Abstract
Background
Wolbachia are endosymbiotic bacteria that are frequently found in arthropods and nematodes. These maternally inherited bacteria manipulate host reproduction by several mechanisms including cytoplasmic incompatibility (CI). CI is the most common phenotype induced by Wolbachia and results in the developmental arrest of embryos derived from crosses between Wolbachia-infected males and uninfected females. Although the molecular mechanisms of CI are currently unknown, several studies suggest that host sperm is modified by Wolbachia during spermatogenesis.
Results
We compared the gene expression of Drosophila melanogaster larval testes with and without the wMel strain of Wolbachia to identify candidate genes that could be involved in the interaction between Wolbachia and the insect host. Microarray, quantitative RT-PCR and in situ hybridization analyses were carried out on D. melanogaster larval testes to determine the effect of Wolbachia infection on host gene expression. A total of 296 genes were identified by microarray analysis to have at least a 1.5 fold change [q-value < 5%] in expression. When comparing Wolbachia-infected flies to uninfected flies, 167 genes were up-regulated and 129 genes down-regulated. Differential expression of genes related to metabolism, immunity, reproduction and other functions were observed. Quantitative RT-PCR (qRT-PCR) confirmed 12 genes are differentially expressed in the testes of the 3rd instar larvae of Wolbachia-infected and uninfected flies. In situ hybridization demonstrated that Wolbachia infection changes the expression of several genes putatively associated with spermatogenesis including JH induced protein-26 and Mst84Db, or involved in immune (kenny) or metabolism (CG4988-RA).
Conclusions
Wolbachia change the gene expression of 296 genes in the larval testes of D. melanogaster including genes related to metabolism, immunity and reproduction. Interestingly, most of the genes putatively involved in immunity were up-regulated in the presence of Wolbachia. In contrast, most of the genes putatively associated with reproduction (especially spermatogenesis) were down-regulated in the presence of Wolbachia. These results suggest Wolbachia may activate the immune pathway but inhibit spermatogenesis. Our data provide a significant panel of candidate genes that may be involved in the interaction between Wolbachia and their insect hosts. This forms a basis to help elucidate the underlying mechanisms of Wolbachia-induced CI in Drosophila and the influence of Wolbachia on spermatogenesis.
Background
Wolbachia are endosymbiotic bacteria that infect a wide range of invertebrates including up to 66% of insect species, as well as spiders, mites and nematodes [1,2]. These intracellular, maternally transmitted bacteria have evolved several strategies such as male killing, feminization, parthenogenesis and cytoplasmic incompatibility (CI) to manipulate insect host reproduction. CI is the most common phenotype in insects and results in the developmental arrest of embryos when Wolbachia-infected males mate with uninfected females or with the females infected by a different Wolbachia strain. Intriguingly, a female infected with the same strain of Wolbachia can rescue embryonic lethality associated with CI. Although several models have been described to explain Wolbachia-induced CI [3-5], the molecular mechanisms of CI are still unclear.
Several studies in Drosophila melanogaster and Drosophila simulans have revealed that extensive chromosome bridging appears during early nuclear division in CI embryos, which implied that the chromosomes did not replicate completely, thus resulting in the defects in chromatin condensation and segregation [6-8]. Therefore, sperm derived from Wolbachia-infected males may be modified during spermatogenesis. In addition, although Wolbachia are present within Drosophila testes in developing spermatocytes and spermatids, Wolbachia are not present in mature sperm [9]. Hence, sperm modification by Wolbachia is likely to occur in the early stages of spermatogenesis. However, studies in other insect species infected with Wolbachia suggest that this may not occur for all strains of Wolbachia that induce CI. For example, Wolbachia-induced CI is close to 100% in the parasitic wasp Nasonia vitripennis but Wolbachia were found in only 28% of developing sperm [10]. An alternative explanation involves the presence of a product produced by Wolbachia that can spread through the testes to Wolbachia-uninfected immature sperm [10,11]. Landmann et al. showed that a delay of histone H3.3 and H4 deposition occurs on the sperm nuclei during formation of male pronuclei in CI embryos [12], which may have resulted in defects of paternal chromosomal replication. Recently, we reported that in both D. melanogaster and D. simulans the expression level of Hira, a gene encoding a chaperone of histone H3.3, is significantly decreased in Wolbachia-infected adult males [13]. In addition, Hira-mutated male flies mimic the CI phenotype resulting in a decreased embryo hatch rate when mated to uninfected females. As there were significantly less female offspring from crosses of Hira-mutated males with Wolbachia-uninfected females [13], we speculated that it was the sperm-carrying X chromosome with mutated Hira that caused embryonic lethality. These results suggest that histone modification in the sperm from Wolbachia-infected males could be involved in embryonic lethality in CI embryos.
Reynolds and Hoffmann previously demonstrated that CI levels dramatically decreased with increasing male age in Drosophila flies infected with Wolbachia [14]. We also recently reported that in both D. melanogaster and D. simulans, 1-day-old males could induce very strong CI, while 5-day-old males were not able to induce CI [13]. These results would suggest that sperm resulting in strong CI is modified by Wolbachia during the early stages of spermatogenesis. An additional study reported that Wolbachia numbers in D. simulans progressively decreased within germ cells from the second larval stage up until adult eclosion [11]. This decrease in Wolbachia numbers may be due to a reduced preference with age for the germ line, resulting in a decreased Wolbachia numbers in spermatocytes and spermatids [11]. Ultimately Wolbachia were eliminated from the mature sperm [11]. Yamada et al. demonstrated that Drosophila male development time could greatly influence the strength of Wolbachia-induced CI. Males that undergo the shortest developmental time in larval stages express strong CI, whereas the males that take a longer time for larval development quickly lose their ability to express CI as adult flies [15]. These results suggest that the regulation of gene expression in spermatogenesis in larval stages may be crucial for the sperm to express the CI phenotype in adult flies. Therefore, in order to investigate the mechanism of CI associated with early modification of sperm by Wolbachia, we selected 3rd instar larval testes for gene expression analysis comparing transcriptional profiles between Wolbachia-infected (Dmel wMel) and uninfected D. melanogaster (Dmel T) larval testes. Differential expression was observed in 296 genes related to reproduction, immunity, metabolism, and other functions. Expression profiles for 12 genes were confirmed by Quantitative Reverse Transcriptase PCR (qRT-PCR) and in situ hybridization was carried out for four genes. Our data provide a comprehensive analysis of differential gene expression between Wolbachia-infected and uninfected larval testes of Drosophila. These results provide important information to help determine the molecular mechanisms involved in Wolbachia's interaction with their hosts and, in particular, the role of Wolbachia on spermatogenesis leading to induction of CI.
