Abstract
Optimization of the surface charges is a promising strategy for increasing thermostability of proteins. Electrostatic contribution of ionizable groups to the protein stability can be estimated from the differences between the pKa values in the folded and unfolded states of a protein. Using this pKa-shift approach, we experimentally measured the electrostatic contribution of all aspartate and glutamate residues to the stability of a thermophilic ribosomal protein L30e from Thermococcus celer. The pKa values in the unfolded state were found to be similar to model compound pKas. The pKa values in both the folded and unfolded states obtained at 298 and 333 K were similar, suggesting that electrostatic contribution of ionizable groups to the protein stability were insensitive to temperature changes. The experimental pKa values for the L30e protein in the folded state were used as a benchmark to test the robustness of pKa prediction by various computational methods such as H++, MCCE, MEAD, pKD, PropKa, and UHBD. Although the predicted pKa values were affected by crystal contacts that may alter the side-chain conformation of surface charged residues, most computational methods performed well, with correlation coefficients between experimental and calculated pKa values ranging from 0.49 to 0.91 (p<0.01). The changes in protein stability derived from the experimental pKa-shift approach correlate well (r = 0.81) with those obtained from stability measurements of charge-to-alanine substituted variants of the L30e protein. Our results demonstrate that the knowledge of the pKa values in the folded state provides sufficient rationale for the redesign of protein surface charges leading to improved protein stability.
Introduction
Improving protein stability is not only a matter of academic curiosity but also has potential biotechnological applications in the engineering of enzymes that are stable and active at elevated temperatures. Protein stability can be rationally improved by optimizing various types of interactions [1]–[9]. One strategy is to optimize charge-charge interactions on the protein surface [10]–[13]. Using this approach Makhatadze and co-workers have successfully increased the thermostability of several proteins [9], [14], [15] including two human enzymes, acylphosphatase and CDC42, without altering their biological activities [14].
The electrostatic contribution of an ionizable group to the Gibbs free energy of unfolding can be estimated from the differences between the pKa values in the folded and unfolded states of a protein [16]–[18]. Direct experimental measurements of the pKa value is afforded by the NMR spectroscopy, although other experimental methods such as potentiometric titration and site-directed mutagenesis (see e.g. [19]–[21]) as well as computational approaches (see e.g. [22]–[39]) can also provide estimates of the pKa values for residues of interest.
We previously demonstrated the stabilizing role of electrostatic interactions in a thermophilic ribosomal protein L30e from Thermococcus celer [40]–[42], and measured the changes in the Gibbs free energy of unfolding (ΔΔGu mut) for 26 charge-to-alanine substituted variants of this protein. We also calculated the energetic contribution of charge-charge interactions using different computational models, and found that these models qualitatively predict the changes in the experimental values of ΔΔGu mut [43]. In this paper, we experimentally determined the values of pKa fold and pKa unfold for aspartic and glutamic acids in the T. celer L30e protein by NMR spectroscopy at two different temperatures (298 K and 333 K). Our results show that the pKa values and, hence, the changes in the electrostatic Gibbs free energy, ΔΔGu charge, are insensitive to temperature changes. Moreover, we showed that the experimental ΔΔGu charge values derived from the pKa-shift analysis correlate strongly (r = 0.81) with ΔΔGu mut values derived from the experimental analysis of stability of the wild type and charge-substituted variants of the L30e protein. This suggests that the knowledge of pKa values can be used as a good predictor for the effects of substitutions in ionizable residues on protein stability. Finally, our experimental pKa values were used as a benchmark to test the robustness of various computational methods of pKa calculation.
Results and Discussion
The 5R→K variant was created as the pseudo-wild-type for NMR experiments
Determination of the pKa values by NMR spectroscopy requires relatively high protein concentrations. In particular, it requires the protein samples to be soluble in a low ionic strength buffer. However, wild-type T. celer L30e crystallized readily in a low ionic strength buffer (e.g. 10 mM sodium citrate/phosphate buffer at pH 6.5) when the protein concentration was above 0.01 mM (Figure S1). The crystals formed under these conditions were used for X-ray diffraction and the structure was solved (PDB: 3N4Y) (Table S1). Structural analysis revealed that Arg-8, Arg-21, Arg-42, Arg-54, and Arg-76 are involved in crystal contacts. To reduce the crystallizability of the L30e protein, we created a quintuple variant, 5R→K, by substituting these five arginine residues with lysine. We chose these lysine substitutions because they conserve the surface charges of the L30e protein and because lysines are less likely to form crystal contacts than arginines [44], [45].
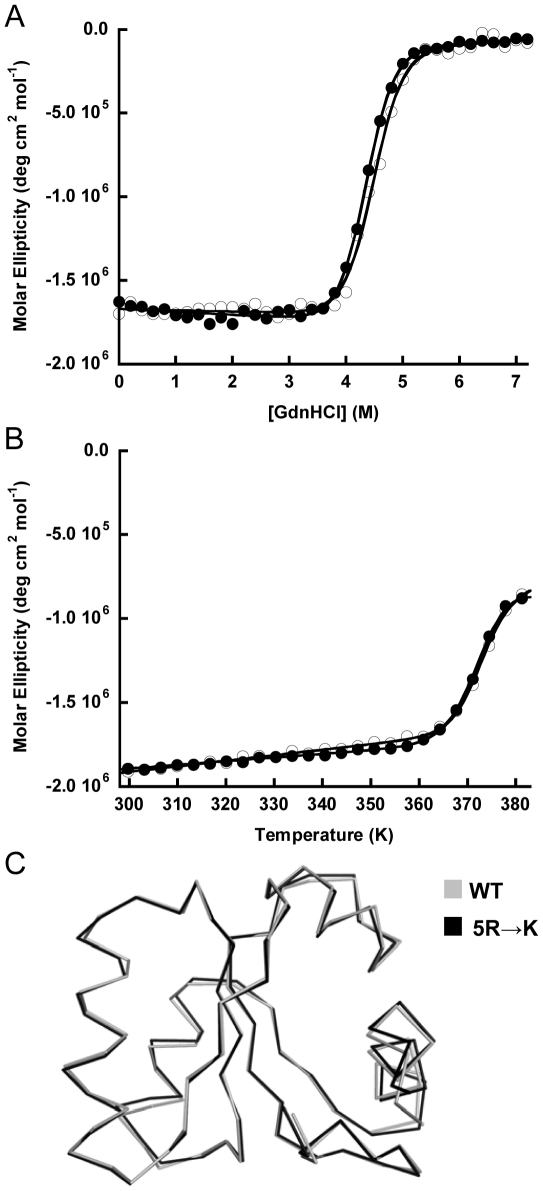
Guanidine-induced and thermal denaturation experiments showed that the 5R→K variant has essentially the same Gibbs free energy of unfolding (ΔGu) and melting temperature (Tm) as the wild-type protein (Figure 1A and 1B). Moreover, the structure of the 5R→K variant (PDB: 3N4Z) (Table S1) is superimposable with the wild-type structure (Calpha r.m.s.d. = 0.47 Å) (Figure 1C). Most importantly, the 5R→K variant was very soluble (>2.6 mM) under the NMR conditions (10 mM sodium citrate/phosphate buffer), and thus we used the 5R→K variant as a pseudo-wild-type, i.e. L30e*, in subsequent NMR experiments.
