Abstract
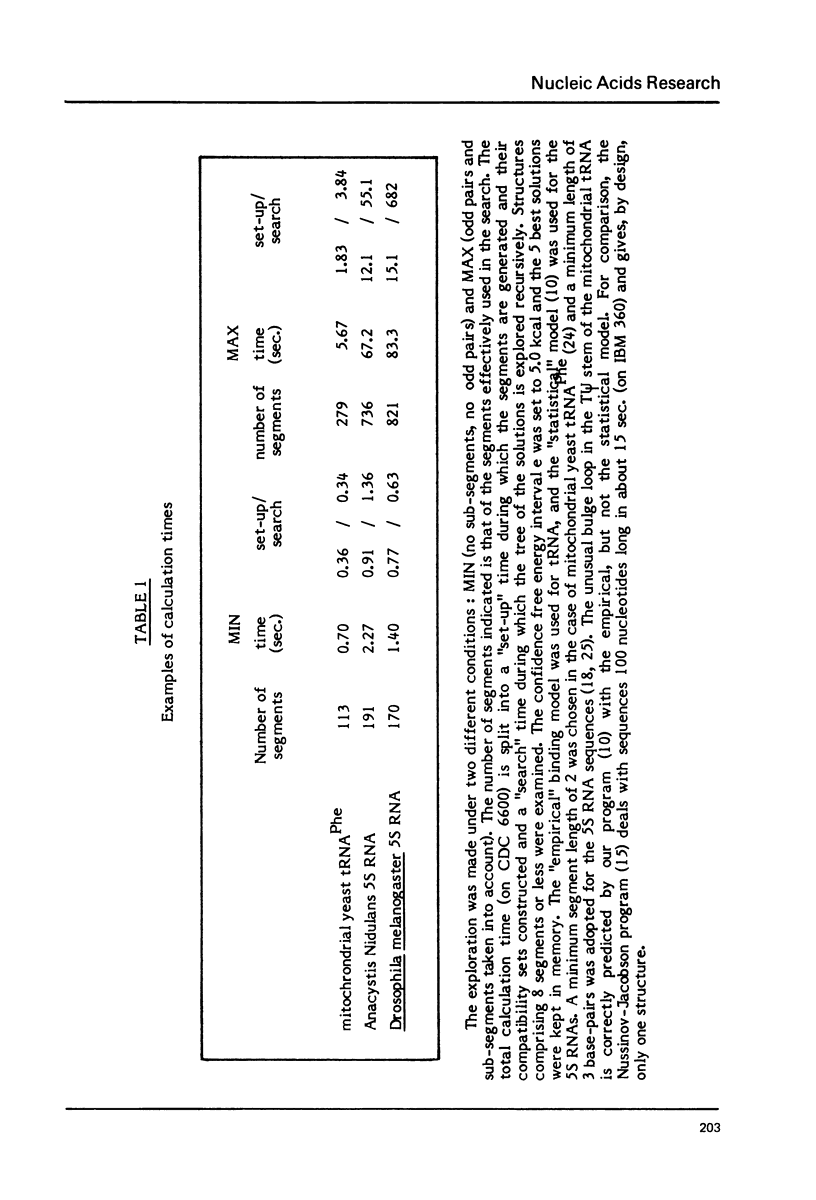
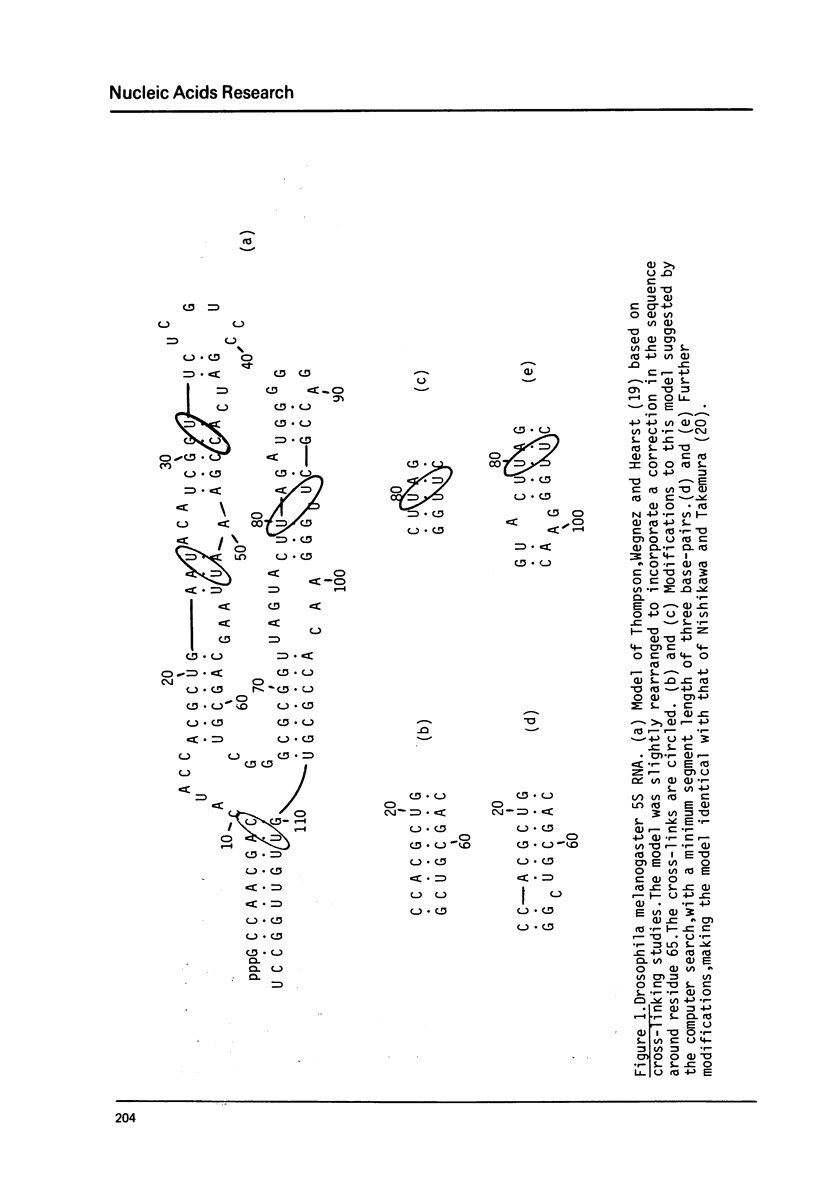
Fast algorithms for analysing sequence data are presented. An algorithm for strict homologies finds all common subsequences of length greater than or equal to 6 in two given sequences. With it, nucleic acid pieces five thousand nucleotides long can be compared in five seconds on CDC 6600. Secondary structure algorithms generate the N most stable secondary structures of an RNA molecule, taking into account all loop contributions, and the formation of all possible base-pairs in stems, including odd pairs (G.G., C.U., etc.). They allow a typical 100-nucleotide sequence to be analysed in 10 seconds. The homology and secondary structure programs are respectively illustrated with a comparison of two phage genomes, and a discussion of Drosophila melanogaster 55 RNA folding.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benhamou J., Jourdan R., Jordan B. R. Sequence of Drosophila 5S RNA synthesized by cultured cells and by the insect at different developmental stages. Homogeneity of the product and homologies with other 5S RNA's at the level of primary and secondary structure. J Mol Evol. 1977 May 13;9(3):279–298. doi: 10.1007/BF01796116. [DOI] [PubMed] [Google Scholar]
- Corry M. J., Payne P. I., Dyer T. A. The nucleotide sequence of 5 S rRNA from the blue-green alga Anacystis nidulans. FEBS Lett. 1974 Sep 15;46(1):63–66. doi: 10.1016/0014-5793(74)80335-2. [DOI] [PubMed] [Google Scholar]
- Fitch W. M. An improved method of testing for evolutionary homology. J Mol Biol. 1966 Mar;16(1):9–16. doi: 10.1016/s0022-2836(66)80258-9. [DOI] [PubMed] [Google Scholar]
- Gingeras T. R., Roberts R. J. Steps toward computer analysis of nucleotide sequences. Science. 1980 Sep 19;209(4463):1322–1328. doi: 10.1126/science.6251542. [DOI] [PubMed] [Google Scholar]