Results
Microarray identification of genes involved in Wolbachia/host interaction
To identify larval testes genes differentially expressed in the presence of Wolbachia, the GeneChip 15 K Drosophila Genome Array (Operon) was used to determine the expression of 17899 transcripts (~13664 genes). Comparison of gene expression profiles revealed significant gene expression differences in 296 candidate genes (at least 1.5 fold, q-value < 5%), with 167 genes up-regulated and 129 genes down-regulated in the presence of Wolbachia infection.
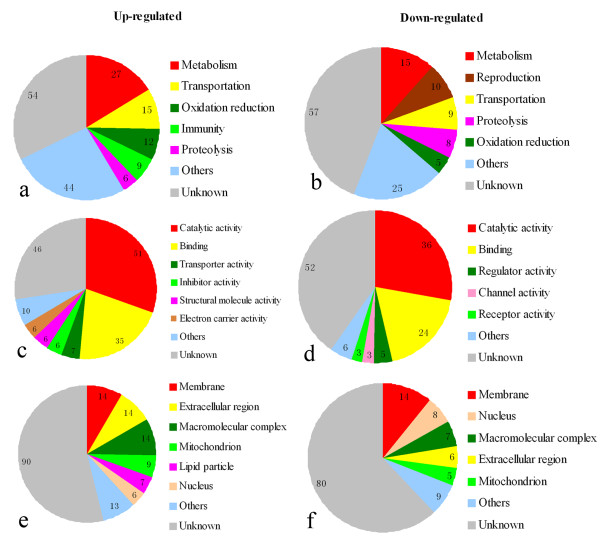
Based on the Gene Ontology (GO)-biological process (BP), molecular function (MF) and cellular component (CC), we observed that among the up-regulated genes with known BP classes, the largest group (27 genes) contained genes involved in metabolism. Genes involved in transportation constituted the second largest group with 15 genes. The third largest group included 12 genes involved with oxidation -reduction. Additionally, 9 genes were identified that are involved in immunity, 6 genes involved in proteolysis and 44 other gene products were identified. The remaining 54 genes (32%) had unknown BP (Additional file 1, Figure 1a). Among the down-regulated transcripts, 15 genes were identified that are involved in metabolism. The second largest group contained 10 genes involved in reproduction. The third largest group consisted of genes involved in transportation (9 genes), followed by 8 genes involved in proteolysis and 5 genes involved in oxidation-reduction. There were 25 transcripts encoding proteins with other functions. The remaining 57 transcripts (44%) encode proteins with unknown BP (Additional file 1, Figure 1b).
Figure 1.
Pie chart representation of gene ontology for genes differentially expressed in microarray analyses according to biological process (a, b), molecular function (c, d) and cellular component (e, f). Gene expression in Wolbachia-infected larval testes was compared to uninfected larval testes of Drosophila melanogaster and the criterion for differential expression was ≥1.5 fold changes with a q-value of <5%.
Most MF classes were involved in catalytic activity (51 genes up-regulated and 36 genes down-regulated) and binding activity (35 genes up-regulated and 24 down-regulated). Additional up-regulated genes were related to transporter activity (7 genes), inhibitor activity (6 genes), structural molecule activity (6 genes) and electron carrier activity (6 genes). Down-regulated genes were also involved in regulator activity (5 genes), channel activity (3 genes) and receptor activity (3 genes) (Additional file 2, Figure 1c, d).
Finally, most CC classes were associated to membranes (14 genes up-regulated and 14 genes down-regulated), extracellular regions (14 genes up-regulated and 6 genes down-regulated), macromolecular complexes (14 genes up-regulated and 7 genes down-regulated), mitochondria (9 genes up-regulated and 5 genes down-regulated), and nucleui (6 genes up-regulated and 8 genes down-regulated). Seven up-regulated genes are involved in lipid particles (Additional file 3, Figure 1e, f).
For both up-regulated and down-regulated genes assigned to BP classes, the majority of identified genes are involved with metabolism, transportation, oxidation-reduction or proteolysis. However, genes involved in immunity were mostly up-regulated (Figure 1a) and genes involved in reproduction were mostly down-regulated (Figure 1b). As shown in table 1, for the genes involved in reproduction, 10 were down-regulated and only 3 were up-regulated. These genes involved in reproduction that are differentially expressed in Wolbachia-infected testes are diverse and include some associated with spermgenesis such as CG5686-RA (Chico), CG32491-RP (mod), CG15179-RA (sunz), CG8827-RB (Ance) and CG17934-RA (Mst84Db).
Table 1.
Classification of genes related to reproduction that are differentially expressed (≥1.5 fold changes, q-value < 5%) in larval testes of Wolbachia-infected flies compared to uninfected flies identified by microarray analyses.
| Relative expression level | Biological function | Transcript | Gene symbol | Fold change |
|---|---|---|---|---|
| Up-regulated | germ cell migration | CG32491-RP | mod (mdg4) | 1.7518 |
| gonad development | CG7194-RA | CG7194 | 1.5730 | |
| male germ-line stem cell division | CG5686-RA | chico | 1.5272 | |
| Down-regulated | ovarian follicle cell development | CG11387-RA | ct | 0.6356 |
| ovarian follicle cell development | CG5993-RA | os | 0.6297 | |
| male genitalia morphogenesis | CG11491-RA | br | 0.6212 | |
| male meiosis | CG15179-RA | sunz | 0.6196 | |
| spermatogenesis | CG9553-RC | chic | 0.6024 | |
| cytoplasmic transport, nurse cell to oocyte | CG8978-RB | Sop2 | 0.6001 | |
| spermatid nucleus differentiation | CG8827-RB | Ance | 0.5681 | |
| reproduction | CG12052-RP | lola | 0.5335 | |
| sperm motility | CG17934-RA | Mst84Db | 0.4979 | |
| male courtship behavior | CG14916-RA | Gr32a | 0.4072 | |
Among immunity genes differentially expressed between Wolbachia-infected and uninfected larval testes, 9 transcripts were up-regulated and only 3 transcripts down-regulated. The immunity genes included some associated with both the cellular and humoral immune response. For example, the antimicrobial peptides (CG1180-RA, CG10810-RA) and the positive regulator (CG16910-RA, kenny) in the IMD signalling pathway (Table 2).
Table 2.