Figure 1. 5R→K variant (L30e*) and wild-type T. celer L30e have similar stabilities and structures.
Protein stabilities and melting temperatures of 5R→K (filled circle) and wild-type T. celer L30e (open circle) were determined in 10 mM citrate/phosphate buffer at pH 6.5. (A) The ΔGu at 298 K of 5R→K and wild-type T. celer L30e were determined by guanidine-induced denaturations to be 46.4±0.3 kJ mol−1 and 47.5±0.3 kJ mol−1 respectively. (B) Tm of 5R→K and wild-type T. celer L30e were determined by thermal denaturations to be 373.2±0.1 K and 372.8±0.1 K respectively. (C) The structure of the 5R→K variant (L30e*) (black) was determined and superimposable upon that of the wild-type T. celer L30e (grey).
Determination of the pKa fold
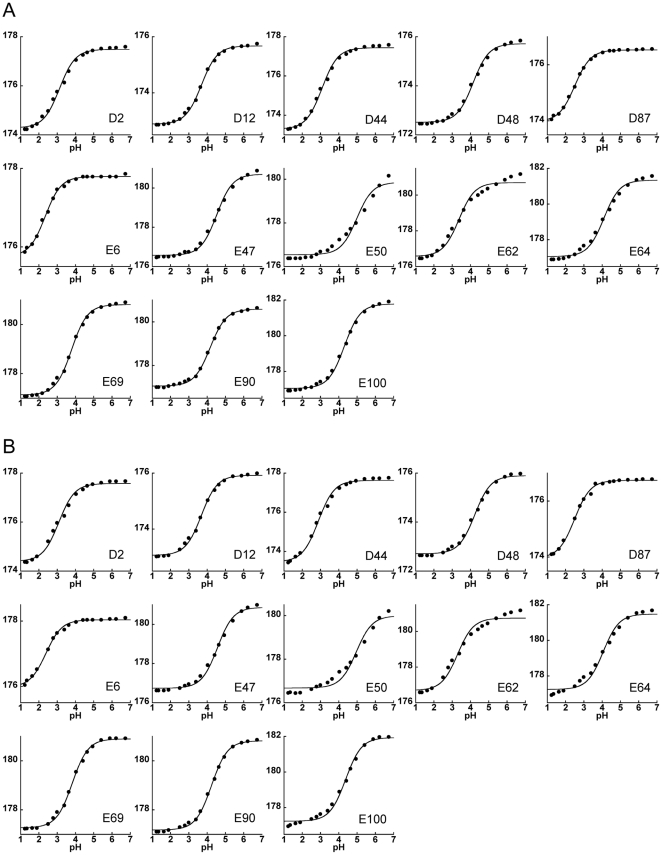
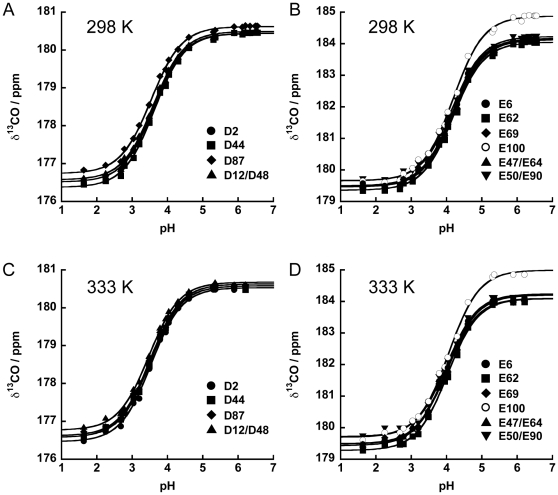
To determine the pKa values of the Asp and Glu residues in the folded protein (pKa fold), the chemical shifts of the side-chain carboxyl groups (δ13CO) in the pH range of 1.2–6.7 were followed by a modified two-dimensional H(CA)CO experiment at two temperatures, 298 K and 333 K (Figure S2A and S2B). The changes in the chemical shifts with pH for all residues except E50 and E62 fit well to a modified Henderson-Hasselbalch equation with Hill coefficient close to 1 (Figure 2). The titration curves for E50 and E62 clearly display biphasic transitions. Interestingly, the position of the major transition of E50 approximately coincides with that of the minor transition of E62, and vice versa (). E50 and E62 are in close proximity in the crystal structure (Figure S3C), and thus these observations suggest that the ionization of E50 and E62 may be coupled [46]. To determine the pKa values of E50 and E62, we used the method of global fitting of titration events (GloFTE) developed by Nielsen and co-workers [38], [47], which allows an estimate of the titration profiles for individual titratable residues (Figure 3D). The pKa fold values of all Asp and Glu determined at 298 K and 333 K are summarized in Table 1. Comparison suggests that that the values of pKa fold obtained at 298 K and 333 K were similar, with >0.2 unit differences observed for two residues, D44 and E64, while the rest of the pKa values remain essentially unchanged (Table 1).
Figure 2. pKa values of all Asp and Glu in L30* were determined.
The side-chain CO chemical shifts of Asp and Glu in native L30e* at pH 1.2–6.7 at (A) 298 K and (B) 333 K were fitted to a standard Henderson-Hasselbalch equation for determination of pKa values. The standard Henderson-Hasselbalch equation described well with all titration curves except those of E50 and E62 at both temperatures.
Figure 3. pKa values of Asp and Glu in denatured L30e* were determined.
At 298 K, the side-chain CO chemical shifts of (A) Asp and (B) Glu obtained in denatured L30e* were fitted by a standard Henderson-Hasselbalch equation for determination of their pKa values. At 333 K, the pKa values of (C) Asp and (D) Glu were also obtained by the same method.