- Godson G. N., Barrell B. G., Staden R., Fiddes J. C. Nucleotide sequence of bacteriophage G4 DNA. Nature. 1978 Nov 16;276(5685):236–247. doi: 10.1038/276236a0. [DOI] [PubMed] [Google Scholar]
- Jacq B. Sequence homologies between eukaryotic 5.8S rRNA and the 5' end of prokaryotic 23S rRNa: evidences for a common evolutionary origin. Nucleic Acids Res. 1981 Jun 25;9(12):2913–2932. doi: 10.1093/nar/9.12.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus I. R., Rosen B., Hepperle R. Secondary structure of Qbeta RNA. J Theor Biol. 1977 Feb 7;64(3):587–595. doi: 10.1016/0022-5193(77)90290-9. [DOI] [PubMed] [Google Scholar]
- MacKay R. M., Doolittle W. F. Nucleotide sequences of Acanthamoeba castellanii 5S and 5.8S ribosomal ribonucleic acids: phylogenetic and comparative structural analyses. Nucleic Acids Res. 1981 Jul 24;9(14):3321–3334. doi: 10.1093/nar/9.14.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin R., Sibler A. P., Schneller J. M., Keith G., Stahl A. J., Dirheimer G. Etablissement de la séquence des nucléotides du phénylalanine-tRNA de mitochondries de Levure. C R Acad Sci Hebd Seances Acad Sci D. 1978 Oct 9;287(8):845–848. [PubMed] [Google Scholar]
- Ninio J. Prediction of pairing schemes in RNA molecules-loop contributions and energy of wobble and non-wobble pairs. Biochimie. 1979;61(10):1133–1150. doi: 10.1016/s0300-9084(80)80227-6. [DOI] [PubMed] [Google Scholar]
- Ninio J. Properties of nucleic acid representations. I. Topology. Biochimie. 1971;53(4):485–494. [PubMed] [Google Scholar]
- Nishikawa K., Takemura S. Nucleotide sequence of 5 S RNA from Torulopsis utilis. FEBS Lett. 1974 Mar 15;40(1):106–109. doi: 10.1016/0014-5793(74)80904-x. [DOI] [PubMed] [Google Scholar]
