HEPES, pH 7.4 stock. Aliquot and store at −20°C.
Creatine phosphate (CP) (Calbiochem): 160 mg/mL stock solution in water. Aliquot and store at −20°C.
Creatine phosphate kinase (CPK) (Calbiochem): 2,000 U/mL stock solution in 20 mM HEPES, pH 7.4 and 50% glycerol. Aliquot and store at −20°C.
ATP regenerating system: freshly prepared from the stock solutions of ATP, CP, and CPK, mixed at a ratio of 1:1:1.
Guanosine triphosphate (GTP) (Calbiochem): 5 mM stock solution in modified TB (lacking EGTA, DTT, and protease inhibitors), filter with a 0.22-μm filter, aliquot and store at −20°C.
Wheat germ agglutinin (WGA) (Sigma): Make a 2 mg/mL stock in modified TB (lacking EGTA and DTT), aliquot and store at −20°C.
Formaldehyde, 37% (Fisher Scientific).
2.6 Immunostaining
Gelatin type B from bovine skin (Sigma): 2% stock solution in PBS, aliquot and store at −20°C.
Purified goat anti-GST antibody (Amersham).
FITC-labeled mouse anti-goat antibody (Pierce).
Solution of Topro-3 (Molecular Probes) or DAPI (Sigma). Aliquot and store at −20°C. Stock solution of DAPI at 1 mg/mL in PBS, aliquot, and store at −20°C.
SlowFade Antifade kit (Molecular Probes).
Coverglasses (22×60 mm; VWR Scientific).
3 Methods
3.1 Cells
For preparation of HeLa cytosol, grow 8 L of HeLa S3 cells at 37°C in spinner flasks, always maintaining a density of 4–10×105 cells/mL. During expansion of cultures, the cell density is checked daily using a hematocytometer and cultures are diluted to a density of 4×105 cells/mL. Starting with 100 mL of culture at 4×105 cells/mL, growth of 8 L of cells should take ~6–8 days. When harvested at ~6–8×105 cells/mL, 8 L of culture yields ~20 mL of packed cells.
NRK cells are used for the nuclear import assay. The cells are grown in NRK medium at 37°C with 5% CO2 in culture flasks. NRK cells are plated on 10-well slides the day before the experiment at a density yielding 12,000 cells/well. Before seeding, the slides are rinsed briefly with a solution of 70% ethanol, dried using Kimwipes, sterilized in a Bunsen burner, and put in the square culture plate using sterile tweezers. The cells should be at ~80% confluence the next day for nuclear import assays.
3.2 Preparation of HeLa Cell Cytosol
Harvest the cells from 8 L of HeLa S3 culture by centrifugation at 1,000×g for 15 min at 4°C in 500-mL conical bottles using a Beckman JS5.2 rotor.
Wash the cells twice with 100 mL of ice-cold PBS per 2 L of original culture, pool the bottles, and centrifuge again.
Resuspend the pellet in ice-cold wash buffer so the final volume reaches 50 mL, transfer the cell suspension to a 50-mL Falcon tube, and centrifuge as before.
Resuspend the cell pellet in an equal volume of cold hypotonic lysis buffer and let the cells swell on ice for 10 min (total volume ~40 mL).
Lyse the cells on ice by adding digitonin to the point where ~90–95% of the cells are permeable to Trypan Blue. To accomplish this, start by adding 120 μL of 10% digitonin solution (25 μL per 109 cells), then add smaller (10 μL) aliquots in a stepwise manner until adequate permeabilization is obtained. After the addition of each aliquot, wait 2–5 min and examine a sample of cells in the microscope to assess Trypan Blue permeability. Stop adding digitonin when 90–95% permeabilization is reached. Avoid adding excess digitonin, as this could interfere with the import assay (see below). Approximately 160 μL of 10% digitonin solution are needed for a 40-mL cell suspension to get suitable permeabilization.
Centrifuge the cells at 1,500×g for 15 min at 4°C to remove permeabilized cells and debris.
Centrifuge the supernatant from the previous step at 15,000×g for 20 min at 4°C in a Beckman JA-20 rotor, collect the supernatant, and centrifuge the latter at 100,000×g for 1 h at 4°C using a Beckman 70Ti rotor.
Dialyze the final supernatant (~20 mL) at 4°C in preboiled dialysis tubing (6,000–8,000 kDa cut-off) in 2 L of transport buffer with three changes of buffer.
After dialysis, the concentration of protein in the cytosol is determined by the Bradford protein assay, and adjusted to 10 mg/mL by dilution with TB or by concentration with a Centricon concentrator (Amersham). Aliquot, freeze in liquid nitrogen, and store at −80°C.
3.3 Preparation of FITC-labeled Transport Cargoes
The FITC-BSA-NLS is prepared in two steps. First, BSA is conjugated with FITC (Section 3.3.1.2), and then a peptide containing the wild-type SV40 NLS is coupled to the FITC-BSA via the N-terminal cysteine on the peptide (Section 3.3.1.3). Since the sulfhydryl group in the peptide tends to be oxidized, it is necessary to reduce the peptide with DTT prior to coupling (Section 3.3.1.1). The FITC-histone H1 is prepared in a single step, following the same procedure as conjugation of BSA with FITC, although the coupling buffer is modified to maintain the solubility of histone H1 (Section 3.3.2).
3.3.1 FITC-BSA-NLS
3.3.1.1 Reduction of Synthetic Peptide
Wash a Sephadex G10 column with 25 mL of 1% v/v acetic acid.
In the meantime, dissolve 10 mg of NLS peptide in 1 mL of 50 mM Hepes, pH 7.4. Add 10 mg of DTT, vortex, and incubate for 1 h at room temperature.
Separate the peptide from free DTT by gel-filtration on the acetic acid-equilibrated G10 column. Load the peptide and elute the column with 1% acetic acid, collecting 0.5-mL fractions; 30 fractions should be enough to elute the NLS peptide and the DTT.