Table 1. pKa values of all Asp and Glu in native L30e* at 298 K and 333 K.
| Residue | pKa(298 K) | pKa(333 K) |
| D2 | 3.13±0.04 | 3.09±0.05 |
| D12 | 3.69±0.02 | 3.69±0.03 |
| D44 | 3.08±0.04 | 2.88±0.04 |
| D48 | 4.14±0.04 | 4.24±0.04 |
| D87 | 2.50±0.02 | 2.49±0.03 |
| E6 | 2.34±0.03 | 2.38±0.04 |
| E47 | 4.53±0.04 | 4.56±0.04 |
| E50 | 5.10±0.07a | 5.00±0.06a |
| E62 | 3.20±0.06a | 3.30±0.06a |
| E64 | 4.10±0.06 | 4.06±0.07 |
| E69 | 3.79±0.03 | 3.82±0.03 |
| E90 | 4.17±0.02 | 4.24±0.02 |
| E100 | 4.31±0.03 | 4.38±0.05 |
pKa values obtained by GloFTE.
Determination of the pKa unfold
To determine the pKa values of the Asp and Glu residues in the unfolded protein (pKa unfold), the chemical shift of the side-chain carboxyl groups in the pH range of 1.2–6.7 in the presence of 5.4 M guanidine-HCl were followed by the modified two-dimensional H(CA)CO experiment (Figure S4A and S4B). The measurements were performed at two temperatures, 298 K and 333 K. The chemical shifts for D12/D48, E47/E64, and E50/E90 were degenerate. All titration curves fit well to a modified Henderson-Hasselbalch equation with Hill coefficient close to 1 (Figure 3). The summary of the obtained values of pKa unfold is given in Table 2. The pKa unfold values were very similar (within ±0.1 units) for each residue type: at 298 K the pKa unfold values for Asp residues were between 3.54 and 3.62, and between 4.16 and 4.19 for Glu residues. Increasing the temperature from 298 K to 333 K resulted in a small but uniform decrease in the pKa unfold values: 3.47–3.53 for Asp and 4.00–4.03 for Glu (Table 2).
Table 2. pKa values of Asp and Glu in unfolded L30e* at 298 K and 333 K.
| Residue | pKa(298 K) | pKa(333 K) |
| D2 | 3.62±0.01 | 3.53±0.02 |
| D44 | 3.62±0.01 | 3.52±0.02 |
| D87 | 3.54±0.02 | 3.47±0.02 |
| D12/D48 | 3.56±0.02 | 3.46±0.02 |
| E6 | 4.17±0.02 | 4.00±0.02 |
| E62 | 4.19±0.02 | 4.03±0.02 |
| E69 | 4.19±0.02 | 4.01±0.03 |
| E100 | 4.19±0.02 | 4.06±0.02 |
| E47/E64 | 4.17±0.02 | 4.00±0.02 |
| E50/E90 | 4.16±0.02 | 4.02±0.03 |
We also determined the pKa values for model pentapeptides (pKa peptide). Model pentapeptides Ac-GGD/EGG-NH2 were titrated with NaOH at 298 K and 333 K in the absence and presence of 5.4 M guanidine hydrochloride. The titration curves are well described by the Henderson-Hasselbalch equation (Figure S5), and the values of pKa peptide obtained are summarized in Table 3. The values of pKa peptide are similar to the values of pKa unfold, suggesting that the guanidine-induced denatured state of L30e* is very close to the random coil. Importantly, the addition of guanidine hydrochloride has no significant effect on the value of pKa peptide (Table 3).
Table 3. pKa values of Asp and Glu in terminal protected peptides at 298 K and 333 K.
| Peptide | [GdnHCl] (M) | 298 K | 333 K |
| Ac-GGDGG-NH2 | 0.0 | 3.69±0.03 | 3.61±0.02 |
| 5.4 | 3.70±0.01 | 3.64±0.02 | |
| Ac-GGEGG-NH2 | 0.0 | 4.17±0.03 | 4.10±0.01 |
| 5.4 | 4.18±0.04 | 4.10±0.02 |
ΔpKa and ΔΔGu charge are insensitive to temperature change
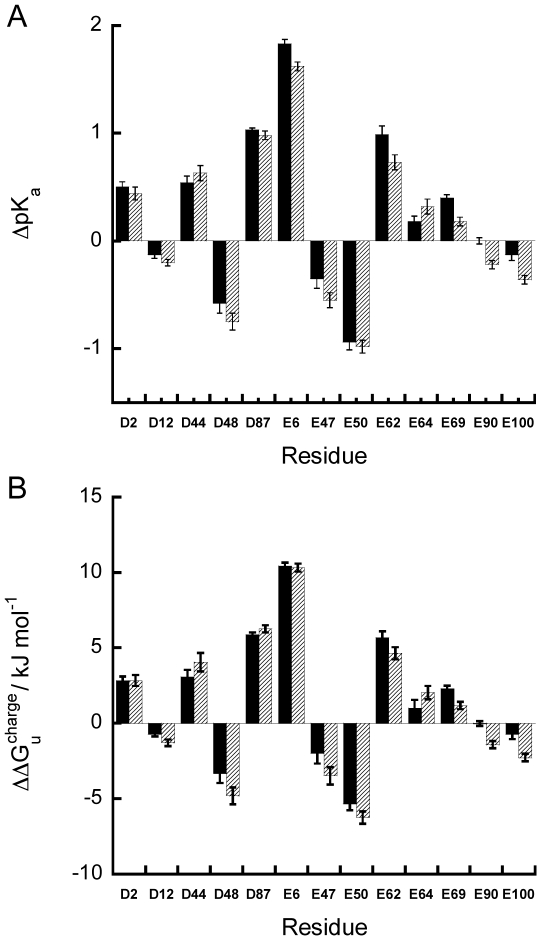
The difference in the pKa values between the folded and unfolded states (ΔpKa = pKa unfold−pKa fold) was used to calculate the contribution of a charged residue to the stability of the T. celer L30e* protein as ΔΔGu charge = 2.303RTΔpKa (Figure 4 and Table 4). The values of ΔΔGu charge range from 10.4 kJ/mol (stabilizing) to −6.3 kJ/mol (destabilizing). In all cases, the values of ΔΔGu charge at 298 K and 333 K were similar, suggesting that the electrostatic contribution of the charged residues to the protein stability is insensitive to temperature changes. Consistent with this observation, we have shown previously by double mutant cycle that the stabilization due to the salt-bridges E6/R92 and E62/K46 were similar at temperatures ranging from 298 to 348 K [42]. Taken together, our results suggest that favorable charge-charge interactions enhance the thermostability of proteins by up-shifting the protein stability curve.
Figure 4. ΔpKa and ΔΔGu charge values of Asp and Glu in L30e* are similar at 298 K and 333 K.