Classification of immune associated genes differentially expressed (≥1
| Relative expression level | Biological function | Transcript | Gene symbol | Fold change |
|---|---|---|---|---|
| Up-regulated | antimicrobial humoral response | CG1180-RA | LysE | 3.3768 |
| hemolymph coagulation | CG15825-RA | Fon | 3.0290 | |
| defense response to fungus | CG10810-RA | Drs | 2.2760 | |
| positive regulation of antibacterial peptide biosynthetic process | CG16910-RA | Key | 1.8311 | |
| response to virus | CG2081-RB | Vago | 1.7291 | |
| defense response | CG2736-RA | CG2736 | 1.5499 | |
| phagocytosis, engulfment | CG9556-RA | alien | 1.5411 | |
| phagocytosis, engulfment | CG5215-RB | Zn72D | 1.5286 | |
| phagocytosis, engulfment | CG10563-RA | l(2)37Cd | 1.5210 | |
| Down-regulated | response to virus | CG11390-RA | PebIII | 0.6570 |
| phagocytosis | CG4280-RB | crq | 0.5416 | |
| phagocytosis, engulfment | CG8189-RB | ATP syn-b | 0.4764 | |
QRT-PCR validation of microarray results
To verify the results of the microarray analyses, 12 differentially expressed genes based on microarray were selected for qRT-PCR to further investigate the expression profiles. The selection criteria for further expression analysis were based on large differences in expression in the microarray analyses in addition to genes associated with reproduction (Table 3). Genes expression measured by qRT-PCR exhibited similar changes as in the microarray analyses, with 7 genes (CG3767-RA, CG16910-RA, CG8627-RA, CG32954-RF, CG12262-RA, CG9081-RA and CG1180-RA) up-regulated and 5 genes (CG12052-RP, CG17934-RA, CG17268-RA, CG8189-RB and CG4988-RA) down-regulated (Figure 2). Among these genes CG3767-RA showed the largest up-regulation and CG4988-RA manifested the largest down-regulation, which was consistent with microarray results. CG3767-RA encodes the juvenile hormone-induced protein 26 (JhI-26), which has recently been identified as a sperm protein of D. melanogaster [16]. Other genes of interest from the qRT-PCR analysis included CG16910 (kenny), which is a key member of insect immune-related Imd pathway, and CG17934 (Mst84Db), which encodes a male specific sperm protein.
Table 3.
Differentially expressed genes selected for qRT-PCR validation based on microarray analyses.
| Relative expression level | Biological function | Transcript | Gene symbol | Fold change |
|---|---|---|---|---|
| Up-regulated | signal transduction | CG3767-RA | JhI-26 | 7.1069 |
| positive regulation of antibacterial peptide biosynthetic process | CG16910-RA | key | 1.8311 | |
| cellular acyl-CoA homeostasis | CG8627-RA | Dbi | 2.33635 | |
| ethanol metabolic process | CG32954-RF | Adh | 4.3842 | |
| fatty acid beta-oxidation | CG12262-RA | CG12262 | 2.1573 | |
| oxidation reduction | CG9081-RA | Cyp4s3 | 2.6798 | |
| antimicrobial humoral response | CG1180-RA | LysE | 3.3768 | |
| Down-regulated | cellular process involved in reproduction | CG12052-RP | lola | 0.5335 |
| sperm motility | CG17934-RA | Mst84Db | 0.4979 | |
| proteolysis | CG17268-RA | Pros28.1A | 0.5457 | |
| phagocytosis, engulfment | CG8189-RB | ATPsyn-b | 0.4764 | |
| hexose metabolic process | CG4988-RA | CG4988 | 0.2999 |
Figure 2.
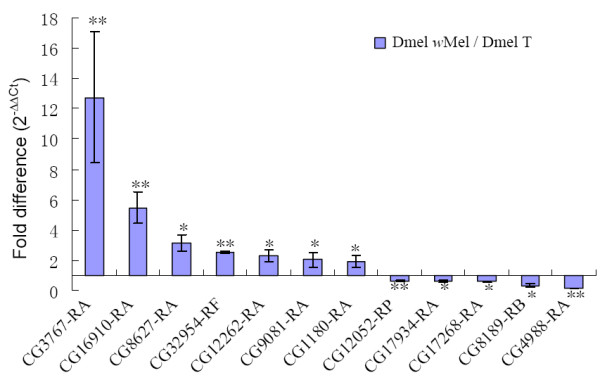
qRT-PCR validation of selected differentially expressed genes identified by microarray analyses. Testes RNA extracts were used for both microarray and qRT-PR analyses to compare the two methods of gene expression. "/" represents the relative value, bars indicate standard error; "*" and "**" indicate significant differences with P < 0.05 and P < 0.01 respectively.
In situ hybridization
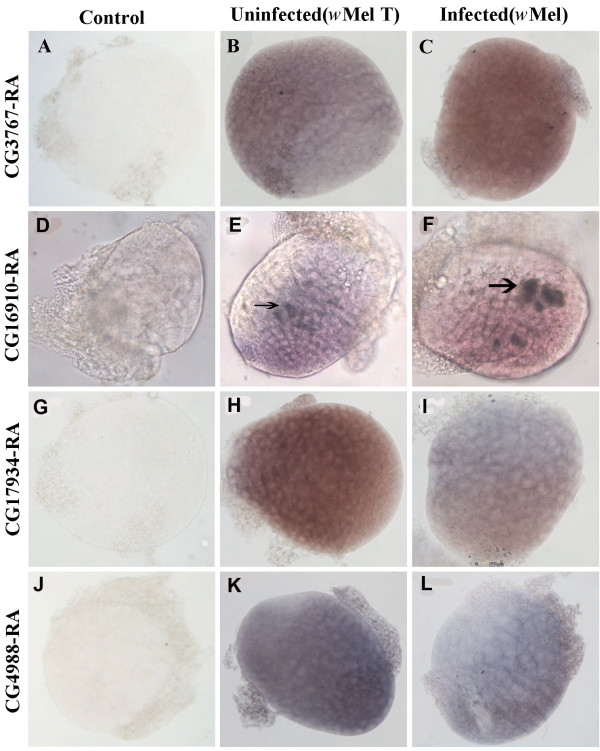
Four candidate genes of interest from qRT-PCR were selected to determine the localization in the testes of Dmel wMel and Dmel T larvae by in situ hybridization using digoxigenin-labelled RNA antisense probes. All four transcripts could be detected in both Dmel wMel and Dmel T larval testes. However, there were much stronger signals detected in Wolbachia-infected larval testes for CG3767-RA and CG16910-RA antisense probes compared to uninfected larval testes (Figure 3B, C, E, F). In contrast, the signals for CG17934-RA and CG4988-RA mRNA were weaker in Dmel wMel larval testes when compared to Dmel T (Figure 3H, I, K, L). Although these results were consistent with both microarray and qRT-PCR results, we did find strong signals in some specific regions of testes from both Dmel wMel and Dmel T larval testes with CG16910-RA antisense probes (Figure 3E, F). In contrast, almost ubiquitous signals were detected in the larval testes in both Dmel wMel and Dmel T with CG3767-RA, CG17934-RA, and CG4988-RA antisense probes. There was no signal detected in control samples when hybridized with sense probes (Figure 3A, D, G, J).