Analyze the fractions using Ellman's reagent. For this, add 10 μL of each fraction to 900μL of Ellman's buffer and 100 μL of Ellman's reagent in an Eppendorf tube and vortex. The sulfhydryl residues yield a bright yellow color. The first yellow peak contains the peptide and the second (brighter) yellow peak contains the DTT.
Pool the fractions containing the peptide, aliquot into eight preweighed Eppendorf tubes, cover the tubes with perforated Parafilm, and dry the peptide using a Speed Vac. Weigh the tubes again (there should be ~1 mg peptide per tube) and store at −80°C.
3.3.1.2 Preparation of FITC-BSA
Dissolve 20 mg of fatty acid-free BSA in 1 mL of Coupling Buffer 1.
Add 200 μL of freshly prepared FITC solution (10 mg/mL FITC isomer I in DMF) to BSA and incubate at room temperature for 1 h in the dark.
In the meantime, wash a PD-10 column with 25 mL of PBS.
Adjust the volume of the FITC-BSA conjugate to 2.5 mL with PBS and load on the column, let the sample run into the column, and elute with 3.5 mL of PBS. Collect the peak of FITC-BSA that elutes prior to the peak of uncoupled FITC.
Dilute the peak of eluted FITC-BSA twofold with PBS to a final concentration of ~2.5 mg/mL.
Freeze 1-mL aliquots in liquid nitrogen and store at −80°C.
3.3.1.3 Conjugation of the NLS Peptide to FITC-BSA
Thaw a 1-mL aliquot of FITC-BSA (Section 3.3.1.2), add 50 μL of freshly prepared 20 mM SMCC and incubate for 30 min at room temperature in the dark.
In the meantime, wash a PD-10 column with 25 mL of PBS.
To remove the unconjugated crosslinker, apply the mixture to the PD-10 column and let the sample run into the column.
Apply 1.5 mL of PBS to the column and allow it to enter.
Elute the column with a further 3.5 mL of PBS. Collect the bright yellow peak in a separate tube (in ~2 mL).
Dissolve 1 mg of lyophilized NLS peptide (from Section 3.3.1.1) with the column eluate and incubate for 2 h at room temperature in the dark.
Meanwhile, equilibrate the PD-10 column with 25 mL of TB.
To remove uncoupled peptide from the FITC-BSA-NLS, apply the mixture to the PD-10 column, let the sample run in and elute with TB. Collect only the bright yellow peak (~2 mL, now at ~1 mg/mL). Freeze 20-μL aliquots in liquid nitrogen and store at −80°C.
3.3.2 FITC-Histone H1
Dilute 3 mg of histone H1 in 1 mL of Coupling Buffer 2 (130 mM sodium carbonate pH 7.0) and mix with 30 μL of FITC dissolved in DMF (2 mg FITC in 50 μL DMF). The concentration of DMF should be < 2% to avoid precipitation of histone H1. Incubate for 1 h at room temperature in the dark.
In the meantime, wash the PD-10 column with 25 mL PBS.
To remove free FITC, load the mixture on the PD-10 column, let it run in, and elute with 5 mL PBS. Collect the first bright yellow peak in a single tube (~4 mL).
Freeze in 20-μL aliquots in liquid nitrogen and store at −80°C.
3.4 Expression and Purification of Cargoes: GST-M9 and GST-IBB
Transform samples of BL21 (DE3) with pGEX2T-IBB and pGEX5X-M9 plasmids.
Grow 2 L of transformed cells in LB medium with ampicillin (50 μg/mL) at 37°C until the cultures reach an OD600 nm of 0.6.
Induce protein expression with 50 μM IPTG for 2 h at 37°C.
Harvest the cells by centrifugation at 1,500×g for 15 min at 4°C. The pellets can be frozen and stored at −80°C before recombinant protein purification (below).
Resuspend the pellets in 30 mL of GST lysis buffer per liter of original culture.
Freeze the suspension in liquid nitrogen and thaw in an ice-water bath.
Sonicate on ice, three times for 30 sec with 30-sec intervals.
Clear the lysate by centrifugation at 100,000×g for 20 min at 4°C in a Beckman Ti 45 rotor and collect the supernatant (cleared lysate).
Incubate the cleared lysate with glutathione–Sepharose beads (1 mL of glutathione beads slurry in PBS (1:1) per liter of original culture) on a rotating wheel for 2 h at 4°C.
Centrifuge at 500×g for 10 min at 4°C. Discard the supernatant.
Wash the beads three times with 50 mL of GST washing buffer.
Elute the recombinant proteins with 2 mL of GST elution buffer.
Check the purity of the GST fusion proteins on an SDS gel. GST-M9 and GSTIBB migrate at ~29 kDa and ~32 kDa, respectively. If the proteins are not pure enough you can add another step of purification by chromatography on a S200 Superdex column.
Dialyze into TB. Determine the concentration of protein using the Bradford protein assay. Freeze in 20-μL aliquots in liquid nitrogen and store at −80°C.
3.5 Expression and Purification of Transport Factors
3.5.1 Importin α
Transform BL21 (DE3) with the plasmid pRSETB-importin α1.
Grow 4 L of transformed bacteria in LB medium with ampicillin (50 μg/mL) at 37°C until cultures reach an OD600 nm of 0.6.
Induce protein expression with 200 μM IPTG for 3 h at 37°C.
Collect the bacteria by centrifugation at 1,500×g for 15 min at 4°C. The pellets can be frozen and stored at −80°C before recombinant protein purification (below).
Resuspend the pellets by pipetting or vortexing in 50 mL of importin α lysis buffer per liter of original culture.
Add lysozyme to 0.5 mg/mL and keep on ice for 20 min.
Add Triton X-100 to 0.2 % v/v final concentration and add fresh PMSF to 1 mM.