(A) ΔpKa and (B) ΔΔGu charge of all Asp and Glu in L30e* at 298 K (black) and 333 K (slash) were calculated. Noted the ΔpKa and ΔΔGu charge at both temperatures are similar.
Table 4. The ΔΔGu ele values of Asp and Glu of L30e* at 298 K and 333 K.
| Residue | ΔΔGu ele (298 K) | ΔΔGu ele (333 K) |
| D2 | 2.84±0.27 | 2.84±0.37 |
| D12 | −0.72±0.18 | −1.44±0.23 |
| D44 | 3.05±0.24 | 4.06±0.28 |
| D48 | −3.29±0.26 | −4.97±0.29 |
| D87 | 5.88±0.14 | 6.26±0.24 |
| E6 | 10.43±0.23 | 10.31±0.26 |
| E47 | −2.05±0.26 | −3.57±0.29 |
| E50 | −5.36±0.40 | −6.25±0.43 |
| E62 | 5.66±0.36 | 4.64±0.41 |
| E64 | 0.40±0.36 | −0.38±0.45 |
| E69 | 2.29±0.19 | 1.17±0.25 |
| E90 | −0.02±0.15 | −1.42±0.23 |
| E100 | −0.68±0.21 | −2.04±0.34 |
Experimental ΔΔGu charge is highly correlated with the mutagenesis data
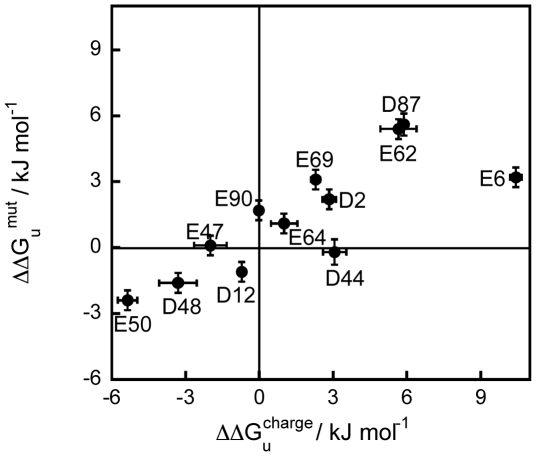
The ΔΔGu charge calculated from the changes in pKa values between the native and unfolded states at 298 K were compared with the ΔΔGu mut obtained from stability measurements of charge-to-alanine substituted variants of the T celer L30e protein, ΔΔGu mut [43] (Figure 5). It was found that the changes in ΔΔGu charge and ΔΔGu mut are highly correlated (r = 0.81, p-value = 0.0015). This observation suggests that ΔΔGu charge can be used to predict the change in free energy of unfolding upon site-directed amino acid substitutions (ΔΔGu mut). Furthermore, since the pKa values in the unfolded state, pKa unfold, for each residue type (Asp and Glu) were virtually the same, the high correlation between ΔΔGu charge and ΔΔGu mut suggests that the knowledge of the pKa values in the native state (pKa fold) is sufficiently rational for amino acid substitutions that will modulate protein stability.
Figure 5. ΔΔGu charge of Asp and Glu correlate with ΔΔGu mut.
ΔΔGu charge values obtained at 298 K from the pKa approach have a very good correlation (r = 0.81) with the ΔΔGu mut values obtained at 298 K from charge-to-alanine mutagenesis.
Comparing experimental and calculated pKa values
The experimental pKa fold values were compared to those calculated by various computational models (H++, MCCE, MEAD, pKD, PropKa, UHBD) (Figure S6). These computational models were selected based on their general availability, i.e. web-based interface or free to download. They are based on different computational algorithms and utilize different models for computing pKas. Separate calculations were done using the two structural models (chain A and B) as given in the PDB entry 3N4Z. The correlation coefficients between the experimental and calculated pKa values as well as overall RMSEs and p-values are summarized in Table 5. Overall, different computational models predict pKas of the acidic residues in the T celer L30e* protein rather well. The p-value analysis indicates the significance of correlations with correlation coefficients ranging from 0.6 to 0.9. This suggests that for the surface residues, most current computational methods have a very good predictive power. Moreover, the correlation analysis also highlights several important aspects.
Table 5. Correlation coefficients between experimental and calculated pKa fold values.
| A | B | Aens | Bens | ||
| H++ | R | 0.91 | 0.50 | 0.80 | 0.76 |
| RMSE | 0.5 | 1.1 | 0.6 | 0.8 | |
| p-value | 0.00004 | 0.1 | 0.002 | 0.004 | |
| MCCE | R | 0.79 | 0.49 | 0.55 | 0.79 |
| RMSE | 0.8 | 1.0 | 1.0 | 0.7 | |
| p-value | 0.002 | 0.1 | 0.06 | 0.002 | |
| MEAD | R | 0.89 | 0.81 | 0.89 | 0.82 |
| RMSE | 0.4 | 0.5 | 0.5 | 0.6 | |
| p-value | 0.0001 | 0.002 | 0.0001 | 0.001 | |
| pKD | R | 0.86 | 0.65 | 0.81 | 0.74 |
| RMSE | 0.6 | 0.8 | 0.6 | 0.7 | |
| p-value | 0.0003 | 0.02 | 0.0009 | 0.01 | |
| ProPKa 2.0 | R | 0.85 | 0.58 | 0.70 | 0.72 |
| RMSE | 0.5 | 0.7 | 0.7 | 0.6 | |
| p-value | 0.0004 | 0.05 | 0.01 | 0.01 | |
| ProPKa 3.0 | R | 0.88 | 0.61 | 0.65 | 0.73 |
| RMSE | 0.6 | 0.8 | 0.8 | 0.7 | |
| p-value | 0.0002 | 0.03 | 0.02 | 0.01 | |
| UHBD | R | 0.84 | 0.59 | 0.81 | 0.77 |
| RMSE | 0.7 | 0.8 | 0.6 | 0.7 | |
| p-value | 0.001 | 0.04 | 0.001 | 0.004 |
The experimental pKa fold values (298 K) were correlated with those derived from computational models. Significant correlations (p<0.05) are indicated in bold.
First, the reliability of prediction depends on the structural model used. It is apparent that in the majority of cases, correlation coefficients for the prediction based on structural model A is much better than the prediction based on the structural model B. Structure A also produces smaller RMSE between calculated and experimental values and is statistically more robust as judged by the lower p-value (Table 5). The backbone and all-atom rmsd between the structural models A and B is 0.66 and 1.24 Å, respectively, which suggests that the position of the side chains due to differences in crystal contact is leading to large differences in the computational predictions. To probe the effect of side chain flexibility on the outcome of pKa predictions, we generated an ensemble of structures starting with either of the chain A or B x-ray models (see Materials and Methods for details). Interestingly, the use of structural ensembles significantly improved correlation coefficients for the predictions based on the chain B. However, the use of structural ensembles generated based on the chain A did not improve the correlation between the experimental and calculated pKa values (Table 5). Without a prior knowledge of experimental pKa values, it is inconclusive to recommend whether it is a good practice to use structure ensembles or just simply use a single model in the crystal structure. Nevertheless, most prediction methods (e.g. MEAD, pKD, ProPKa, UHBD) gave significant correlation (p-value<0.05) regardless of the structural model used (Table 5).