Figure 3.
In situ hybridization in Drosophila melanogaster larval testes for four differentially expressed genes. CG3767-RA (A-C), CG16910-RA (D-F), CG17934-RA (G-I) and CG4988-RA (J-L) from Wolbachia-infected (C, F, I, L) and uninfected larvae (B, E, H, K). Panels (A, D, G, J) are negative controls using sense probes. The arrows indicate the specific region of signals. All testes were dissected from the 3rd instar larvae and were magnified 200 × under the microscope.
Discussion
Previous studies revealed that the strength of CI in Wolbachia-infected Drosophila was dramatically reduced with both male age [13,14] and increased larval stage development [15]. Therefore, Wolbachia action during spermatogenesis in larval stages of Drosophila may be important for induction of CI. In this study, we selected 3rd instar Drosophila larval testes to investigate the effect of Wolbachia on early spermatogenesis and provide a basis to understand the underlying mechanisms of CI. A genome-wide analysis of the Wolbachia/host interaction in Drosophila larval testes was conducted and a number of genes showing differentially expression between Wolbachia infected and uninfected larval testes were identified. These genes are involved in diverse functions including reproduction, oxidation-reduction, immunity, transportation and metabolism.
The majority of genes involved in reproduction were down-regulated in the presence of Wolbachia, including CG8827-RB (Ance) and CG12052-RP (Lola). Some of these genes have previously been reported to be associated with the interaction between Wolbachia and their insect hosts. For example, angiotensin converting enzyme (Ance) gene has been demonstrated to be down-regulated in the adult testes of Wolbachia-infected flies relative to uninfected flies in both D. melanogaster and D. simulans [17]. This suggests that Ance gene may play a potential role in the interaction between Wolbachia and their hosts. There is also further evidence that Ance is involved in spermatid differentiation [18]. A potential role of Ance in CI was also shown with mutation affecting CI levels [17]. The Longitudinals lacking (Lola) gene is associated with cellular processes involved in reproduction. Lola mutant flies showed a block in developmental nurse cell death and abnormal nuclear organization [19]. During programmed cell death in Drosophila ovaries, chromatin condensation is not complete in Lola-mutant flies [19]. In the present study, Wolbachia infection results in significant down-regulation of Lola gene expression in larval testes, which in turn may induce abnormal chromatin condensation during spermatogenesis.
An additional reproduction gene identified to be differentially expressed, CG32491-RP, is an alternative splice product of mod (mdg4) previously observed in vitro [17]. Soltani-Bejnood et al. found a mutant in the common region of mod (mdg4) expressed in Drosophila spermatocytes but confined to cytoplasm during prophase I and demonstrated that the common region of mod (mdg4) plays an important role in homolog segregation during Drosophila male meiosis [20]. Therefore, we can speculate that Wolbachia may affect Drosophila spermatogenesis by regulating the expression of mod (mdg4). Another gene, CG17934-RA (Mst84Db), encodes for a male specific sperm protein. Kuhn et al. previously demonstrated that Mst84Db is expressed exclusively in the male germ line and associated with sperm motility [21]. Further studies revealed that there was not only a translational repression element but also a transcriptional control element within the Mst84Db gene [22], suggesting that Mst84Db could also be regulated at the transcriptional level. Our data obtained from microarray, qRT-PCR and in situ hybridization showed that the Mst84Db gene was down-regulated in Wolbachia-infected larval testes relative to uninfected testes, suggesting that Mst84Db could be inhibited at the transcriptional level by Wolbachia directly or indirectly in testes. Recent work revealed that there are multiple altered sperm structures during the late stage of spermatogenesis in Wolbachia-infected males [11]. Therefore it is likely that Wolbachia infection inhibits the transcription of Mst84Db during spermatogenesis resulting in modified sperm.
In addition to genes involved in reproduction, microarray analyses revealed differential expression of genes involved in insect immunity between Wolbachia-infected and uninfected D. melanogaster larval testes. One of the first lines of defence of insects against invading microbes is the generation of reactive oxygen species (ROS). However, high concentration of ROS may lead to oxidative stress and potentially damage lipids, nucleic acids and proteins, thus resulting in a reduction in insect lifespan [23]. Wolbachia has been shown to disturb the cellular physiology of its insect host especially via the generation of oxidative stress [24]. Correspondingly, an increase of antioxidant expression in mosquito cells is induced by Wolbachia, which could be an adaptation to symbiosis [24]. Moreover, Wolbachia that infects Asobara tabida interferes with iron, which limits oxidative stress and cell death, thus promoting its survival within host cells [25]. Recently fecundity of Wolbachia-uninfected A. tabida females was correlated with variable expression of genes regulating iron homeostasis and oxidative stress [26]. Our microarray data also show elevated expression levels in Wolbachia-infected larval testes for multiple genes involved in oxidation-reduction. Up-regulated gene expression was observed for CG9081-RA (Cyp4s3), which is involved in the oxidation-reduction process, CG6770-RA which is associated with response to oxidative stress, and CG12262-RA which is correlated with fatty acid beta-oxidation. This suggests the possibility that Wolbachia might regulate the redox reaction by various pathways to neutralize the potentially deadly ROS and thus maintain the Wolbachia/host symbiotic relationship.