Sonicate on ice, three times for 30 sec with 30-sec intervals.
Clear the lysate by centrifugation at 100,000×g for 20 min at 4°C in a Beckman Ti 45 rotor and collect the supernatant (cleared lysate).
Add imidazole, pH 7.0 from a 1 M stock solution to the cleared lysate to a final concentration of 10 mM, then add to the Talon beads previously equilibrated with importin α lysis buffer (0.5 mL resin per liter of original culture).
Incubate for 1–2 h on a rotating wheel at 4°C.
Load the beads into a column.
Wash the column with importin α lysis buffer containing 10 mM imidazole until no more protein is released. Check the protein concentration of the eluate using the Bradford protein assay. You will need ~20 mL to wash the column fully.
Elute the importin α with five column volumes of importin α lysis buffer containing 100 mM imidazole.
Dialyze into TB containing an additional 390 mM potassium acetate (500 mM total) and 5% v/v glycerol (see Note 1).
Check the purity of the importin α on an SDS gel. Importin α migrates at ~60 kDa. Determine the final concentration using the Bradford protein assay (see Note 2), freeze in 20-μL aliquots in liquid nitrogen and store at −80°C.
3.5.2 Importin β
Transform ER2566 cells with the pTYB4-importin β1 construct. Grow the cells in LB medium with ampicillin (100 μg/mL).
Grow 8 L of transformed bacteria in LB medium with ampicillin (100 μg/mL) at 37°C until cultures reach an OD600 nm of 0.4. Cool cultures to room temperature.
Induce with 1 mM IPTG for 4 h at 20–25°C.
Harvest the cells by centrifugation at 3,000×g for 15 min at 4°C.
Wash the pellets with 100 mL of PBS containing 0.2 mM TCEP. The pellets can be frozen and stored at −80°C before recombinant protein purification (below).
Resuspend the pellets by pipetting or vortexing in 40 mL of importin β lysis buffer per liter of original culture (See Note 3).
Sonicate on ice, three times for 30 sec with 30-sec intervals.
Clear the lysate by centrifugation at 70,000×g for 20 min at 4°C in a Beckman Ti 45 rotor and collect the supernatant (cleared lysate).
In the meantime, wash 9 mL of packed chitin beads. Use 1 mL of beads per liter of original culture. Wash two times with 40 mL of importin β lysis buffer. Spin down the beads at 500×g for 5 min at 4°C.
Add the cleared lysate to the beads and incubate for 2–3 h on a rotating wheel at 4°C.
Pour the cleared lysate/bead mixture into a column at 4°C and allow to pack by gravity flow.
Wash the resin with 10 column volumes of importin β lysis buffer.
Pass three column volumes of importin β elution buffer over the column. Stop the column flow and incubate overnight at 4°C.
Elute importin β from the column with the addition of three columns volume of importin β elution buffer.
Add 50 μL of chitin beads (equilibrated in importin β elution buffer) to the total eluate and incubate for 15 min on a rotating wheel to remove the excess tag. Remove the beads by centrifugation at 500×g for 5 min at 4°C.
Add 50 μL of Q Sepharose Fast Flow beads (equilibrated in importin β elution buffer) to the supernatant to remove nucleic acids, which selectively bind to Q Sepharose Fast Flow beads in importin β elution buffer. Incubate for 15 min on a rotating wheel and remove the beads by centrifugation at 500×g for 5 min at 4°C.
Dialyze the supernatant against TB with 0.01 mM CHAPS (see Note 4).
Check the purity of the importin β on an SDS gel. Importin β migrates at ~97 kDa. Determine the protein concentration using the Bradford protein assay. Freeze in 50-μL aliquots in liquid nitrogen and store at −80°C.
3.5.3 Importin 7
For the expression of importin 7, we use TG-1 or M15 Escherichia coli cells. Both contain the Lac repressor for tightly induced expression, since low expression of importin 7 is toxic. After transformation, grow TG-1 cells in LB medium with ampicillin (50 μg/mL) or M15 cells in LB with kanamycin (50μg/mL) and ampicillin (50 μg/mL). Expand cultures to 8 L at 37°C until they reach an OD600 nm of 0.6.
Induce with 500 μM IPTG for 5 h at 20°C. Bacteria are chilled in the cold room before induction to decrease the temperature from 37°C to 20°C.
Harvest the cells at 1,500×g for 15 min at 4°C.
Wash the cells with importin 7 phosphate buffer.
Resuspend the pellet by pipetting or vortexing in 30 mL of importin 7 phosphate buffer per liter of original culture.
Sonicate on ice, three times for 30 sec with 30-sec intervals.
Clear the lysate by centrifugation at 17,000×g for 20 min at 4°C in a Beckman Ti 45 rotor and collect the supernatant (cleared lysate)
Incubate the cleared lysate with Talon beads (equilibrated with importin 7 phosphate buffer) for 2 h at 4°C on a rotating wheel. Use 0.5 mL of beads per liter of original culture.
Pour the supernatant and bead mixture into a column.
Wash the beads with five column volumes of importin 7 phosphate buffer.
Elute with three column volumes of importin 7 phosphate buffer containing < 20 mM imidazole (see Note 5). Collect 0.5-mL fractions.
Check the purity of the importin 7-containing fractions on an SDS gel. Importin 7 migrates at ~120 kDa on an SDS gel. The eluate contains most of the importin 7 but is substantially contaminated with proteins with a Mr lower than 50 kDa. To eliminate the latter, pool fractions containing importin 7 and use an Amicon Ultra centrifugal filter unit (Millipore) with a cut-off of 50 kDa to remove the small proteins and to exchange the phosphate buffer with TB. If a further purification step is needed, chromatography on an S200 column can be employed. Measure the protein concentration using the Bradford assay. Aliquot, freeze in liquid nitrogen, and store at −80°C.