Second, the standard errors of the prediction of pKa values using ensemble structures are relatively large in units of pH (∼0.5–0.8). However, if one thinks in the units of energy, this translates into error in ΔΔG determination of ∼2–4 kJ/mol. This highlights the difficulties in pKa predictions. One needs to be able to perform energy calculations with the accuracy better than ∼2–4 kJ/mol which is a rather difficult considering that 1 kT at 25°C is ∼2.5 kJ/mol, only twice smaller!.
Third, both physics based continuum electrostatics models and empirical models perform equally well. This is probably not surprising considering that all residues in T celer L30e* are located on the protein surface, and thus the contribution of solvation is largely reduced as compared to the fully buried residues. In the past, empirical models were sometimes preferred over continuum models because of the speed of calculations. However, developments in the computational algorithms and an increase in cpu power have essentially erased these differences in the speed and the use of web-based servers have made pKa calculations broadly accessible.
Fourth, there are two residues that are located in the “high charge clusters”. These are residues E50 and E62. Experimental pKa measurements show a biphasic titration profile indicating linkage in the titration properties of these residues. These effects are reasonably well predicted by the some of the computational algorithms. For example, H++ predicts strong interactions between E50 and E62, consistent with the experimental observations that there is a linkage in the titration properties of these two residues. In addition, H++ predicts the interactions of E50 with K46 and the interactions of E62 with K46/R39, which may also be responsible for abnormal titration. Using double mutant analysis we have already shown that E62/K46 pair has the energy of interactions of 3.6 kJ/mol [42]. This suggests future experiments that will measure the titration properties of the basic residues or the effect of mutations in these residues on the titration properties of E50 and E62.
Concluding remarks
The strategy of improving the thermostability of proteins by optimizing surface charges relies on a robust method of pKa prediction and the fact that mutagenesis results can be well predicted by the pKa-shift approach. In this study, we showed that ΔΔGu charge derived from the experimental pKa values correlates strongly with ΔΔGu mut due to charge-to-alanine substitutions. This observation suggests that the knowledge of the pKa values (obtained either by experimental or computational methods) is sufficiently rational for the optimization of protein surface charges. Our experimental pKa values were also served as a benchmark to test the robustness of various computational methods of pKa prediction. Most current computational methods gave good prediction of pKa values. The encouraging fact is that all of these methods are generally available, either via a web-interface or free to download, making rational redesign of the protein surface charges accessible to all protein engineers.
Materials and Methods
Cloning, expression and purification
The DNA coding for the 5R→K variant of L30e was synthesized by Mr. Gene GmbH (http://mrgene.com) and was sub-cloned in the expression vector pET3d (Novagen). All unlabeled and 13C/15N labeled protein samples were expressed and purified as described previously [41], [43].
Measurement of thermodynamics stability
Thermal-induced denaturation experiments were performed in 10 mM citrate/phosphate buffer at pH 6.5 using 20 µM protein concentration. The melting temperature (Tm) of the protein was determined as described [42]. Guanidine-induced denaturation experiments at 298 K were performed in the same buffer. The Gibbs free energy of unfolding (ΔGu) was determined as described [40].
Crystallization and structure determination
Crystals of wild-type T. celer L30e were grown by dialyzing 0.1 mM protein into 10 mM citrate/phosphate buffer pH 6.5 at 277 K. Crystals of the 5R→K variant were grown by the sitting-drop-vapor-diffusion method using 1.6 M Na/K phosphate buffer at pH 7.5 and 289 K. Data collection and structure determination were performed as described [42].
Determination of pKa
For the folded state of the 5R→K variant, sequential assignment of backbone resonances was obtained by Cα and Cβ connectivities generated by the HNCACB and CBCA(CO)NH experiments at pH 6.7 at 298 K and 333 K. The side-chain resonances were obtained from 15N-TOCSY-HSQC, HC(CCO)NH, (HC)C(CO)NH, and HCCH-TOCSY experiments. The side-chain carboxyl carbon of Asp/Glu (δexp) at pH 6.7 at 298 K and 333 K were assigned using a modified three dimensional HCACO experiment, which correlates side-chain COγ/δ with Hβ/γ and Cβ/γ resonances [48]. The assignments of the δexp at other pH were obtained by tracing the peak shift in the modified two dimensional H(CA)CO experiments collected in the pH range of 1.2–6.7.
For the pKa determination in the unfolded state of the 5R→K protein, the protein was denatured in 5.4 M guanidine-HCl for all NMR experiments. Sequential assignment of backbone resonances pH 6.4 at 298 K and 333 K was obtained by the connectivities of Cα and Cβ generated by HNCACB and of d NN(i,i+1) NOEs generated by the modified HSQC-NOESY-HSQC experiment [49]. The assignments of side-chain resonances and the δexp at pH 6.4 at 298 K and 333 K were obtained as described above. The δexp in the pH range of 6.4–1.6 were also obtained by tracing the gradual peak shift associated with the pH change. The pH of all protein samples were measured by glass pH-electrode (Beckman Coulter φ510) before NMR experiments. The pH of all samples was re-measured after performing each NMR experiments, and the variation of the pH was less than 0.05 units.
pKa fold and pKa unfold were determined by fitting the δexp into a modified Henderson-Hasselbalch equation: δexp = [δA+δB10n(pH–pKa)]/[1+10n(pH–pKa)], where δA and δB are chemical shifts for the protonated and deprotonated residue respectively. The Hill coefficient, n, was set to be a free parameter during data-fitting. With the exception of the titration profiles for E50 and E62 in the folded state, the Hill coefficient in all cases was close to 1. According to F-test, a simpler model with Hill coefficient set equal to 1 was used in the data fitting.