Up-regulation of immune genes may represent 'detection' of Wolbachia as occurs in the relationship between the primary endosymbiont of the weevil Sitophilus zeamais through up-regulation of three local immune genes in the bacteriome [27]. We identified multiple up-regulated genes involved in immunity in Wolbachia-infected larval testes relative to uninfected ones, including two genes encoding antimicrobial peptides (Lysozyme E and Drosomycin). In addition, the CG16910-RA (kenny) gene was also up-regulated in Wolbachia-infected testes and located in specific regions of larval testes (Figure 3F). Previous studies have shown that IKKβ (encoded by ird5) and IKKγ (encoded by kenny) constitute the Drosophila IKK complex which directly phosphorylates Relish in the Imd immune signalling pathway [28]. The phosphorylated Relish then recruits RNA polymerase II and induces the expression of antimicrobial peptides. Furthermore, ird5 has been shown to be down-regulated while Relish and some antimicrobial peptides (attacin A, B, C, D and diptericin B) are up-regulated in Wolbachia-infected Drosophila S2 cells [17]. These experiments suggest that high levels of kenny gene expression in Wolbachia-infected testes probably contributes to phosphorylate Relish and thus induces the production of antimicrobial peptides in testes. Earlier work demonstrated that Wolbachia had no effects on the transcription of three antimicrobial peptide marker genes in adults of two insect species, D. simulans and Aedes albopictus, naturally infected with Wolbachia [29]. Therefore, induction of the insect immune system appears to not occur with naturally occurring Wolbachia infections. However, several lines of evidence have shown that Wolbachia infection does increase resistance against pathogens, such as viruses and filarial nematodes in both naturally infected hosts and artificially transinfected hosts [30-33]. Moreover, Wolbachia infection has been demonstrated in both its original host D. melanogaster and a novel mosquito host (Aedes aegypti) to be able to increase the levels of melanization which is a major component of the insect immune system [34]. Cook and McGraw raised two alternate explanations that do not involve an immune response: Wolbachia might mediate modification of the membrane, thus prevent entry of pathogens into host cells, or there may be direct competition between Wolbachia and pathogens for an intracellular resource [35]. Clearly the mechanism by which Wolbachia is able to increase pathogen resistance of insect hosts needs to be further investigated. Further experiments are required to determine if naturally occurring avirulent or low-density Wolbachia strains can activate the insect host immune response.
Our microarray analyses also highlight a large number of differentially expressed genes that code for proteins involved in metabolism including CG32954-RF (coding for alcohol dehydrogenase), CG8627-RA (involved in cellular acyl-CoA homeostasis), and CG8782-RA (coding Ornithine aminotransferase precursor). CG4988-RA, which codes for aldose 1-epimerase, was found to exhibit a lower expression level in Wolbachia-infected larval testes suggesting Wolbachia infection could decrease hexose metabolism. Up-regulation expression of CG2718-RB, which codes for glutamine synthetase 1, suggests that glutamate synthesis is increased in Wolbachia-infected testes. As glutamate importers were identified in the sequenced genome of wMel [36], it is reasonable to suggest that Wolbachia may use host glutamate as an important component for a variety of metabolic pathways. Indeed, the genome sequence of the wMel strain revealed that Wolbachia does not contain the complete set of metabolic pathways present in free-living bacteria [36]. Wolbachia probably only use a limited number of substrates and synthesizes very few metabolic intermediates. The successful survival and proliferation of endoysmbiotic Wolbachia in many host species may be due to the effect of Wolbachia on host metabolism to obtain most of the energy by importation of amino acids and other metabolites. It is likely that Wolbachia affects the expression of its host genes involved in metabolic pathway indirectly, namely, Wolbachia presumably consumes metabolites from the host, and then the host has to up-regulate the expression of metabolic related genes to increase the biosynthesis of that metabolite. In addition, the wMelPop strain of Wolbachia was shown to increase both locomotor activity and metabolic rate in Aedes aegypti [37], suggesting Wolbachia can manipulate host metabolism by inducing changes in expression levels of host metabolic genes.
Microarray data also reveals numerous differently expressed genes that are involved in transportation, including CG4450-RA (Shawl), which is involved in transmembrane transport and potassium ion transport, and CG13795-RA, which is related to neurotransmitter transport. As discussed previously, the transportation of host metabolites appears to be critical to the survival of Wolbachia in insect hosts. Interestingly, studies in Nasonia vitripennis wasps have revealed although Wolbachia are found in only ~28% of developing sperm, all sperm are modified. In the beetle Chelymorpha alternans, Wolbachia can modify up to 90% of sperm, but are never observed within the developing sperm or within the surrounding cyst cells, though they are abundant within the outer testis sheath [10]. These observations suggest Wolbachia may produce some factors that can cross multiple tissue membrane barriers to effect developing spermatids. Wolbachia is known to possess a type IV secretion system, which is likely used for exporting molecules into host cells [38]. Although the molecules that Wolbachia secretes into host cells are currently unknown, it is reasonable to suggest that various transporters may be required to establish molecules in locations in which they interact with the insect host.
This interaction between Wolbachia and the insect host may also be influenced by differential expression of other genes with unknown functions. For example, Juvenile hormone (JH) has been shown to play a key role in regulating both development and reproduction in insects [39,40]. In D. melanogaster, Dubrovsky et al. demonstrated that JH could induce the gene CG3767-RA (JhI-26) rapidly and specifically [41]. High expression levels of JhI-26 were observed in adult male accessory glands, although it was absent during the period from late third instar larvae to eclosion [41]. Recently JhI-26 has been identified as a sperm protein of D. melanogaster [16], implying that it could be associated with sperm function. In our study, we found that Wolbachia infection results in ~10 fold increase of JhI-26 transcription in later larval testes. This high level of JhI-26 gene expression may inhibit testes development in the later larval stages. Therefore, sperm produced by Wolbachia-infected young males that develop fastest in larval stage may be mostly immature. Yamada et al. observed that male Drosophila developmental time influences the strength of Wolbachia-induced CI. Male flies that have the shortest development time in larval stages express very strong CI, while males that spend a longer time in larval development quickly lose their ability to express CI [15]. A longer larval development time may allow the testes to develop completely resulting in young males with fully mature sperm not able to induce CI. Furthermore, studies in Nasonia CI embryos have shown a delay in nuclear envelope breakdown and Cdk1 (cyclin-dependent kinase 1) activation in the male pronucleus relative to the female pronucleus [42]. During the first mitotic division, when the maternal chromosomes enter the metaphase/anaphase transition, the improperly condensed paternal chromosome remnants fail to segregate and remain arrested in metaphase [7]. This is probably due to a delay in recruiting the replication-independent histone H3.3/H4 complex to the male pronucleus [12]. The delay in development of male pronucleus or paternal chromosomes may be involved in inhibiting the production of fully mature sperm. Therefore, it is possible that Wolbachia may mimic the function of JH to induce the expression of JhI-26. As high expression of JhI-26 may inhibit testes development in the larval stages, male flies that develop fastest may produce incompletely matured sperm. Immature sperm may then result in the delayed development of the male pronucleus and high embryonic lethality associated with CI upon fertilization of eggs from Wolbachia-uninfected females.
Conclusions
In this study, microarray, qRT-PCR and in situ hybridization were used to show numerous differentially expressed genes between testes of Wolbachia-infected and uninfected Drosophila melanogaster larvae. The genes showing variable expression levels are involved in diverse functions including metabolism, immunity and reproduction. The data presented here provides a significant panel of candidate genes to investigate the underlying mechanisms of the Wolbachia/host interaction. In particular, the genetic basis of CI and the role of Wolbachia in spermatogenesis in Drosophila. We are currently undertaking additional experiments with several candidate genes using overexpression or RNA interference to determine their function to provide further insight into this complex endosymbiotic relationship.