3.5.4 Transportin
Transportin is expressed using BL21 (DE3) cells. Grow the cells in LB medium with kanamycin (50 μg/mL).
Grow 2 L of transformed bacteria in LB at 37°C until cultures reach an OD600 nm of 0.7.
Cool cultures to room temperature.
Induce with 1 mM IPTG at 17°C for 4 h. Bacteria are chilled in the cold room before induction to decrease the temperature to 17°C.
Collect the bacteria by centrifugation at 3,000×g for 15 min at 4°C. The pellet can be frozen in liquid nitrogen and stored at −80°C before recombinant protein purification (below).
Resuspend the pellet by pipetting or vortexing in transportin lysis buffer. Add 50 mL of transportin lysis buffer per liter of culture.
Freeze at −80°C for 1 h and thaw the lysate in an ice-water bath. This step of freeze–thaw increases the lysis yield.
Sonicate on ice, three times for 30 sec with 30-sec intervals.
Clear the lysate by centrifugation at 17,000×g for 20 min at 4°C in a Beckman Ti 45 rotor and collect the supernatant (cleared lysate).
Incubate the cleared lysate with Talon beads (preequilibrated with transportin washing buffer) for 2 h at 4°C on a rotating wheel. Add 0.75 mL of beads per liter of original culture.
Pour the supernatant and beads into a column.
Wash the beads with five column volumes of transportin washing buffer and 10 mM imidazole.
Elute with three column volumes of transportin elution buffer containing 100 mM imidazole. Collect 1-mL fractions in Eppendorf tubes.
Check the transportin-containing fractions and the purity of transportin on a SDS gel. The transportin migrates at ~101 kDa. Pool the transportin-containing fractions and dialyze into TB containing 0.05 mM CHAPS. The small amount of detergent from transportin does not interfere with nuclear import. Freeze in 20-μL aliquots in liquid nitrogen and store at −80°C. If a higher degree of purity is needed, transportin can be chromatographed on a Superdex S200 column.
3.5.5 Ran
BL21 (DE3) cells are used for the expression of Ran. After transformation with the Ran expression vector, the bacteria are grown in LB medium with ampicillin (50 μg/mL). Typically 2 L of cells are used for protein purification.
Grow at 37°C until cultures reach an OD600 nm of 0.6.
Induce the rest of the culture with 0.5 mM IPTG at 37°C for 2 h (see Note 6).
Harvest the cells by centrifugation at 3,000×g for 15 min at 4°C. The pellets can be frozen in liquid nitrogen and stored at −80°C before recombinant protein purification (below).
Thaw the pellet from 1 L of culture with 50 mL of Ran lysis buffer in an ice-water bath.
Resuspend the pellet by vortexing. Incubate for 30 min on ice during cell lysis and vortex again.
Clear the lysate by centrifugation at 70,000×g for 20 min at 4°C in a Beckman Ti 45 rotor. Collect the supernatant (cleared lysate).
Keep an aliquot of the cleared lysate.
For the first purification step, pass the cleared lysate over a DEAE-cellulose column. Prepare the column during the lysis or the day before (store at 4°C), containing 10 mL bed volume per liter of original culture. The column is prewashed with Ran lysis buffer.
Collect and save the flow-through and wash with 10 mL of Ran lysis buffer.
Run a 12.5% SDS gel to compare 10 μL of the cleared lysate and 10 μL of the flow-through from the DEAE column. Ran should migrate as a double band close to a 25-kDa marker and should be present in large amounts in the cleared lysate and the flow-through (see Note 7). Most contaminants adsorb to the DEAE column, so Ran in the flow-through should be significantly more enriched than in the cleared lysate.
Take the flow-through and while stirring at 4°C, slowly add solid ammonium sulfate to 65% saturation to precipitate the protein (40.4 g/100 mL). Let stir for at least 1 h after dissolving (or overnight, for convenience).
Centrifuge at 70,000×g for 20 min at 4°C and resuspend the pellet in 6 mL of TB (filtered on a 0.22-μm filter) per liter of original culture.
Clarify the resuspended pellet by centrifuging at 100,000×g for 10 min at 4°C.
Pass the supernatant through a 0.22-μm syringe filter (Nalgene; #190-2520) to remove any precipitate.
Load the filtered supernatant on a S200 gel filtration column (26/60). The column and pumps are first washed with TB. Use a flow rate of 2 mL/min and collect 3-mL fractions.
Check the content and purity of fractions that have absorbance at 280 nm on an SDS gel. Pool the Ran-containing fractions that lack most contaminants.
Determine the concentration of Ran using the Bradford protein assay. You should get around 15 mg Ran per liter of original culture. Freeze in 50-μL aliquots in liquid nitrogen and store at −80°C.
If the S200 fractions are not sufficiently pure (see Note 8), add another purification step involving an SP-Sepharose cation-exchange column. Exchange the buffer in the Ran pool to SP buffer using a PD-10 column. Load 5 mL onto the column pre-equilibrated with buffer SP, wash the column until the eluate has a baseline absorbance at 280 nm, and then load the sample. Elute with a 50–500 mM gradient of KCl in SP buffer. Check the content of the fractions on an SDS gel, pool the fractions containing pure Ran, and precipitate with ammonium sulfate as described previously (see Note 9). Resuspend in TB to ~2 mg/mL, determine the concentration of Ran using Bradford protein assay, aliquot, and freeze in liquid nitrogen and store at −80°C.
3.5.6 NTF2
The plasmid coding for NTF2 is transformed into BL21 (DE3) pLysS cells. Transformants are grown in LB containing ampicillin (50 μg/mL) and chloramphenicol (10 μg/mL) overnight at 37°C.
Grow cultures at 37°C to an OD600 nm of 0.6. Typically, we prepare 4 L of culture.
Induce with 0.4 mM IPTG for 2 h at 37°C (see Note 10).