For E50 and E62, their pKa fold were determined by global fitting of titrational event (GloFTE) using pKaTool [38], [47]. The pH step was set to 0.1 and the Monte Carlo step number was set to 300. The titration curves were calculated based on the explicit evaluation of the Boltzmann sum. The pKa fold was determined by finding the pH where the titratable group is half-protonated [38].
pH titration of side-chain carboxyl groups in model peptides
Termini protected glycine-based pentapeptides Ac-GGD/EGG-NH2 were purchased from GL Biochem (Shanghai). 40.2 mg lyophilized peptide powder of Ac-GGDGG-NH2 and 41.6 mg lyophilized peptide powder Ac-GGEGG-NH2 were dissolved in 10 ml nano-pure water using 10 ml volumetric flask. Before pH titration, the solution was equilibrated at 298 K and 333 K for 15 minutes. The pH value of the solution was continuously measured by the calibrated pH meter using glass electrode (Beckman Coulter φ510). Commercially purchased 1.0 M NaOH (Sigma) was used for the titration. 10 µl of NaOH was added to the solution stepwise until a pH value greater than 8.0 was reached.
pKa peptide were determined by fitting the NaOH concentration in the solution into a modified Henderson-Hasselbalch equation: Cexp = 1000×10−pH−[10−pKa×Cpeptide/(10−pH+10−pKa)]−Coffset, where Cexp is the NaOH concentration in the solution in mM, Cpeptide is the peptide concentration, and Coffset is the OH− concentration in the solution before a given titration step.
Calculation of pKas
We used seven different software packages that predict the pKas of amino acid residues. In all cases the default parameter sets were used. The coordinate files were supplied in the PDB format using individual models A and B from the X-ray structure of L30e* protein (PDB:3N4Z). To probe the effects of side-chain conformational flexibility, these two structural models were used as a starting template for generating a structural ensemble within the MODELER environment [50]. Calculations were performed on each individual structure of the ensemble, and the results were averaged.
MEAD v2.2.9 [23] (http://hospital.stjude.org/mead_filerequest/request.html) solves linearized Poisson-Boltzmann to perform various electrostatic calculations including pKas of amino acid residues in proteins.
H++ [24] (http://biophysics.cs.vt.edu/) uses the MEAD engine [23] to solve the Poisson-Boltzmann equation, but in addition uses a “smeared charge” concept that places partial charges on several atoms in the ionizable side chain instead of one single atom.
pKD [25] (http://enzyme.ucd.ie/cgi-bin/pKD/server_start.cgi) uses the Poisson-Boltzmann solver DELPHI II [26] together with the optimization of a hydrogen bonding network as implemented in the WHATIF software package [27].
PROPKA_2.0 [29] and PROPKA_3.0 [28] (http://enzyme.ucd.ie/cgi-bin/pKD/server_start.cgi) estimate the shift in the pKa arising from hydrogen bonds, relative burial and coulombic interactions. These contributions are parametrized to fit experimentally measured ΔpKa deposited into pKa databases (e.g. http://www.sci.ccny.cuny.edu/~mcce/index.php and http://www.ddg-pharmfac.net/ppd/PPD/pKahomepage.htm).
UHBD [30] (http://mccammon.ucsd.edu/uhbd.html) uses the finite difference Poisson–Boltzmann (FDPB) method to calculate electrostatic interactions. Energies of interactions were combined with the hybrid method to calculate pKas [31].
MCCE Multi Conformer Continuum Electrostatics (version 2.5) software package uses Monte-Carlo sampling of different side-chain rotamers in conjunction with the FDPB calculations using the DELPHI software package and PARSE solvation [32].
Supporting Information
Wild-type T. celer L30e crystallized in low ionic strength buffer (10 mM citrate/phosphate buffer, pH 6.5.
(PDF)
Assignment of side-chain carboxyl carbon of Asp and Glu in native L30e*.
(PDF)
Titration of Glu-50 and Glu-62 was coupled. Two transitions were observed in the titration curves of Glu-50 and Glu-62 at (A) 298 K and (B) 333 K, of which the major transition of one residue corresponds to the minor transition of another. This observation suggests that the protonation of Glu-50 and Glu-62 are coupled, as these two residues are in close proximity to each other (C). The pKa values of Glu-50 and Glu-62 at (D) 298 K and (E) 333 K were determined by fitting their titration curves simultaneously by the method of global fitting of titration events (GloFTE).
(PDF)
Assignment of side-chain carboxyl carbon of Asp and Glu in unfolded L30e*.
(PDF)
Determination of pKapeptide. Titration curves of model peptides (A, C, E, G) Ac-GGDGG-NH2 and (B, D, F, H) Ac-GGEGG-NH2 in the presence of (A–D) 5.4 M and (E–H) 0 M guanidine HCl (A, B, E, F) 298 K and (C, D, G, H) 333 K. All of the titration data were fitted to the standard Henderson-Hasselbalch equation for determination of the pKa values of Asp and Glu in model peptides.
(PDF)
Correlation of experimental and predicted pKa values. pKa values were calculated using several different software packages indicated on each plot. Left panels for each method are based on chain A of the crystal structure of L30e* (PDB code: 3N4Z), right panels for each method are based on chain B. Blue circles represent the results of based on x-ray model (A or B). Red squares represent the results of ensemble calculations based on structures A or B. Red and blue lines are the corresponding linear correlations. Thin black line shows the perfect correlation. See Materials and Methods section for the details of the calculations.
(PDF)
Statistics for structure determination of wild-type T. celer L30e and 5R→K variant.