- Nussinov R., Jacobson A. B. Fast algorithm for predicting the secondary structure of single-stranded RNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6309–6313. doi: 10.1073/pnas.77.11.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipas J. M., McMahon J. E. Method for predicting RNA secondary structure. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2017–2021. doi: 10.1073/pnas.72.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queen C. L., Korn L. J. Computer analysis of nucleic acids and proteins. Methods Enzymol. 1980;65(1):595–609. doi: 10.1016/s0076-6879(80)65062-9. [DOI] [PubMed] [Google Scholar]
- Sege R., Söll D., Ruddle F. H., Queen C. A conversational system for the computer analysis of nucleic acid sequences. Nucleic Acids Res. 1981 Jan 24;9(2):437–444. doi: 10.1093/nar/9.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A computer program to search for tRNA genes. Nucleic Acids Res. 1980 Feb 25;8(4):817–825. [PMC free article] [PubMed] [Google Scholar]
- Staden R. Sequence data handling by computer. Nucleic Acids Res. 1977 Nov;4(11):4037–4051. doi: 10.1093/nar/4.11.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studnicka G. M., Eiserling F. A., Lake J. A. A unique secondary folding pattern for 5S RNA corresponds to the lowest energy homologous secondary structure in 17 different prokaryotes. Nucleic Acids Res. 1981 Apr 24;9(8):1885–1904. doi: 10.1093/nar/9.8.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studnicka G. M., Rahn G. M., Cummings I. W., Salser W. A. Computer method for predicting the secondary structure of single-stranded RNA. Nucleic Acids Res. 1978 Sep;5(9):3365–3387. doi: 10.1093/nar/5.9.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Wegnez M. R., Hearst J. E. Determination of the secondary structure of Drosophila melanogaster 5 S RNA by hydroxymethyltrimethylpsoralen crosslinking. J Mol Biol. 1981 Apr 15;147(3):417–436. doi: 10.1016/0022-2836(81)90493-9. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezenbeek P. M., Hulsebos T. J., Schoenmakers J. G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980 Oct;11(1-2):129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]