Collect the bacteria by centrifugation at 6,000×g for 15 min at 4°C. The pellets can be frozen in liquid nitrogen and stored at −80°C before recombinant protein purification (below).
The cell pellets from each 1 L of culture are resuspended in 50 mL of TNE containing 0.2 % v/v Triton X-100, 1 mM PMSF, 5 mM DTT, and 1 μg/mL each of aprotinin, leupeptin, and pepstatin (see Note 11).
Freeze by putting the resuspended cell suspensions at −20°C or in liquid nitrogen, and thaw in an ice-water bath.
Sonicate on ice, three times for 30 sec with 30-sec intervals.
Clear the lysate by centrifugation of the sample, which will be viscous, at 70,000×g for 20 min at 4°C in a Beckman Ti 45 rotor.
Precipitate the proteins in the cleared lysate by slowly adding solid ammonium sulfate, while stirring, to 50% saturation (29.5 g/100 mL) and stir for 1 h at 4°C after total dissolution.
Collect the precipitate by centrifugation at 70,000×g for 15 min at 4°C in a Beckman Ti 45 rotor, and resuspend the pellet in TB.
Clarify the sample by centrifugation at 140,000×g for 30 min in a Beckman TLA 100.3 rotor.
Chromatograph the clarified sample on a S200 HR column that has been equilibrated with two column volumes of TB. The composition of fractions is checked by analysis on an SDS gel and the fractions containing the peak of NTF2 are pooled. NTF2 migrates at ~28 kDa.
NTF2 is further purified using Q Sepharose anion-exchange column. The column is pre-equilibrated with two columns volumes of Q buffer. Dilute the pooled fractions from the S200 HR column 200-fold in buffer Q and incubate batch-wise with the resin on a rotating wheel for 2 h at 4°C.
Pour the sample into a column and wash with one column volume of Q buffer.
Elute NTF2 with Q buffer containing 150 mM KCl. Collect 1-mL fractions and check their composition on an SDS gel. Pool the fractions containing NTF2 and dialyze against TB. Determine the protein concentration using the Bradford assay. You should get ~2 mg/mL (~8 mg per liter of original culture). Freeze in 10-μL aliquots in liquid nitrogen and store at −80°C.
3.6 Nuclear Import Assay
We describe implementation of the nuclear import assay with permeabilized NRK cells, but the assay can easily be adapted to other adherent cell lines. Whatever cells are used, in our experience the most homogeneous and reproducible nuclear import is obtained when cells are at 50–80% confluence (see Note 12). The assay can be carried out on cover slips, but 10-well slides are much easier to manipulate. Furthermore, their use provides the capacity for simple implementation of 10 or more nuclear import assays at the same time (see Note 13). The transport buffer and all reaction mixtures are prepared before the beginning of the assay. The reaction mixture containing fluorescently labeled cargo should be kept in the dark.
3.6.1 Import Reaction Mixtures
The simplest nuclear import assay involves the incubation of permeabilized cells with a reaction mix containing cargo (e.g., FITC-BSA-NLS), cytosol, and “energy,” i.e., an ATP-regenerating system and GTP (+Cytosol +E). Several negative controls should be performed to demonstrate that accumulation of cargo in the nucleus is physiologically relevant, i.e., that it depends on energy, exogenous nuclear transport factors (which are provided by cytosol), and passage through the NPC (which is inhibited by WGA).These negative controls are: cargo alone in TB (−Cytosol −E); cargo with an ATP-regenerating system and GTP (−Cytosol +E); and cytosol with an ATP-regenerating system, GTP and WGA (+Cytosol +E +WGA) (see Fig. 11.1). The concentration of cytosol can be adjusted (see Note 14). The standard transport mix for wells of a 10-well slide is a 50-μL solution containing 1.5 μM cargo (e.g., FITC-BSA-NLS), cytosol to a final protein concentration of 4 mg/mL , an ATP-regenerating system (1 mM ATP, 1 mg/mL CP, and 15 U/mL CPK), and 0.1 mM GTP. When included, WGA is used at 0.8 mg/mL.
To study the nuclear import of cargo with recombinant import factors, the reaction mix contains cargo together with the import receptor, an ATP regenerating system, GTP, Ran, and NTF2 (+Receptor +E). Recommended negative controls are cargo alone in TB (−Receptor −E); cargo with an ATP-regenerating system, GTP, Ran, and NTF2 (−Receptor +E); and cargo with receptor, an ATP-regenerating system, GTP, Ran, NTF2, and WGA (+Receptor +E +WGA) (see Fig. 11.2). The reaction mix is a 50-μL solution containing appropriate concentrations of cargo, receptor, and Ran (see below) together with an ATP-regenerating system (1 mM ATP, 1 mg/mL CP, and 15 U/mL CPK), 0.1 mM GTP, and 1 μM NTF2. When used, WGA is present at 0.8 mg/mL. The concentration of each cargo, receptor, and Ran should be titrated to determine concentrations that yield optimal import (see Note 15) (See Fig. 11.3a, b). To analyze the nuclear import of FITC-BSA-NLS, we use 2 μM FITC-BSA-NLS, 1.5 μM importin α, and 1 μM importin β; to analyze GST-IBB, we use 100 nM GST-IBB and 120 nM importin β; to analyze GST-M9, we use 100 nM GST-M9 and 500 nM transportin; and to analyze FITC-Histone H1, we use 0.4 μM FITC-Histone H1 and 0.3 μM importin 7, or 0.3 μM importin β, or a combination of 0.3 μM importin 7 and 0.3 μM importin β.
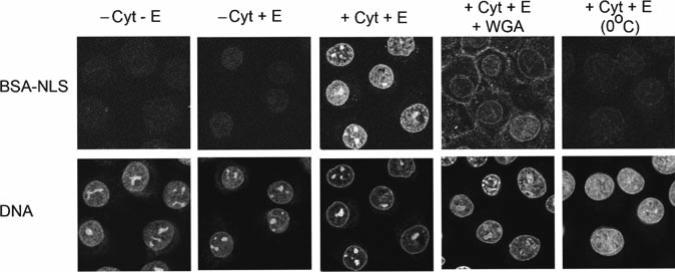
Fig. 11.1.