(PDF)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by the General Research Fund of Research Council of Hong Kong (project no. 479307). Research on electrostatic interactions in GIM lab is supported by a grant from the National Science Foundation (MCB-0110396). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Akasako A, Haruki M, Oobatake M, Kanaya S. Conformational stabilities of Escherichia coli RNase HI variants with a series of amino acid substitutions at a cavity within the hydrophobic core. J Biol Chem. 1997;272:18686–18693. doi: 10.1074/jbc.272.30.18686. [DOI] [PubMed] [Google Scholar]
- 2.Fu H, Grimsley GR, Razvi A, Scholtz JM, Pace CN. Increasing protein stability by improving beta-turns. Proteins. 2009;77:491–498. doi: 10.1002/prot.22509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grimsley GR, Shaw KL, Fee LR, Alston RW, Huyghues-Despointes BM, et al. Increasing protein stability by altering long-range coulombic interactions. Protein Sci. 1999;8:1843–1849. doi: 10.1110/ps.8.9.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson RW, Nicholson EM, Thapar R, Klevit RE, Scholtz JM. Increased helix and protein stability through the introduction of a new tertiary hydrogen bond. J Mol Biol. 1999;286:1609–1619. doi: 10.1006/jmbi.1999.2574. [DOI] [PubMed] [Google Scholar]
- 5.Street AG, Mayo SL. Computational protein design. Structure. 1999;7:R105–109. doi: 10.1016/s0969-2126(99)80062-8. [DOI] [PubMed] [Google Scholar]
- 6.Pokala N, Handel TM. Protein design–where we were, where we are, where we're going. J Struct Biol. 2001;134:269–281. doi: 10.1006/jsbi.2001.4349. [DOI] [PubMed] [Google Scholar]
- 7.Dantas G, Kuhlman B, Callender D, Wong M, Baker D. A large scale test of computational protein design: folding and stability of nine completely redesigned globular proteins. J Mol Biol. 2003;332:449–460. doi: 10.1016/s0022-2836(03)00888-x. [DOI] [PubMed] [Google Scholar]
- 8.Kajander T, Cortajarena AL, Regan L. Consensus design as a tool for engineering repeat proteins. Methods Mol Biol. 2006;340:151–170. doi: 10.1385/1-59745-116-9:151. [DOI] [PubMed] [Google Scholar]
- 9.Schweiker KL, Zarrine-Afsar A, Davidson AR, Makhatadze GI. Computational design of the Fyn SH3 domain with increased stability through optimization of surface charge charge interactions. Protein Sci. 2007;16:2694–2702. doi: 10.1110/ps.073091607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loladze VV, Ibarra-Molero B, Sanchez-Ruiz JM, Makhatadze GI. Engineering a thermostable protein via optimization of charge-charge interactions on the protein surface. Biochemistry. 1999;38:16419–16423. doi: 10.1021/bi992271w. [DOI] [PubMed] [Google Scholar]
- 11.Sanchez-Ruiz JM, Makhatadze GI. To charge or not to charge? Trends Biotechnol. 2001;19:132–135. doi: 10.1016/s0167-7799(00)01548-1. [DOI] [PubMed] [Google Scholar]
- 12.Schweiker KL, Makhatadze GI. A computational approach for the rational design of stable proteins and enzymes: optimization of surface charge-charge interactions. Methods Enzymol. 2009;454:175–211. doi: 10.1016/S0076-6879(08)03807-X. [DOI] [PubMed] [Google Scholar]
- 13.Schweiker KL, Makhatadze GI. Protein stabilization by the rational design of surface charge-charge interactions. Methods Mol Biol. 2009;490:261–283. doi: 10.1007/978-1-59745-367-7_11. [DOI] [PubMed] [Google Scholar]
- 14.Gribenko AV, Patel MM, Liu J, McCallum SA, Wang C, et al. Rational stabilization of enzymes by computational redesign of surface charge-charge interactions. Proc Natl Acad Sci U S A. 2009;106:2601–2606. doi: 10.1073/pnas.0808220106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strickler SS, Gribenko AV, Gribenko AV, Keiffer TR, Tomlinson J, et al. Protein stability and surface electrostatics: a charged relationship. Biochemistry. 2006;45:2761–2766. doi: 10.1021/bi0600143. [DOI] [PubMed] [Google Scholar]
- 16.Lumb KJ, Kim PS. Response: how much solar radiation do clouds absorb? Science. 1996;271:1137–1138. doi: 10.1126/science.271.5252.1137. [DOI] [PubMed] [Google Scholar]
- 17.Yang AS, Sharp KA, Honig B. Analysis of the heat capacity dependence of protein folding. J Mol Biol. 1992;227:889–900. doi: 10.1016/0022-2836(92)90229-d. [DOI] [PubMed] [Google Scholar]
- 18.Bosshard HR, Marti DN, Jelesarov I. Protein stabilization by salt bridges: concepts, experimental approaches and clarification of some misunderstandings. J Mol Recognit. 2004;17:1–16. doi: 10.1002/jmr.657. [DOI] [PubMed] [Google Scholar]
- 19.Dwyer JJ, Gittis AG, Karp DA, Lattman EE, Spencer DS, et al. High apparent dielectric constants in the interior of a protein reflect water penetration. Biophysical Journal. 2000;79:1610–1620. doi: 10.1016/S0006-3495(00)76411-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fitch CA, Karp DA, Lee KK, Stites WE, Lattman EE, et al. Experimental pK(a) values of buried residues: analysis with continuum methods and role of water penetration. Biophys J. 2002;82:3289–3304. doi: 10.1016/s0006-3495(02)75670-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitten ST, Garcia-Moreno B. pH dependence of stability of staphylococcal nuclease: Evidence of substantial electrostatic interactions in the denatured state. Biochemistry. 2000;39:14292–14304. doi: 10.1021/bi001015c. [DOI] [PubMed] [Google Scholar]
- 22.Jimenez-Cruz CA, Makhatadze GI, Garcia AE. Protonation/deprotonation effects on the stability of the Trp-cage miniprotein. Physical Chemistry Chemical Physics. 2011;13:17056–17063. doi: 10.1039/c1cp21193e. [DOI] [PubMed] [Google Scholar]
- 23.Bashford D, editor. An object-oriented programming suite for electrostatic effects in biological molecules. Berlin: Springer; 1997. pp. 233–240. [Google Scholar]
- 24.Gordon JC, Myers JB, Folta T, Shoja V, Heath LS, et al. H++: a server for estimating pK(a)s and adding missing hydrogens to macromolecules. Nucleic Acids Research. 2005;33:W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tynan-Connolly BM, Nielsen JE. pKD: re-designing protein pK(a) values. Nucleic Acids Research. 2006;34:W48–W51. doi: 10.1093/nar/gkl192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicholls A, Honig B. A Rapid Finite-Difference Algorithm, Utilizing Successive over-Relaxation to Solve the Poisson-Boltzmann Equation. Journal of Computational Chemistry. 1991;12:435–445. [Google Scholar]
- 27.Vriend G. What If - a Molecular Modeling and Drug Design Program. Journal of Molecular Graphics. 1990;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 28.Olsson MHM, Sondergaard CR, Rostkowski M, Jensen JH. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pK(a) Predictions. Journal of Chemical Theory and Computation. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 29.Bas DC, Rogers DM, Jensen JH. Very fast prediction and rationalization of pK(a) values for protein-ligand complexes. Proteins-Structure Function and Bioinformatics. 2008;73:765–783. doi: 10.1002/prot.22102. [DOI] [PubMed] [Google Scholar]