Methods
Drosophila melanogaster flies were reared on a standard cornmeal/yeast diet at 25°C with a photoperiod of 8L:16D (light:dark) and under non-crowded conditions (200 ± 10 eggs per 50 ml vial of media in 150 ml conical flask) [15]. The Wolbachia-infected Drosophila strain designated Dmel wMel (D. melanogaster Brisbane nuclear background with introgressed wMel from YW) was kindly provided by Prof. O'Neill S (Monash University, Australia). Wolbachia-uninfected Dmel wMel T was subsequently generated by tetracycline treatment following established protocols [43] and confirmed to be Wolbachia-free by PCR with Wolbachia surface protein gene (wsp) primers (Additional file 4) (data not shown).
Microarray analysis
Approximately 80 pairs of testes were dissected from 3rd instar larvae (72 h after hatch) of both Dmel wMel and Dmel T and total RNA was extracted by using Trizol (Invitrogen) according to the manufacturer's instructions. Total RNA was used to generate the Cy5/Cy3 labelled DNA by using a cRNA amplification labelled kit (CapitalBio). Briefly, the 1st-strand cDNA was reverse transcribed with T7 Oligo (dT) primer, then the 2nd-strand was synthesized and purified as the template for cRNA transcription with T7 Enzyme Mix. Two micrograms of cRNA was reverse transcribed to cDNA with random primers and the cDNA was used as the template for Cy5/Cy3 labelled DNA synthesis with Cy5/Cy3-dCTP. The Cy5/Cy3 labelled DNA in 80 μL hybridization solution (3 × SSC, 0.2% SDS, 5 × Denhart's, 25% formamide) was hybridized to the 15 K Drosophila Genome Array (Operon). After hybridized at 42°C overnight, washed with 2 × SSC (plus 0.2% SDS) and 0.2 × SSC for 5 min, respectively, the GeneChips were used for scanning using LuxScan 10 KA (CapitalBio). Data was normalized and analyzed by using LuxScan 3.0 software (CapitalBio). Three biological replicates were carried out in this study. Statistical analysis was performed and differential expressed genes were selected with the criteria of a fold-change of ≥ 1.5 and a q-value (%) of <5%. The q-value was calculated using CapitalBio® Molecule Annotation System V4.0.
MAPPfinder [44] was used to enrich, rank and classify the differently expressed genes based on Gene Ontology (GO) [45] information of each gene. All gene ontology terms were further corrected manually based on FlyBase (http://flybase.org/; FB2011_06, released June 24th, 2011) according to their Gene Ontology (GO)-biological process (BP), molecular function (MF) and cellular component (CC).
Quantitative real-time PCR (qRT-PCR)
To further investigate the differentially expressed genes identified by microarray analysis, 12 genes (7 up-regulated and 5 down-regulated) were selected for qRT-PCR analysis. Specific primers for the 12 genes and rp49 (ribosome protein 49 reference gene) were designed based on sequences from flybase database (Additional file 4).
Approximately 2 μg of total RNA was used to synthesize the first strand cDNA by using M-MLV reverse transcriptase (Promega) with the primer oligo-anchor R (5'-GACCACGCGTATCGATGTCGACT16(A/C/G)-3') according to manufacture's instruction. The cDNA was diluted (1:20) for use as a template and qRT-PCR was performed on a Miniopticon system (Bio-Rad). A reaction volume of 20 μL was used containing 10 μl of Platinum SYBR Green qRCR superMix (Takara) containing 2 × SYBR Premix Ex Taq, 2 μl of dilluted cDNA and 4 μl of each primer (1 μM). The qRT-PCR cycling program was 95°C for 2 min, followed by 40 cycles of 95°C for 10 s, 61°C for 20 s and 72°C for 20 s, and then a melting curve was constructed from 55°C to 98°C. The relative expression of each gene comparing Wolbachia-infected testes to uninfected testes was normalized against the reference gene (rp49) by using the 2-ΔΔCT calculation method: ΔΔCT = (CT, Target-CT, rp49)wMel-(CT, Target-CT, rp49)wMel T [46]. Student t-tests were then done to compare differences with significant values when P < 0.05.
In situ hybridization
Fragments of four transcripts (CG3767-RA, CG16910-RA, CG17934-RA and CG4988-RA) were amplified with specific primers (Additional file 4) and then subcloned into pGEM-T Easy vector (Promega Biosciences, Madison, WI), respectively. Subsequently, the recombinant plasmids were linearized and in vitro transcribed using T7/Sp6 polymerase (Roche, Boehringer Mannheim, Mannheim, Germany) to synthesize the Dig-tagged antisense or sense probes, respectively.
The in situ hybridization was carried out according to the method described previously with minor modifications [47]. In order to avoid loss of samples testes were exposed but still attached to the body of the 3rd instar larvae. The testes were first fixed in 4% paraformaldehyde at 4°C overnight and then incubated with proteinase K (10 mg/ml) at 37°C for 30 min. After prehybridization for 4 h at 65°C, samples were hybridized with Dig-labelled antisense probes at 65°C overnight and washed with SSC buffer (2 ×, 1 ×, and 0.1 ×) sequentially and then incubated with anti-Dig phosphatase AB (1:1000, Roche) overnight at 4°C. Finally, NBT/BCIP (Beyotime, Shanghai, China) was used for color development. Testes were peeled off in glycerin and visualized with a Leica DM 4000 B microscope (Leica, Germany). Sense probes were used as negative control.
Authors' contributions
YZ and J-LW carried out microarray experiments and data analysis. YZ, CL and C-PW performed qRT-PCR and in situ hybridization. J-LW, YZ, TW drafted the manuscript. Y-FW contributed experimental conception/design, data analysis and manuscript drafting. All authors read and approved the final manuscript.
Supplementary Material
All the enriched biological process (BP) classes based on Gene Ontology (GO) term generated from differently expressed genes induced by Wolbachia infection.
All the enriched molecular function (MF) classes based on Gene Ontology (GO) term generated from differently expressed genes induced by Wolbachia infection.
All the enriched cellular component (CC) classes based on Gene Ontology (GO) term generated from differently expressed genes induced by Wolbachia infection.
Primers for qRT-PCR or RT-PCR.
Contributor Information
Ya Zheng, Email: zhengyahb@163.com.
Jia-Lin Wang, Email: galin66@163.com.
Chen Liu, Email: weinasidetianshi@yahoo.com.cn.
Cui-Ping Wang, Email: wangcuiping725@163.com.
Thomas Walker, Email: tom.walker@monash.edu.
Yu-Feng Wang, Email: yfengw@mail.ccnu.edu.cn.