Nuclear import of FITC-labeled BSA-NLS in digitonin-permeabilized NRK cells supplemented with cytosol. The assays contained 2 μM FITC-labeled BSA-NLS and were incubated at 30°C for 30 min, except as noted, with the following conditions: in the absence of an ATP-regenerating system and GTP (“energy” [E]) and without cytosol (−Cyt −E); in presence of energy but without cytosol (−Cyt +E); with energy and 4 μg/μL of cytosol (+Cyt +E); with energy and cytosol except the reaction was incubated at 0°C (+Cyt +E [0°C]), or with energy, 4 μg/μL of cytosol, and WGA (+Cyt +E +WGA). The cells were examined with a Bio-Rad 1024 laser scanning confocal microscope using a ×63 objective, and the images were collected with Bio-Rad Lasersharp 2000 software. Fluorescent cargo is imaged in the top row, and DNA staining with Topro-3 is shown in the bottom row
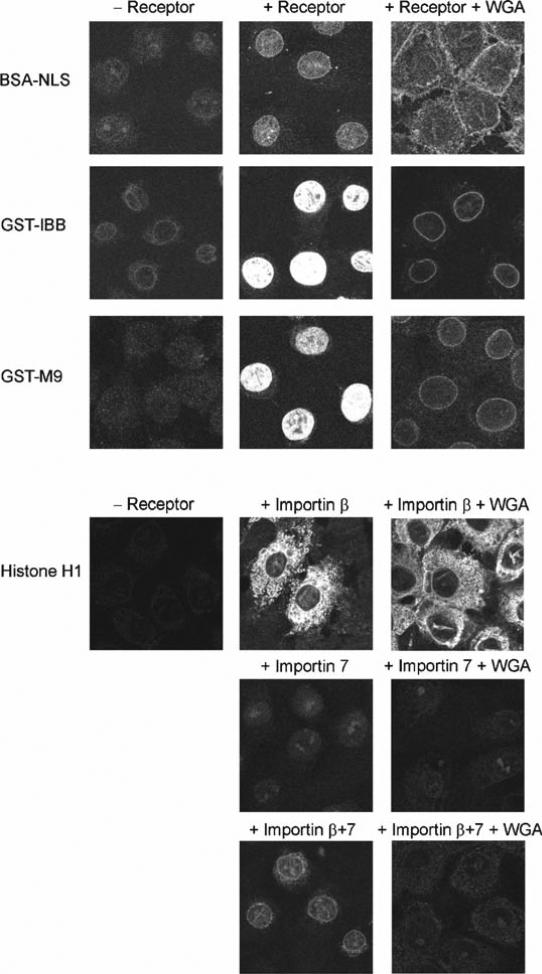
Fig. 11.2.
Nuclear import assays of various cargoes in digitonin-permeabilized NRK cells reconstituted with recombinant import factors. The localization of each cargo was visualized in the absence of receptor (−Receptor), in the presence of receptor (+Receptor) and in the presence of receptor and WGA (+Receptor +WGA). The assays involved the following concentrations of cargoes and receptors: for BSA-NLS: 2 μM FITC labeled-BSA-NLS, 1.5 μM importin α, and 1 μM importin β; for GST-IBB: 100 nM GST-IBB and 120 nM importin β; for GST-M9: 100 nM GSTM9 and 500 nM transportin; and for histone H1: 0.4 μM FITC-labeled histone H1 and either 0.3μM importin 7, 0.3 μM importin β, or both 0.3 μM importin 7 and 0.3 μM importin β. GST-IBB and GST-M9 were detected with an anti-GST antibody. Images were obtained as in Fig. 11.1
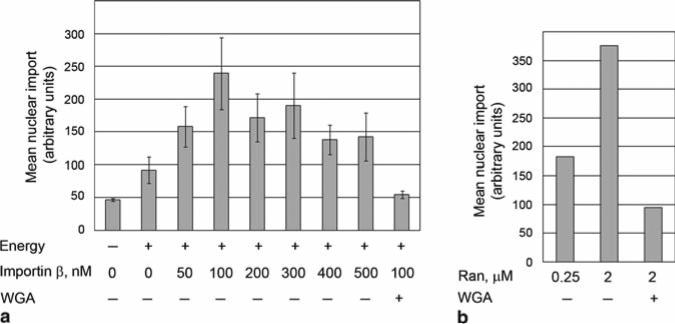
Fig. 11.3.
Titration of recombinant factors to optimize nuclear import in digitonin-permeabilized NRK cells. The nuclear import assays were performed with reactions reconstituted with recombinant transport factors and cargoes. GST-IBB and GST-M9 were detected with an anti-GST antibody. The cells were examined using a LEICA DM IRE2 microscope and a ×40 objective. The pictures were collected and the mean nuclear intensity was quantified using SimplePCI software (Compix, Sewickley, PA, USA). Error bar represents the standard deviation of the mean nuclear fluorescence found in three different fields, each containing 30–50 cells. a Titration of importin β for the nuclear import of GST-IBB. The nuclear import assay was performed in presence of 1 μM NTF2, 2 μM Ran, an ATP-regenerating system and GTP (Energy), WGA, 100 nM GST-IBB, and different concentrations of importin β as indicated. b Titration of Ran for the nuclear import of GST-M9 by transportin. The reactions contained 1 μM NTF2, 0.25 or 2 μM Ran, an ATP-regenerating system, and GTP, WGA (in some cases), 100 nM GST-M9, and 500 nM transportin
3.6.2 Nuclear Import Assay in Permeabilized NRK Cells
Plate NRK cells on 10-well slides the day before the experiment.