- 30.Madura JD, Davis ME, Gilson MK, Wade RC, Luty BA, et al. Biological Applications of Electrostatic Calculations and Brownian Dynamics Simulations. Reviews in Computational Chemistry V. 1994;5:229–267. [Google Scholar]
- 31.Gilson MK. Multiple-site titration and molecular modeling: two rapid methods for computing energies and forces for ionizable groups in proteins. Proteins. 1993;15:266–282. doi: 10.1002/prot.340150305. [DOI] [PubMed] [Google Scholar]
- 32.Gunner MR, Zhu X, Klein MC. MCCE analysis of the pK(a) s of introduced buried acids and bases in staphylococcal nuclease. Proteins. 2011;79:3306–3319. doi: 10.1002/prot.23124. [DOI] [PubMed] [Google Scholar]
- 33.Shan J, Mehler EL. Calculation of pK(a) in proteins with the microenvironment modulated-screened coulomb potential. Proteins. 2011;79:3346–3355. doi: 10.1002/prot.23098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Machuqueiro M, Baptista AM. Is the prediction of pK(a) values by constant-pH molecular dynamics being hindered by inherited problems? Proteins. 2011;79:3437–3447. doi: 10.1002/prot.23115. [DOI] [PubMed] [Google Scholar]
- 35.Khandogin J, Brooks CL., III Constant pH molecular dynamics with proton tautomerism. Biophys J. 2005;89:141–157. doi: 10.1529/biophysj.105.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unni S, Huang Y, Hanson RM, Tobias M, Krishnan S, et al. Web Servers and Services for Electrostatics Calculations with APBS and PDB2PQR. Journal of Computational Chemistry. 2011;32:1488–1491. doi: 10.1002/jcc.21720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warshel A, Sharma PK, Kato M, Parson WW. Modeling electrostatic effects in proteins. Biochimica Et Biophysica Acta-Proteins and Proteomics. 2006;1764:1647–1676. doi: 10.1016/j.bbapap.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen JE. Analysing the pH-dependent properties of proteins using pKa calculations. J Mol Graph Model. 2007;25:691–699. doi: 10.1016/j.jmgm.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 39.Nielsen JE. Analyzing Enzymatic Ph Activity Profiles and Protein Titration Curves Using Structure-Based Pk(a) Calculations and Titration Curve Fitting. Methods in Enzymology: Computer Methods. 2009;Vol 454, Pt A 454:233–258. doi: 10.1016/S0076-6879(08)03809-3. [DOI] [PubMed] [Google Scholar]
- 40.Lee CF, Allen MD, Bycroft M, Wong KB. Electrostatic interactions contribute to reduced heat capacity change of unfolding in a thermophilic ribosomal protein l30e. J Mol Biol. 2005;348:419–431. doi: 10.1016/j.jmb.2005.02.052. [DOI] [PubMed] [Google Scholar]
- 41.Wong KB, Lee CF, Chan SH, Leung TY, Chen YW, et al. Solution structure and thermal stability of ribosomal protein L30e from hyperthermophilic archaeon Thermococcus celer. Protein Sci. 2003;12:1483–1495. doi: 10.1110/ps.0302303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chan CH, Yu TH, Wong KB. Stabilizing salt-bridge enhances protein thermostability by reducing the heat capacity change of unfolding. PLoS One. 2011;6:e21624. doi: 10.1371/journal.pone.0021624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee CF, Makhatadze GI, Wong KB. Effects of charge-to-alanine substitutions on the stability of ribosomal protein L30e from Thermococcus celer. Biochemistry. 2005;44:16817–16825. doi: 10.1021/bi0519654. [DOI] [PubMed] [Google Scholar]
- 44.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 45.Czepas J, Devedjiev Y, Krowarsch D, Derewenda U, Otlewski J, et al. The impact of Lys→Arg surface mutations on the crystallization of the globular domain of RhoGDI. Acta Crystallogr D Biol Crystallogr. 2004;60:275–280. doi: 10.1107/S0907444903026271. [DOI] [PubMed] [Google Scholar]
- 46.Shrager RI, Cohen JS, Heller SR, Sachs DH, Schechter AN. Mathematical models for interacting groups in nuclear magnetic resonance titration curves. Biochemistry. 1972;11:541–547. doi: 10.1021/bi00754a010. [DOI] [PubMed] [Google Scholar]
- 47.Sondergaard CR, McIntosh LP, Pollastri G, Nielsen JE. Determination of electrostatic interaction energies and protonation state populations in enzyme active sites. J Mol Biol. 2008;376:269–287. doi: 10.1016/j.jmb.2007.09.070. [DOI] [PubMed] [Google Scholar]
- 48.Yamazaki T, Nicholson LK, Wingfield P, Stahl SJ, Kaufman JD, et al. 1994. NMR and X-ray Evidence That the HIV Protease Catalytic Aspartyl Groups Are Protonated in the Complex Formed by the Protease and a Non-Peptide Cyclic Urea-Based Inhibitor.
- 49.Wong KB, Freund SM, Fersht AR. Cold denaturation of barstar: 1H, 15N and 13C NMR assignment and characterisation of residual structure. J Mol Biol. 1996;259:805–818. doi: 10.1006/jmbi.1996.0359. [DOI] [PubMed] [Google Scholar]
- 50.Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Wild-type T. celer L30e crystallized in low ionic strength buffer (10 mM citrate/phosphate buffer, pH 6.5.
(PDF)
Assignment of side-chain carboxyl carbon of Asp and Glu in native L30e*.
(PDF)
Titration of Glu-50 and Glu-62 was coupled. Two transitions were observed in the titration curves of Glu-50 and Glu-62 at (A) 298 K and (B) 333 K, of which the major transition of one residue corresponds to the minor transition of another. This observation suggests that the protonation of Glu-50 and Glu-62 are coupled, as these two residues are in close proximity to each other (C). The pKa values of Glu-50 and Glu-62 at (D) 298 K and (E) 333 K were determined by fitting their titration curves simultaneously by the method of global fitting of titration events (GloFTE).
(PDF)
Assignment of side-chain carboxyl carbon of Asp and Glu in unfolded L30e*.
(PDF)
Determination of pKapeptide. Titration curves of model peptides (A, C, E, G) Ac-GGDGG-NH2 and (B, D, F, H) Ac-GGEGG-NH2 in the presence of (A–D) 5.4 M and (E–H) 0 M guanidine HCl (A, B, E, F) 298 K and (C, D, G, H) 333 K. All of the titration data were fitted to the standard Henderson-Hasselbalch equation for determination of the pKa values of Asp and Glu in model peptides.
(PDF)
Correlation of experimental and predicted pKa values. pKa values were calculated using several different software packages indicated on each plot. Left panels for each method are based on chain A of the crystal structure of L30e* (PDB code: 3N4Z), right panels for each method are based on chain B. Blue circles represent the results of based on x-ray model (A or B). Red squares represent the results of ensemble calculations based on structures A or B. Red and blue lines are the corresponding linear correlations. Thin black line shows the perfect correlation. See Materials and Methods section for the details of the calculations.
(PDF)
Statistics for structure determination of wild-type T. celer L30e and 5R→K variant.
(PDF)