Acknowledgements
We thank Professor Scott O'Neill (Monash University, Australia) for kindly providing Dmel wMel flies. This work was supported by the National Nature Science Foundation of China (30970405) and the key project of Chinese ministry of education (109118) to Y-FW.
References
- Werren JH, Baldo L, Clark ME. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 2008;6:741–751. doi: 10.1038/nrmicro1969. [DOI] [PubMed] [Google Scholar]
- Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH. How many species are infected with Wolbachia?--A statistical analysis of current data. FEMS Microbiol Lett. 2008;281:215–220. doi: 10.1111/j.1574-6968.2008.01110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtzis K, Dobson SL, Braig HR, O'Neill SL. Rescuing Wolbachia have been overlooked. Nature. 1998;391:852–853. doi: 10.1038/36017. [DOI] [PubMed] [Google Scholar]
- Poinsot D, Charlat S, Merçot H. On the mechanism of Wolbachia-induced cytoplasmic incompatibility: confronting the models with the facts. Bioessays. 2003;25:259–265. doi: 10.1002/bies.10234. [DOI] [PubMed] [Google Scholar]
- Bossan B, Koehncke A, Hammerstein P. A new model and method for understanding Wolbachia-induced cytoplasmic incompatibility. PLoS One. 2011;6:e19757. doi: 10.1371/journal.pone.0019757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassy CW, Karr TL. Cytological analysis of fertilization and early embryonic development in incompatible crosses of Drosophila simulans. Mech Dev. 1996;57:47–58. doi: 10.1016/0925-4773(96)00527-8. [DOI] [PubMed] [Google Scholar]
- Callaini G, Dallai R, Riparbelli MG. Wolbachia-induced delay of paternal chromatin condensation does not prevent maternal chromosomes from entering anaphase in incompatible crosses of Drosophila simulans. J Cell Sci. 1997;110:271–280. doi: 10.1242/jcs.110.2.271. [DOI] [PubMed] [Google Scholar]
- Tram U, Fredrick K, Werren JH, Sullivan W. Paternal chromosome segregation during the first mitotic division determines Wolbachia-induced cytoplasmic incompatibility phenotype. J Cell Sci. 2006;119:3655–3663. doi: 10.1242/jcs.03095. [DOI] [PubMed] [Google Scholar]
- Clark ME, Veneti Z, Bourtzis K, Karr TL. Wolbachia distribution and cytoplasmic incompatibility during sperm development: the cyst as the basic cellular unit of CI expression. Mech Dev. 2003;120:185–198. doi: 10.1016/S0925-4773(02)00424-0. [DOI] [PubMed] [Google Scholar]
- Clark ME, Bailey-Jourdain C, Ferree PM, England SJ, Sullivan W, Windsor DM, Werren JH. Wolbachia modification of sperm does not always require residence within developing sperm. Heredity. 2008;101:420–428. doi: 10.1038/hdy.2008.71. [DOI] [PubMed] [Google Scholar]
- Riparbelli MG, Giordano R, Callaini G. Effects of Wolbachia on sperm maturation and architecture in Drosophila simulans Riverside. Mech Dev. 2007;124:699–714. doi: 10.1016/j.mod.2007.07.001. [DOI] [PubMed] [Google Scholar]
- Landmann F, Orsi GA, Loppin B, Sullivan W. Wolbachia-mediated cytoplasmic incompatibility is associated with impaired histone deposition in the male pronucleus. PLoS Pathog. 2009;5:e1000343. doi: 10.1371/journal.ppat.1000343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Ren PP, Wang JL, Wang YF. Wolbachia-induced cytoplasmic incompatibility is associated with decreased hira expression in male Drosophila. PLoS One. 2011;6:e19512. doi: 10.1371/journal.pone.0019512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds KT, Hoffmann AA. Male age, host effects and the weak expression or non-expression of cytoplasmic incompatibility in Drosophila strains infected by maternally transmitted Wolbachia. Genet Res. 2002;80:79–87. doi: 10.1017/s0016672302005827. [DOI] [PubMed] [Google Scholar]
- Yamada R, Floate KD, Riegler M, O'Neill SL. Male development time influences the strength of Wolbachia-induced cytoplasmic incompatibility expression in Drosophila melanogaster. Genetics. 2007;177:801–808. doi: 10.1534/genetics.106.068486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasbrough ER, Dorus S, Hester S, Howard-Murkin J, Lilley K, Wilkin E, Polpitiya A, Petritis K, Karr TL. The Drosophila melanogaster sperm proteome-II (DmSP-II) J Proteomics. 2010;73(11):2171–2185. doi: 10.1016/j.jprot.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Xi Z, Gavotte L, Xie Y, Dobson SL. Genome-wide analysis of the interaction between the endosymbiotic bacterium Wolbachia and its Drosophila host. BMC Genomics. 2008;9:1. doi: 10.1186/1471-2164-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst D, Rylett CM, Isaac RE, Shirras AD. The Drosophila angiotensin -converting enzyme homologue Ance is required for spermiogenesis. Dev Biol. 2003;254:238–247. doi: 10.1016/S0012-1606(02)00082-9. [DOI] [PubMed] [Google Scholar]
- Bassa BP, Cullenb K, McCallb K. The axon guidance gene lola is required for programmed cell death in the Drosophila ovary. Dev Biol. 2007;2:771–785. doi: 10.1016/j.ydbio.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltani-Bejnood M, Thomas SE, Villeneuve L, Schwartz K, Hong CS, McKee BD. Role of the mod (mdg4) common region in homolog segregation in Drosophila male meiosis. Genetics. 2007;176:161–180. doi: 10.1534/genetics.106.063289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn R, Kuhn C, Börsch D, Glätzer KH, Schäfer U, Schäfer M. A cluster of four genes selectively expressed in the male germ line of Drosophila melanogaster. Mech Dev. 1991;35:143–151. doi: 10.1016/0925-4773(91)90064-D. [DOI] [PubMed] [Google Scholar]
- Gigliotti S, Balz V, Malva C, Schäfer MA. Organisation of regulatory elements in two closely spaced Drosophila genes with common expression characteristics. Mech Dev. 1997;68:101–113. doi: 10.1016/S0925-4773(97)00137-8. [DOI] [PubMed] [Google Scholar]
- Dröge W. Oxidative stress and aging. Adv Exp Med Biol. 2003;543:191–200. doi: 10.1007/978-1-4419-8997-0_14. [DOI] [PubMed] [Google Scholar]