To begin the assay, move the slides to a prechilled metal block on ice.
Add some cold TB to dilute the NRK medium and prevent the cells from drying out.
Draw boundaries around each well with a hydrophobic pen to keep the assay solution in each well separate.
Wash three times with cold TB to remove NRK medium and anything that remains from the hydrophobic pen. A wash step involves applying a ~100μL drop of TB to each well using a plastic transfer pipet, and removing the liquid by vacuum aspiration (see Note 16).
Permeabilize the cells with 50 μL of digitonin solution (0.005% or 50 μg/mL)/ well for 5 min at room temperature (see Note 17). The 10% w/v digitonin stock is diluted to 0.005% in TB. The slide is removed from the prechilled block to the bench for 5 min and put back on the block for the washes.
Wash three times with ice-cold TB to remove the digitonin.
Incubate the cells with TB alone or with solutions containing WGA for 15 min at 30°C on a prewarmed metal block in a humidified chamber or on ice.
Wash five times with TB to remove the cytosol (see Note 18).
Incubate the cells with 50 μL of import reaction mix for 30 min at 30°C on a prewarmed metal block in a humidified chamber, or for 30 min on ice (see Note 19).
Wash three times with TB.
Fix the cells for 10 min at room temperature with 3.7% formaldehyde in PBS.
Wash three times with PBS.
Proceed directly to immunostaining if this is needed for cargo detection. Alternatively, slides may be stored overnight at 4°C in PBS with 0.05% w/v NaN3 before immunostaining.
3.6.3 Immunostaining to Detect GST-Cargoes
All steps are performed at room temperature.
Wash the wells three times with PBS.
Permeabilize for 10 min with 50 μL of PBS solution containing 0.2% v/v Triton X-100.
Wash three times with PBS.
Pre-block wells with 50 μL of PBS solution containing 0.3% w/v gelatin for 5 min.
Aspirate the gelatin/PBS and incubate for 1 h with 25 μL of purified goat anti-GST antibody diluted 1:100 in PBS with 0.3% gelatin.
Wash three times with PBS to remove unbound antibody.
Wash once with PBS containing 0.3% gelatin.
Aspirate and incubate in the dark for 1 h with 25 μL of FITC-labeled mouse anti-goat antibody diluted 1:100 in PBS with 0.3% gelatin.
Wash three times with PBS to remove unbound antibody.
Stain the DNA with DAPI diluted 1:5,000 in PBS from a stock solution of 1 mg/mL, or with Topro-3 diluted 1:500 in PBS, for 15 min in the dark (see Note 20).
Wash three times with PBS to remove the unbound dye.
Apply SlowFade Antifade or an equivalent antifade mounting reagent to the slide.
Put a cover glass (22×60 mm) over the entire slide and aspirate away excess mounting reagent from the edges of the cover glass.
Seal with nail varnish.
Acknowledgments
The writing of this chapter was supported by the National Institutes of Health (NIH) grant AI55729 to LG. AC was supported by a fellowship from the French Foundation for Medical Research (FRM), SPE20041102385. We are grateful to Geza Ambrus-Aikelin for comments on the manuscript. We thank the postdocs of the Gerace lab for their protocols.
Footnotes
Importin α precipitates if its concentration is too high; make sure its concentration is ≤0.5 mg/mL before dialysis.
In the purification of importin α, the protein concentration should be determined by the Bradford assay since imidazole absorbs at 280 nm.
For the importin β lysis buffer, it is important to use TCEP instead of DTT or β-mercaptoethanol, since these latter reducing agents are not compatible with the binding of the intein tag. Moreover, lysozyme cannot be used in the initial lysis of the bacteria since it inactivates chitin beads.
The addition of a small amount of CHAPS to the dialysis buffer for importin β helps to increase protein yield and does not interfere in the nuclear import assay (the final concentration of CHAPS in the assay will be ~1μM).
His-tagged importin 7 does not bind tightly to the Talon beads, and is eluted by the addition of 1 mM imidazole.
Most of the recombinant Ran is not soluble, but the level of expression is so high that even with this low solubility, ~30% of Ran is in the soluble pool.
Sometimes β lactamase from the bacteria, which is about 30–32 kDa, is strongly induced during the induction of Ran. Expanding cultures using colonies from freshly transformed plates without letting the bacteria reach the stationary phase helps to reduce the levels of β lactamase.
The final column is necessary if β lactamase is present at significant levels in the Ran peak from the S200 column.
To exchange the buffer to TB after the cation exchange column we recommend precipitating Ran with ammonium sulfate added to 65% saturation, since Ran precipitates during dialysis to TB if the pH before dialysis is below 6.8.
Though only 10–20% of NTF2 remains soluble under these preparative conditions, our attempts to recover active NTF2 from the insoluble fraction have been unsuccessful.
For the NTF2 lysis buffer, lysozyme should be avoided because its size (~20 kDa) is too close to that of the NTF2 dimer (28 kDa) and it could contaminate the NTF2 from the S200 column.
The optimal seeding density for the nuclear import assay can vary between different cell lines. The most important point is that the cells should not be confluent. The efficiency and specificity of plasma membrane permeabilization with digitonin is essential for the success of import assays, and have to be checked for each cell line and for each seeding. We recommend testing a range of digitonin concentrations from 0.002 to 0.008%. The integrity of the nuclear envelope can be tested using antibodies against histones or lamins, as well as with large FITC–dextrans (150 kDa). Antibodies and large dextrans are excluded from intact nuclei, while small FITC–dextrans (e.g., 9 kDa) have access (5, 19).
We suggest starting with one or two slides to become familiar with the procedure. We recommend analyzing the nuclear import of FITC-BSA-NLS in the absence or presence of cytosol to check the procedure, before starting experiments with different cargoes or recombinant receptors.
The concentration of cytosol that gives optimal nuclear import can vary with different cargoes, with different concentrations of the same cargo, and with different batches of cargo. An excess of cytosol can have an inhibitory effect on transport. A high concentration of WGA is typically used, but its concentration can be adjusted according to the specific cells used in the assay. The concentrations of cytosol and WGA that we commonly use in the nuclear import assay are 4 mg/mL and 0.8 mg/mL, respectively.
Each cargo and recombinant receptor has to be titrated to obtain optimal levels of nuclear import (see Fig. 11.3). Cargo is titrated to determine the concentration that yields the optimal ratio between nuclear import and background (0°C control, no receptor, or no cytosol controls). The fluorescently labeled cargoes typically are used at a concentration of 0.5–2 μM to be readily detected, whereas unlabeled cargoes detected by immunofluorescence staining with anti-GST antibody can be analyzed from 100 nM. Nuclear import involving receptors is saturable. In addition, if the concentration of receptor is too high compared with the Ran concentration, nuclear import can be a limiting factor for nuclear import with recombinant receptors, so its concentration should also be optimized.
The washes are performed well by well and one well at a time. It is very important to keep a thin layer of liquid on top of the slides to prevent the cells from drying out. Therefore, the addition and aspiration of solutions on one well should be closely coordinated. If the cells become dry during the nuclear import assay, import is nonspecifically inhibited and strong cytoplasmic staining can be obtained.
The concentration of digitonin for cell permeabilization should be tested for each new supply of digitonin and for each new cell line examined. The appropriate concentration permeabilizes the plasma membrane but not the nuclear envelope, and yields cytosol-dependent nuclear import of FITCBSA-NLS.
This step is essential to remove the endogenous cytosolic factors. We recommend increasing the number of washes from three to five. Alternatively, the cells can be incubated in presence of TB with energy or in presence of TB and RanQ69L for 15 min at 30°C. The presence of residual receptors in the permeabilized cells can induce nuclear import of cargo in the presence of energy alone.
Nuclear import is already detectable after 2 min at 30°C, and low levels of transport can be observed even at 10°C. Optimal import conditions usually involve 30 min incubation at 30°C. The permeabilized cells tend to become detached from cover glasses after ~60 min, and ATP supplies can also be depleted by this time. Nuclear import involving receptors is temperature dependent and is totally inhibited at 0°C. To perform the negative control of nuclear import at 0°C, we keep the cells on ice during the preincubation and incubation periods.
DAPI and Topro-3 are commonly used to stain DNA for fluorescence microscopy. The choice between these two dyes is dictated by the filters available on your microscope. Marking the nuclear space by fluorescence staining is a useful alternative to other techniques for visualizing the nucleus (e.g., phase contrast microscopy).
References
- 1.Fahrenkrog B, Koser J, Aebi U. The nuclear pore complex: a jack of all trades? Trends Biochem. Sci. 2004;29:175–182. doi: 10.1016/j.tibs.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Mosammaparast N, Pemberton LF. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004;14:547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Macara IG. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pemberton PL, Paschal B. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- 5.Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell. Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan RC, Forbes DJ. In vitro study of nuclear assembly and nuclear import using Xenopus egg extracts. In: Liu JX, editor. Xenopus protocols. Humana Press; Totowa, NJ: 2005. pp. 289–300. [DOI] [PubMed] [Google Scholar]
- 7.Colbeau A, Nachbaur J, Vignais PM. Enzymatic characterization and lipid composition of rat liver subcellular membrane. Biochim. Biophys. Acta. 1971;249:462–492. doi: 10.1016/0005-2736(71)90123-4. [DOI] [PubMed] [Google Scholar]
- 8.Melchior F. Nuclear protein import in a permeabilized cell assay. In: Clegg RA, editor. Protein targeting protocols. Humana Press; Totowa, NJ: 1998. pp. 265–273. [DOI] [PubMed] [Google Scholar]
- 9.Kehlenbach RH, Gerace L. Analysis of nuclear protein import and export in vitro using fluorescent cargoes. In: Manser E, Leung T, editors. GTPase protocols. Humana Press; Totowa, NJ: 2002. pp. 231–245. [DOI] [PubMed] [Google Scholar]
- 10.Paraskeva E, Izaurralde E, Bischoff FR, Huber J, Kutay U, Hartmenn E, Luhrmann R, Gorlich D. CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol. 1999;145:255–264. doi: 10.1083/jcb.145.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalderon D, Roberts BL, Richardson WD, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 12.Dingwall C, Laskey RA. Nuclear targeting sequences-a consensus? Trends Biochem. Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 13.Lyman SK, Guan T, Bednenko J, Wodrich H, Gerace L. Influence of cargo size on Ran and energy requirements for nuclear protein import. J. Cell Biol. 2002;159:55–67. doi: 10.1083/jcb.200204163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weis K, Mattaj IW, Lamond AI. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 15.Jakel S, Albig W, Kutay U, Bischoff FR, Schwamborn K, Doenecke D, Gorlich D. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 1999;18:2411–2423. doi: 10.1093/emboj/18.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollard VW, Michael WM, Nakielny S, Siomi MC, Wang F, Dreyfuss G. A novel receptor-mediated nuclear import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 17.Melchior F, Paschal B, Evans J, Gerace L. Inhibition of nuclear protein import by nonhydrolyzable analogues of GTP and identification of the small GTPase Ran/TC4 as an essential transport factor. J. Cell Biol. 1993;123:1649–1659. doi: 10.1083/jcb.123.6.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paschal B, Gerace L. Identification of NTF2, a cytosolic factor for nuclear import that interacts with nuclear pore complex protein p62. J. Cell Biol. 1995;129:925–937. doi: 10.1083/jcb.129.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paine PL, Moore LC, Horowitz SB. Nuclear envelope permeability. Nature. 1975;254:109–114. doi: 10.1038/254109a0. [DOI] [PubMed] [Google Scholar]