- Brennan LJ, Keddie BA, Braig HR, Harris HL. The endosymbiont Wolbachia pipientis induces the expression of host antioxidant proteins in an Aedes albopictus cell line. PLoS One. 2008;3:e2083. doi: 10.1371/journal.pone.0002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer N, Voronin D, Charif D, Mavingui P, Mollereau B, Vavre F. Wolbachia interferes with ferritin expression and iron homeostasis in insects. PLoS Pathogens. 2009;5:e1000630. doi: 10.1371/journal.ppat.1000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer N, Dedeine F, Charif D, Finet C, Allemand R, Vavre F. Do variable compensatory mechanisms explain the polymorphism of the dependence phenotype in the Asobara tabida-wolbachia association? Evolution. 2010;64(10):2969–2979. doi: 10.1111/j.1558-5646.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- Anselme C, Pérez-Brocal V, Vallier A, Vincent-Monegat C, Charif D, Latorre A, Moya A, Heddi A. Identification of the weevil immune genes and their expression in the bacteriome tissue. BMC Biol. 2008;6:43. doi: 10.1186/1741-7007-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertürk-Hasdemir D, Broemer M, Leulier F, Lane WS, Paquette N, Hwang D, Kim CH, Stöven S, Meier P, Silverman N. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci USA. 2009;106:9779–9784. doi: 10.1073/pnas.0812022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourtzis K, Pettigrew MM, O'Neill SL. Wolbachia neither induces nor suppresses transcripts encoding antimicrobial peptides. Insect Mol Biol. pp. 635–639. [DOI] [PubMed]
- Hedges LM, Brownlie JC, O'Neill SL, Johnson KN. Wolbachia and Virus Protection in Insects. Science. 2008;322:722. doi: 10.1126/science.1162418. [DOI] [PubMed] [Google Scholar]
- Moreira LA, Iturbe-Ormaetxe I, Jeffery JA, Lu G, Pyke AT, Hedges LM, Rocha BC, Hall-Mendelin S, Day A, Riegler M, Hugo LE, Johnson KN, Kay BH, McGraw EA, van den Hurk AF, Ryan PA, O'Neill SL. A Wolbachia Symbiont in Aedes aegypti limits infection with dengue, chikungunya, and plasmodium. Cell. 2009;139:1268–1278. doi: 10.1016/j.cell.2009.11.042. [DOI] [PubMed] [Google Scholar]
- Kambris Z, Cook PE, Phuc HK, Sinkins SP. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science. 2009;326:134–136. doi: 10.1126/science.1177531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian G, Xu Y, Lu P, Xie Y, Xi Z. The endosymbiotic bacterium Wolbachia induces resistance to dengue virus in Aedes aegypti. PLoS pathog. 2011;6(4):e1000833. doi: 10.1371/journal.ppat.1000833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P, Kenny N, Eyles D, Moreira LA, O'Neill SL, Asgari S. Infection with the wMel and wMelPop strains of Wolbachia leads to higher levels of melanization in the hemolymph of Drosophila melanogaster, Drosophila simulans and Aedes aegypti. Dev Comp Immunol. 2011;35(3):360–365. doi: 10.1016/j.dci.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Cook PE, McGraw EA. Wolbachia pipientis: an expanding bag of tricks to explore for disease control. Trends Parasitol. 2010;26(8):373–375. doi: 10.1016/j.pt.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Wu M, Deboy R, Brownlie JC, McGraw EA, Martin W, Esser C, Ahmadinejad N, Wiegand C, Madupu R, Beanan MJ, Brinkac LM, Daugherty SC, Durkin AS, Kolonay JF, Nelson WC, Mohamoud Y, Lee P, Berry K, Young MB, Utterback T, Weidman J, Nierman WC, Paulsen IT, Nelson KE, Tettelin H, O'Neill SL, Eisen JA. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PloS Biol. 2004;2:327–341. doi: 10.1371/journal.pbio.0020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans O, Caragata EP, McMeniman CJ, Woolfit M, Green DC, Williams CR, Franklin CE, O'Neill SL, McGraw EA. Increased locomotor activity and metabolism of Aedes aegypti infected with a life-shortening strain of Wolbachia pipientis. J Exp Biol. 2009;212:1436–1441. doi: 10.1242/jeb.028951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Sasaki T, Ishikawa H. Genes for the type IV secretion system in an intracellular symbiont, Wolbachia, a causative agent of various sexual alterations in arthropods. J Bacteriol. 2000;182:6529–6531. doi: 10.1128/JB.182.22.6529-6531.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruntenko NE, Karpova EK, Adonyeva NV, Chentsova NA, Faddeeva NV, Alekseev AA, Rauschenbach IY. Juvenile hormone, 20-hydroxyecdysone and dopamine interaction in Drosophila virilis reproduction under normal and nutritional stress conditions. J Insect Physiol. 2005;51:417–425. doi: 10.1016/j.jinsphys.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Taub-Montemayor TE, Min KJ, Chen Z, Bartlett T, Rankin MA. JH III production, titers and degradation in relation to reproduction in male and female Anthonomus grandis. J Insect Physiol. 2005;51:427–434. doi: 10.1016/j.jinsphys.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Dubrovsky EB, Dubrovskaya VA, Bilderback AL, Berger EM. The isolation of two juvenile hormone-inducible genes in Drosophila melanogaster. Dev Biol. 2000;224:486–495. doi: 10.1006/dbio.2000.9800. [DOI] [PubMed] [Google Scholar]
- Tram U, Sullivan W. Role of delayed nuclear envelope breakdown and mitosis in Wolbachia induced cytoplasmic incompatibility. Science. 2002;296:1124–1126. doi: 10.1126/science.1070536. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Turelli M. Unidirectional incompatibility in Drosophila simulans: inheritance, geographic variation and fitness effects. Genetics. 1988;119:435–444. doi: 10.1093/genetics/119.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlquist KD, Salomonis N, Vranizan K, Lawlor SC, Conklin BR. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 2002;31(1):19–20. doi: 10.1038/ng0502-19. [DOI] [PubMed] [Google Scholar]
- Gene Ontology. http://www.geneontology.org
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Morris CA, Benson E, White-Cooper H. Determination of gene expression patterns using in situ hybridization to Drosophila testes. Nat Protoc. 2009;4:1807–1819. doi: 10.1038/nprot.2009.192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All the enriched biological process (BP) classes based on Gene Ontology (GO) term generated from differently expressed genes induced by Wolbachia infection.
All the enriched molecular function (MF) classes based on Gene Ontology (GO) term generated from differently expressed genes induced by Wolbachia infection.
All the enriched cellular component (CC) classes based on Gene Ontology (GO) term generated from differently expressed genes induced by Wolbachia infection.
Primers for qRT-PCR or RT-PCR.